1. Introduction
The aquaculture industry is considered the fastest-growing industry in several countries worldwide and represents about 17% of global protein intake as reported by the FAO [
1]. The increased demand for fish in the world market has led to production intensification with the increase in stocking densities being associated with stress factors and fish susceptibility to pathogens [
2]. Therefore, in recent years strategies based on the manipulation of intestinal microbiota balance have been proposed to improve fish survival and growth [
3]. Among these strategies, probiotics selection with mechanisms of action such as modulatory effect on the intestinal microbiota, the production of antimicrobial metabolites, vitamins, and enzymes, and immune regulation have been employed.
Numerous studies have highlighted the significance of the composition of intestinal bacteria for animal health due to its crucial role in protecting against infectious diseases and in maintaining the host’s immune and metabolic homeostasis [
3,
4,
5]. Due to the complexity of the intestinal microbiota of humans and other animals, probiotics composed of more than one strain have mainly been developed with aerobic bacteria and yeast [
6,
7]. Even though most probiotic studies have been conducted to evaluate aerobic bacteria, it is known, mostly by culture-independent studies, that anaerobic bacteria are significant members of the fish intestinal microbiota.
Tsuchiya et al. [
8] isolated vitamin B12-producing
Cetobacterium somerae from the intestines of freshwater fish: goldfish, common carp, and Mozambique tilapia. Later, Ramírez et al. [
9] reported
C.
somerae as a major component of the intestinal microbiota of the giant Amazonian freshwater fish,
Arapaima gigas, using sequenced microbial DNA-based techniques.
Likewise, Melo-Bolivar et al. [
10] reported the significant presence of Fusobacteria, represented mainly by
Cetobacterum species, in
O.
niloticus microbial content from two different farms, and also in a continuous-flow competitive exclusion culture from the intestinal content of Nile tilapia juveniles using a metataxonomic DNA-based approach.
In addition, LaFrentz et al. [
11] reported a
C.
somerae genome sequence from intestinal isolates of pond-raised channel catfish,
Ictalurus punctatus. Recently, Xie et al. [
12] evaluated the effects of stabilized
C.
somerae XMX-1 fermentation products on gut and liver health and zebrafish survival during a viral challenge;
C.
somerae XMX-1, was isolated from zebrafish intestines, cultured in anaerobic conditions, and added to the diet in a four-week feeding trial. It was found that dietary administration of
C.
somerae (XMX-1) improved the gut and liver health of zebrafish, reducing the intestinal inflammatory score, reducing proinflammatory cytokines, and increasing the antiviral gene expression; it also altered the composition of gut microbiota, reducing proteobacteria and increasing Firmicutes and Actinobacteria.
Zhang et al. [
13] found that
Cetobacterium was the core genus in the foregut, midgut, and hindgut of tilapia. They isolated
Cetobacterium sp. NK01 from Nile tilapia foreguts, and sequenced the whole genome of the isolate, which indicated it to be a novel candidate species of the
Cetobacterium sp. The genome analysis showed the production of amino acids, participating in various metabolic activities, and synthesizing vitamins, which indicated that
Cetobacterium plays a key role in fish nutrition. However, the functions of
Cetobacterium in the fish gut need to be further explored through in vivo and in vitro experiments [
12,
13].
In summary, probiotics are widely used in aquafeeds and exhibit beneficial effects in fish by improving host health and resistance to pathogens. Nevertheless, probiotics applied to aquaculture are mostly from terrestrial sources rather than the host animal and are mostly aerobic [
14].
The purpose of the work was to isolate and characterize anaerobic bacteria from the gastrointestinal tract of Nile tilapia (O. niloticus) and to evaluate the probiotic potential in vitro.
4. Discussion
In the present study, we isolated the bacterial strain C33 from the intestinal content of Nile tilapia. It was previously found, in a continuous-flow competitive exclusion culture (CFCEC) obtained from Nile tilapia intestinal content, that Fusobacteria, primarily represented by the
Cetobacterum genus, were highly abundant in the first days of the CFCEC [
10].
