Abstract
Strain Q11T of an irregular coccoid Gram-positive bacterium, aerobic and non-motile, was isolated from Meconopsis integrifolia seeds. Strain Q11T grew optimally in 1% (w/v) NaCl, pH 7, at 30 °C. Strain Q11T is most closely related to Flexivirga, as evidenced by 16S rRNA gene analysis, and shares the highest similarity with Flexivirga aerilata ID2601ST (99.24%). Based on genome sequence analysis, the average nucleotide identity and digital DNA–DNA hybridization values of strains Q11T and D2601ST were 88.82% and 36.20%, respectively. Additionally, strain Q11T showed the abilities of nitrogen fixation and indole acetic acid production and was shown to promote maize growth under laboratory conditions. Its genome contains antibiotic resistance genes (the vanY gene in the vanB cluster and the vanW gene in the vanI cluster) and extreme environment tolerance genes (ectoine biosynthetic gene cluster). Shotgun proteomics also detected antibiotic resistance proteins (class A beta-lactamases, D-alanine ligase family proteins) and proteins that improve plant cold tolerance (multispecies cold shock proteins). Strain Q11T was determined to be a novel species of the genus Flexivirga, for which the name Flexivirga meconopsidis sp. nov. is proposed. The strain type is Q11T (GDMCC 1.3002T = JCM 36020 T).
1. Introduction
The genus Flexivirga, a member of the Phylum Actinobacteria, was first described by Anzai et al. [1]. The List of Prokaryotic Names with Standing in Nomenclature (https://lpsn.dsmz.de/search?word=Flexivirga, accessed on 8 October 2023) contains six species with legitimately recognized names, i.e., Flexivirga alba [1], Flexivirga endophytica [2], Flexivirga lutea [3], Flexivirga oryzae [4], Flexivirga caeni [5], and Flexivirga aerilata [6], which were isolated from the leaves of basil [2], the feces of ibis [3], the soil of a rice paddy [4], activated sludge [5], and an automobile air conditioning system [6], respectively. All members of the genus Flexivirga are described as Gram-positive bacteria, aerobic, non-motile, and with a high G + C content in their DNA (~67 mol%).
Meconopsis integrifolia is mostly distributed in the Tibetan Plateau. Many researchers found that this medicinal plant contains large amounts of bioactive compounds such as flavonoids and alkaloids, which are used in the pharmaceutical and healthcare industries [7]. However, the survival and growth of Meconopsis integrifolia have been significantly challenged by environmental changes, habitat loss, overexploitation, and difficulties in artificial cultivation [8].
Endophytic bacteria are symbiotic and beneficial organisms that colonize plants without causing disease; they are also plant growth-promoting bacteria (PGPB) [9]. PGPB promote plant growth and development and improve plant adaptability to the environment by secreting various hormones, facilitating the uptake of dissolved minerals, and carrying out nitrogen fixation [10,11]. Some endophytes, such as Paenibacillus polymyxa SK1 [12], Bacillus cereus N5 [13], Burkholderia phytofirmans PsJN [14], along with Kocuria sp. TRI2-1, Micromonospora sp. TSI4-1, and Sphingomonas sp. MG-2 [15], exhibit the ability to fix nitrogen, dissolve phosphate, produce IAA, and promote plant growth.
Therefore, this study aimed to characterize the new species Flexivirga meconopsidis Q11T, which inhabits the seeds of the medicinal plant Meconopsis integrifolia from the Qinghai–Tibet Plateau, by genome and proteomics analysis. The growth-promoting ability of strain Q11T was identified by analyzing IAA production, phosphorus dissolution, and nitrogen fixation, and the effect of strain Q11T on plant growth was further confirmed by pot experiments. As far as we know, this is the first report of a species of the genus Flexivirga that can promote plant growth.
2. Materials and Methods
2.1. Isolation and Cultivation
Strain Q11T was isolated from the seeds of Meconopsis integrifolia using Beef Extract Peptone Agar medium (BA). The surface of the seeds was disinfected [16], followed by crushing the seeds in a ceramic bowl and diluting them with sterile water. Each diluted sample (100 µL) was separately spread on BA medium. After being cultured at 30 °C for 3 days, a white colony Q11T was isolated and purified. Strain Q11T was stored at −80 °C in sterile 60% (v/v) glycerol.
2.2. 16S rRNA Gene Phylogeny
We performed 16S rRNA gene amplification using the process outlined by Li et al. [17]. The sequence was determined by the Life Technologies Company (Shenzhen, China). The sequence was uploaded to the EzBioCloud database for identification. By using the CLUSTAL_W program in MEGA (Version 7.0) software for sequence alignment, the neighbor-joining (NJ) [18], maximum likelihood (ML) [19], and maximum parsimony (MP) [20] methods were applied to analyze the genetic relationship among the strains, and a phylogenetic tree was constructed [21]. By performing 1000 repeated bootstrap analyses, the phylogenetic distance was calculated using the Kimura two-parameter model [22].