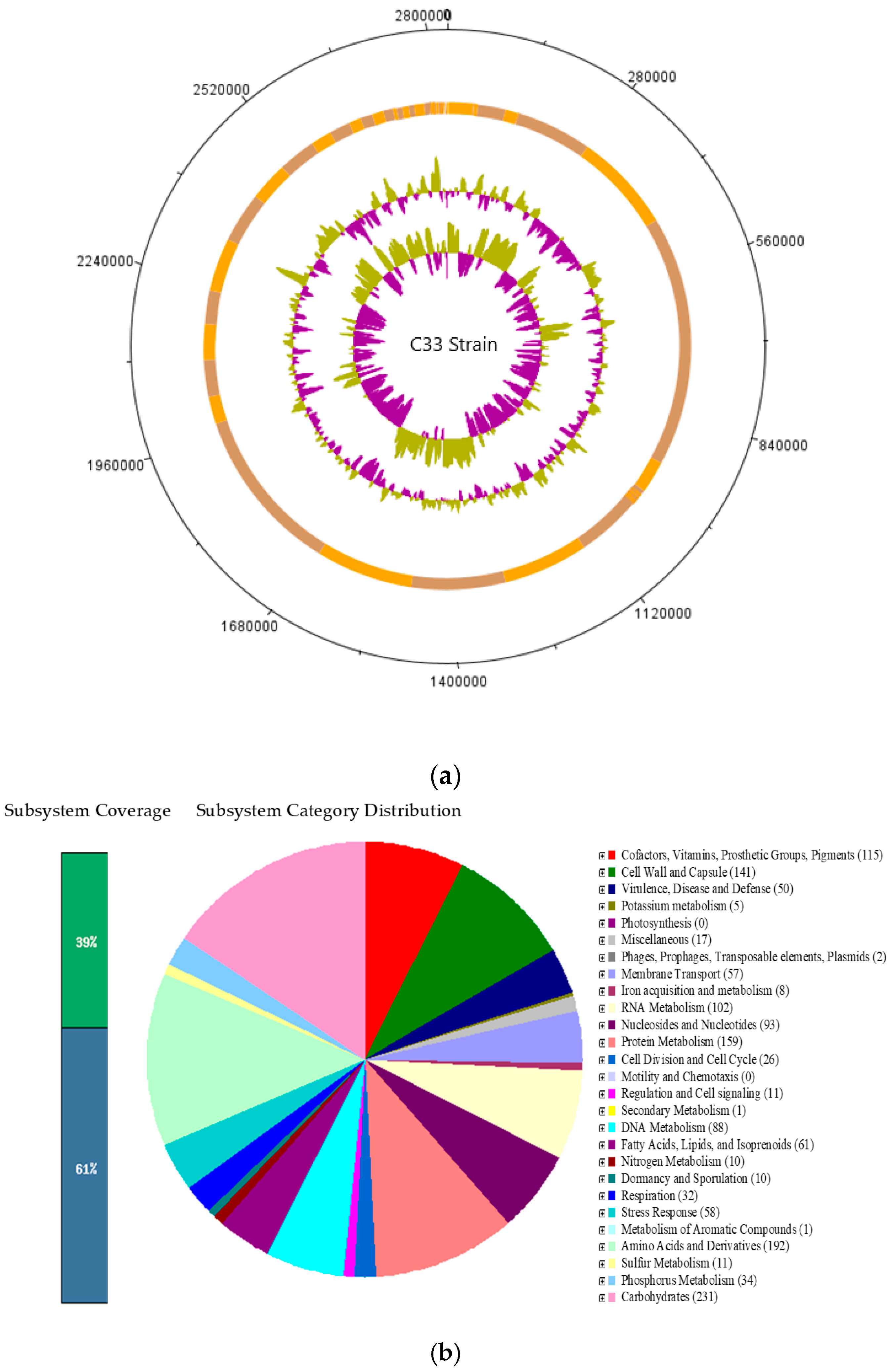
C33 is an anaerobic gram-negative bacterium, that, according to the genome annotation, was identified as Cetobacterium sp. nov C33. Since it was found to be a dominant species, it in vitro analyses that could determine its potential use as probiotic were conducted.
The results of the phenotypic characterization of C33 showed gelatinase negative activity, which is favorable since it has been suggested that the presence of this activity in probiotic microorganisms could be detrimental to the health of the host due to the possible damage it can cause in the extracellular protein matrices of intestinal tissue [
42]. In addition, the positive activity of esterase is related to the breaking activity of the ester bonds of polysaccharides, favoring the action of hydrolases of high molecular weight compounds such as carbohydrates and proteins. Likewise, the positive activity of acid phosphatase is important for the degradation of organic phosphorus found primarily in plant and animal protein sources [
43]. In addition to these characteristics, the C33 strain has the possibility of using different energy sources such as glucose, sucrose, maltose, salicin, mannitol, and trehalose, a fact that may favor the fish’s nutrition, improving the absorption of nutrients by solubilizing the elements of the diets through extracellular enzymes; this facilitates the absorption of individual molecules through the intestinal epithelium of the animal host [
44,
45] and provides enzymes that the animal host does not have [
44,
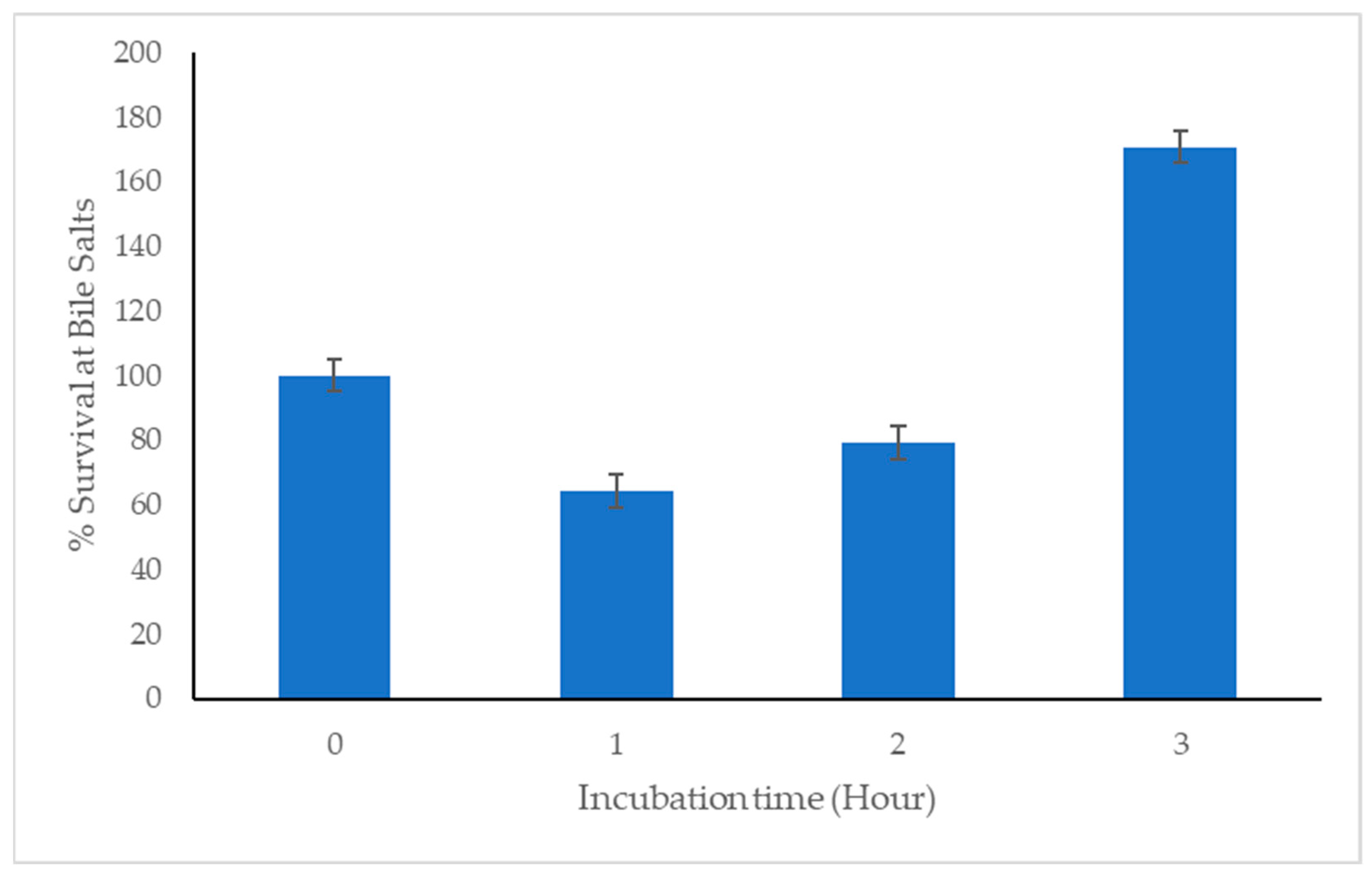
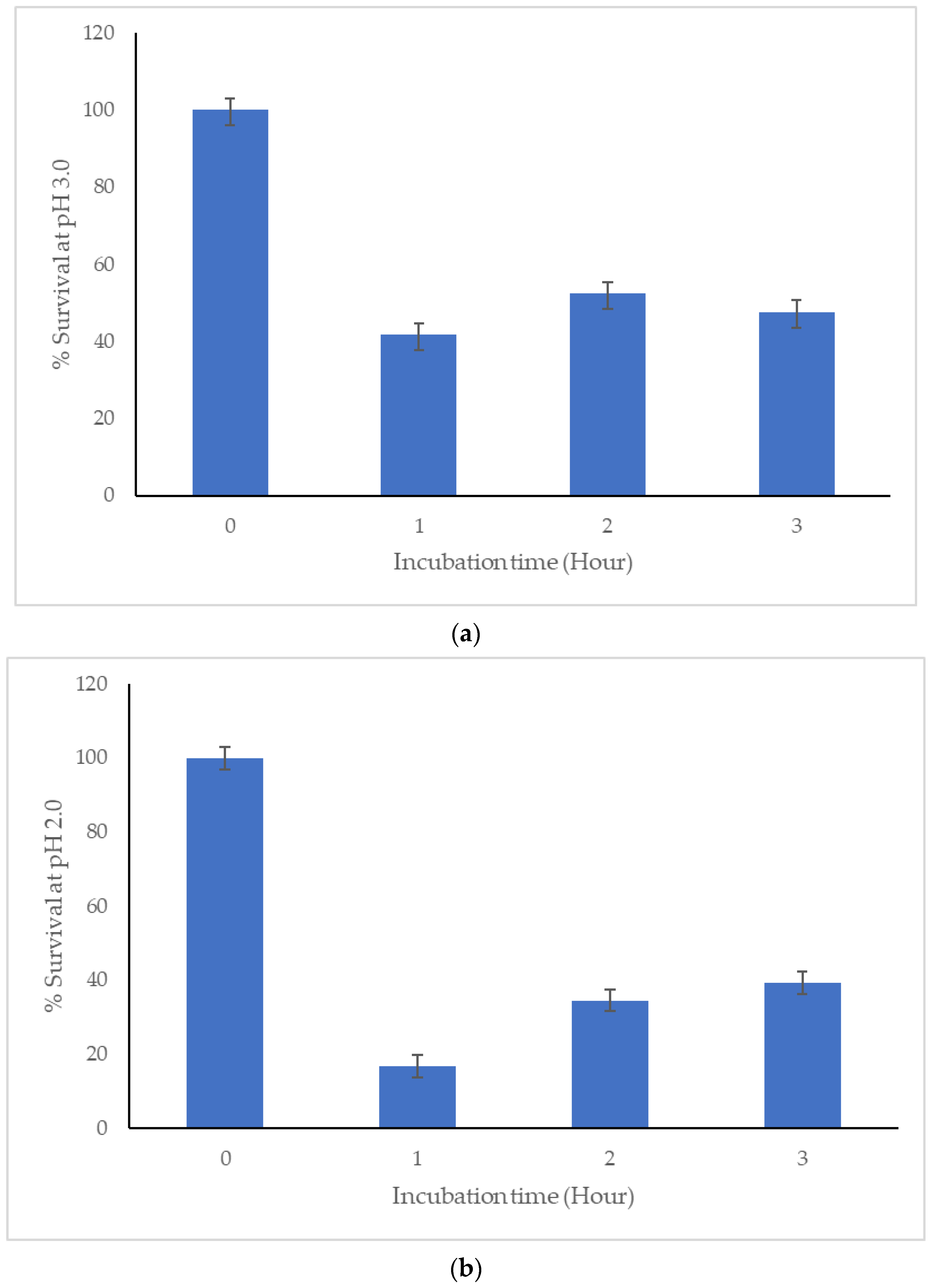
46]. Furthermore, we have identified the C33 strain as a gamma (γ) hemolytic bacterium. In addition, for a bacterium to qualify as a probiotic, it must be able to survive and ideally colonize the intestine. This involves overcoming the stress generated by the low pH of the stomach and contact with bile salts, which are inhibitory for multiple microorganisms because they cause cell lysis [
47,
48,
49,
50,
51]. Here we report the significant capacity of C33 to survive even at pH 2.0.
On the other hand, after passing through the digestive tract of the host, for a strain candidate to be used as a probiotic it must be able to colonize the gut. The C33 strain showed hydrophobicity percentages higher than 50% with a non-polar solvent (chloroform), a result that indicates a hydrophobic character of the cell surface, a factor that contributes to the interaction of the bacteria with the cells of the gastrointestinal tract. This indicates the potential ability of this isolate to colonize and persist in the gastrointestinal tract [
6,
41].
Antibiotic resistance and a growing reluctance to administer antibiotics have led to an increase in the use of probiotics [
6,
52]. Bacterial antibiotic resistance mechanisms can be innate, natural, or acquired. There is no horizontal transferability of the intrinsic resistance; nevertheless, the acquired resistance can be gained by mutations or the acquisition of genes through mobile genetic elements into their genomes [
6,
53,
54]. The resistance to antimicrobial agents in a potential bacterial probiotic should be considered with caution. On the other hand, some authors mention that resistance to given antibiotics may be acceptable because, in the event of the use of any of these compounds being required in the fish culture, the probiotic will not be eliminated [
6,
55].
Nevertheless, the cutoff value of all the antibiotics tested in C33 indicated that this bacterium is sensitive. Additionally, in the RASfinder genome annotation, no resistance genes were found for any antibiotics (amoxicillin, ertapenem, aztreonam, amoxicillin, ampicillin, piperacillin, cefixime, ceftriaxone, ticarcillin, penicillin, cefotaxime, temocillin, metronidazole, doxycycline, minocycline, tigecycline, teicoplanin, and vancomycin). Due to the potential migration of antibiotic resistance factors, we consider that it is better to use a probiotic free of any acquired antibiotic resistance to avoid further changes in the microbial community induced by the overuse of the probiotic bacteria.
C33 presents with the plasmid rep7a, belonging to the group of glutathione S-transferases (GST), which are dimeric proteins that can conjugate glutathione (GSH) with a variety of compounds containing electrophilic centers. On the other hand, C33 also presented with the rep7a gene, the tetA and tetB genes related with tetracycline resistance, and the Lnu(C) gene that confers resistance to lincomycin [
56].
C33 has two ribosomal sactipeptides (peptides cross-linked from sulfur to carbon alpha thioether) belonging to RiPP, that show various biological activities such as antibacterial properties and post-translationally modified peptides (RiPPs) [
57]. Nevertheless, this condition requires further research for better understanding of its bacterial physiology.
The C33 strain has a gene encoding the Linocin M18 bacteriocin; a protein that forms nano compartments within the pathogenic bacterium, it also contains ferritin-like proteins or peroxidases and enzymes involved in the oxidative stress response. Various authors have reported that Linocin M18 has bacteriostatic activity against strains of
Arthrobacter,
Bacillus,
Brevibacterium,
Corynebacterium, and
Listeria [
58]. In addition, C33 presents with genes encoding Zoocin A, a peptidoglycan hydrolase, which, combined with lauricidin, a cell membrane active lipid, has been reported to selectively suppress the growth of
Streptococcus mutans [
59].
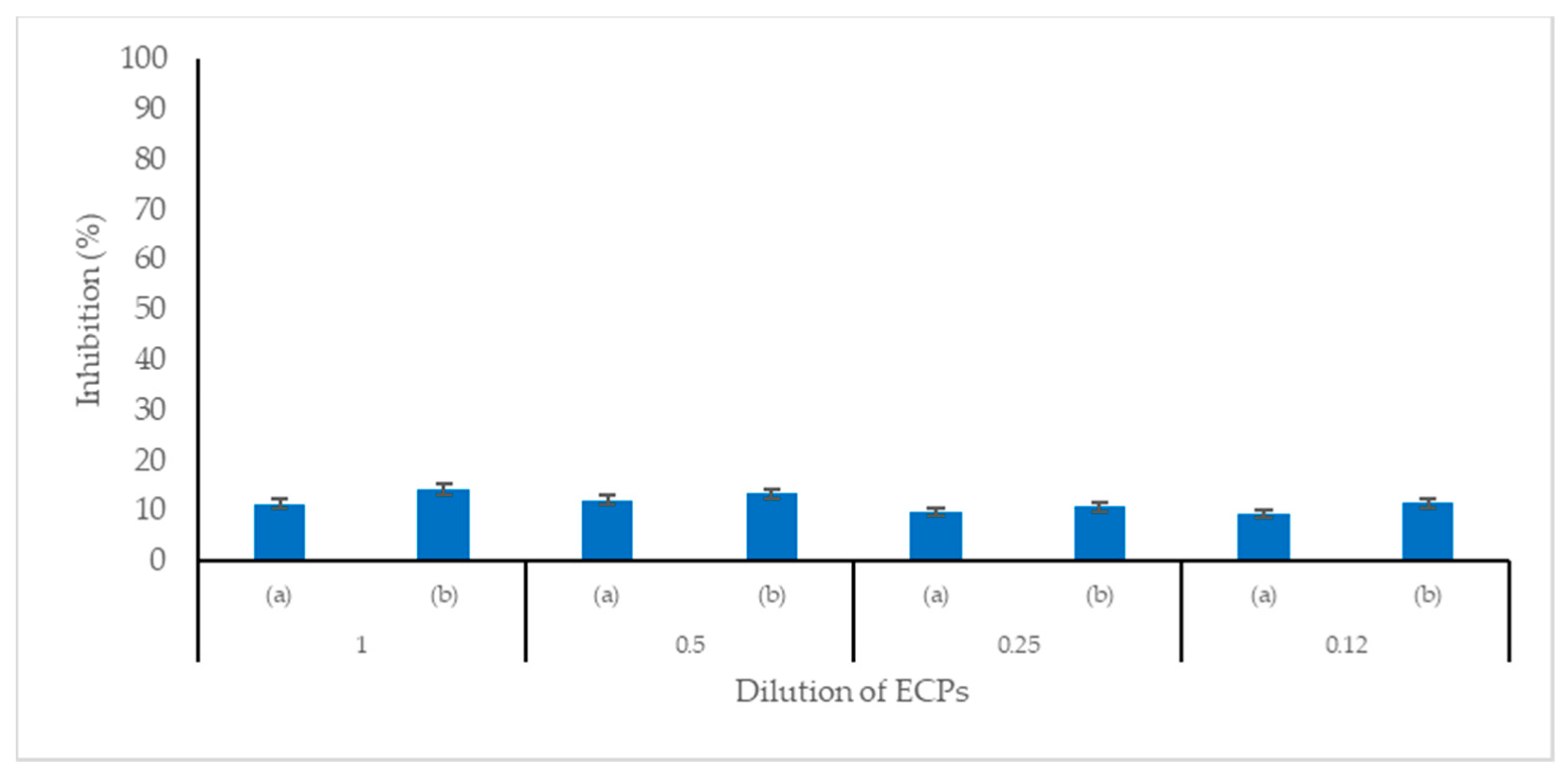
Here cell-free extracellular products (ECPs) of
Cetobacterium sp. C33 are found to have antimicrobial activity against
S.
agalactiae and
A.
hydrophila, two pathogenic bacteria responsible for high mortality in Nile tilapia cultures and substantial economic losses in tilapia farms [
48,
60,
61,
62].
Probiotic use has been reported as an alternative to the use of antibiotics which has caused antimicrobial resistance in the aquaculture industry [
63]. They contribute to natural resistance and a higher survival rate of the fish [
64,
65].
C33 has the capacity to biosynthesize amino acids, such as glutamine, glutamate, aspartate, asparagine, polyamine, methionine, threonine, homoserine, lysine, tryptophan, phenylalanine, tyrosine, proline, glycine, alanine, and serine, that benefit fish growth [
66]. In addition, biotin production is important for the synthesis of fatty acids, the oxidation of carbohydrates, and the synthesis of purines. Biotin deficiency causes loss of appetite, dark coloration, and seizures in fish. Thiamine acts in Nile tilapia as a coenzyme in various metabolic decarboxylation reactions (pyruvic acid, alpha-ketoglutaric acid). Thiamine deficiency in fish results in weakness, terminal convulsions, degeneration, and fish fin paralysis. Tetrapyrrole compounds like heme, cobalamin, etc., have multiple essential functions in fish. Its synthesis begins with the formation of aminolaevulinic acid from glutamine [
67]. Riboflavin and FAD are components of two enzymes (FMN and FAD) that are oxidases and reductases that participate in the metabolic degradation of proteins, carbohydrates, and lipids. Its deficiency in tilapia generates loss of appetite, dark skin, cataracts, and photophobia. Deficiencies in B vitamins, such as folate and vitamin B12, affect the offspring’s resistance and fertility [
68].
In general, the C33 strain breaks down and uses Mannose, Chitin, N-acetylglucosamine, Sucrose, Maltose, Maltodextrin, Lactose, Galactose, Lactate, Glycerol, Glycerol-3-phosphate, D-ribose utilization, L-ascorbate, and Fructose. The genus
Cetobacterium provides extracellular enzymes to degrade complex carbohydrates [
69]. The presence of
Cetobacterium may be associated with better glucose utilization [
70] and could improve glucose homeostasis and increase insulin expression [
71]. The above supports the role of
Cetobacterium in carbohydrate regulation.
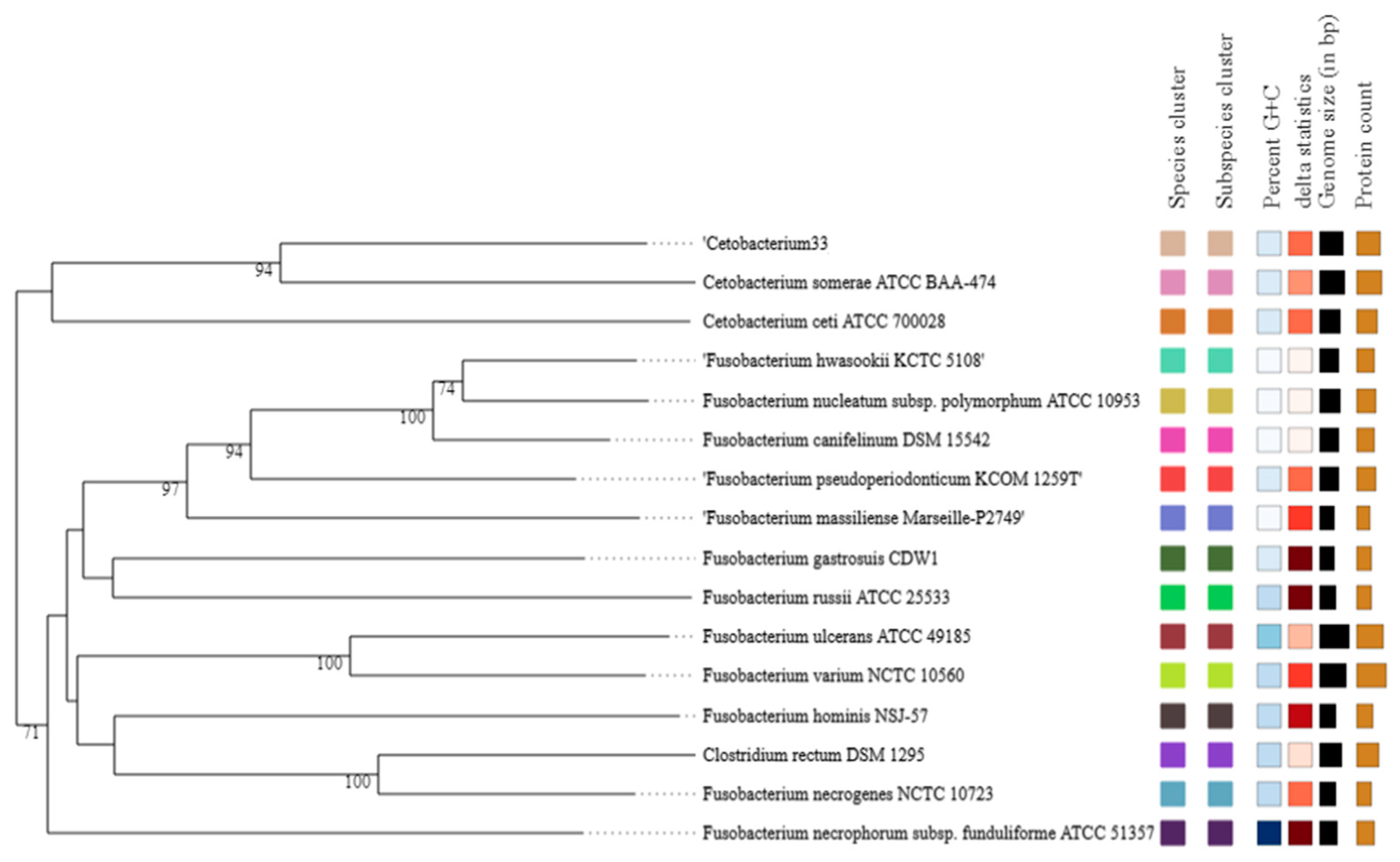
Finally, by comparing the sequencing results of the whole genome with the NCBI database, it was found that the C33 strain can be classified within the genus
Cetobacterium. Similarly, the phylogenetic analysis indicates that the C33 strain has the greatest similarity with the species
Cetobacterium somerae ATCC BAA-474, with a tetranucleotide signature correlation index (Tetra) result of 0.98712, a value that was less than 0.99 (Minimum value to consider it to be the same species [
72]), which suggests that it can be considered a new species. In addition, the C33 strain differs from
C.
somerae WAL 14325T
Cetobacterium sp. NK01 and
C.
ceti M 3333T in other aspects, such as no indole production, positive glucosidase reaction, positive enzyme activities for esterase (C4) (2-naphthyl butyrate) and acid phosphatase (2-naphthyl phosphate), and finally, it did not present any vitamin B12 production.
Consequently, the physiological characteristics and the phylogenetic analysis suggest that the C33 strain represents a new species of the genus Cetobacterium with high probiotic potential for Nile tilapia cultures based on the in vitro analysis shown here. Nevertheless in vivo experiments should be conducted in other studies to verify the effects on immune regulation, microbiota modulation fish growth, and resistance.