2.3. Genome Features
The genomic DNA of the strain Q11T was collected [23]. The genome sequence was completed on the Illumina MiSeq platform by Guangzhou Magi company (Guangzhou, China). The obtained sequence was submitted to NCBI, and we obtained the genome number JAOBQJ000000000. The average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values of strain Q11T and its closest members (F. aerilata ID2601ST, F. caeni BO-16T, F. endophytica KCTC 39536T, and F. oryzae R1T) were compared by OrthoANIu and the genome-to-genome distance calculator. In addition, gene analysis and annotation were carried out by the RAST server (version 2.0) [24] and Proksee [25]. The gene clusters of secondary metabolism biosynthesis in bacterial genome were analyzed by the antiSMASH program (version 7.0.1) [26,27].
2.4. Physiology
The morphology of strain Q11T was observed by transmission electron microscopy (Hitachi 7500) [28]. Strain Q11T was cultured under various conditions using BA to examine its physiological properties. Strain Q11T was subjected to Gram staining [29]. Cell motility was observed by the semi-solid puncture method [30]. To determine the culture conditions for the growth and development of the strain, the growth status of the strain was observed at different temperatures (10–50 °C), different pH values (pH = 4.0–11.0), and different NaCl concentrations (0, 0.5, and 1–10% at intervals of 1.0%, w/v) [31]. The pH value was adjusted in BA liquid medium [32]. Several key characteristics were studied according to the bacterial identification manual [33], using the methyl red test and the Voges–Proskauer test and determining the occurrence of catalases and oxidases and the hydrolysis of Tween 20, 40, 60, and 80 (final concentration 1%), starch (0.2%, w/v), and gelatin (10%, w/v). Further physiological and biochemical properties of the cells were identified by cultivating them at 30 °C and using the API ZYM, 20NE, and 50CH test strips (bioMérieux) as described by the manufacturer.
2.5. Chemotaxonomy
Well-grown cells of strain Q11T cultured on BA liquid medium for 24 h at 30 °C were used for fatty acid analysis. The fatty acids were identified by the Sherlock microbial identification system (MIDI) and GC (6890 N, Agilent), and the peaks were identified using the RTSBA6 database [34]. Respiratory quinones of strain LT1T were extracted from freeze-dried cells [35] and analyzed by LC-MS (Agilent 1260) [36]. Polar lipids were extracted from freeze-dried bacteria [37] and separated by two-dimensional TLC (MERCK, Silica gel 60) [36]. Total lipids were measured in a molybdophosphoric acid hydrate ethanol solution, amino lipids using the ninhydrin reagent, phospholipids using the Zinzadze reagent, and glycolipids using the α-naphthol reagent [38].
2.6. Shotgun Proteomics Analyses
The global profile of the protein/polypeptide complement in strain Q11T cells and supernatants was detected by shotgun proteomics [39,40]. The cells were washed with sterile water 3 times, frozen in liquid nitrogen for 10 min, and transported to Shanghai Paisenuo Biology Co., Ltd. (Shanghai, China) at −80 °C for detection. The samples were analyzed by an ORBITRAP ECLIPSE mass spectrometer (Thermo Scientific, Waltham, MA, USA) and a FAIMSPro™ Interface instrument (Thermo Scientific) using the NanosprayFlex™ (NSI, London, UK) ion source and setting the ion spray voltage to 2.0 kV, in the data-dependent acquisition mode, and the full scanning range was m/z 350–1500. The data were obtained by searching the database with Proteome Discoverer 2.4 software.
2.7. Determination of the Growth-Promoting Ability
The ability of strain Q11T to produce indole acetic acid (IAA) was evaluated [41], and the yield of IAA was quantitatively at the 535 nm wavelength. The nitrogen fixation ability of strain Q11T was evaluated [42], and the nitrogenase activity of the strain was determined by a nitrogenase enzyme-linked immunoassay kit (LMAI Bio, Shanghai, China). Chrome Azurol S (CAS) agar medium was used to determine whether strain Q11T had the ability to produce siderophores [43]. Strain Q11T was inoculated in organophosphorus (Mongina medium, Hopebiol, Qingdao, China) and inorganic phosphorous media and observed for the appearance of a phosphorus solubilization halo around the cells [44].
2.8. Potting Test
Strain Q11T was inoculated in LB liquid medium for 24 h, washed three times with sterile water (centrifuged at 8000 r/min for 5 min), and finally diluted with sterile water to OD600 = 1. Based on existing research [8], the potting experiment is difficult to perform due to the obstacles posed by the germination of Meconopsis integrifolia; so, we chose corn seeds for the plant growth promotion experiments (Cultivar Zhengdan 958). We used black soil, pH 6.01 ± 0.11, with an organic matter content of 26.45 ± 0.86 g/kg; other soil properties are shown in Table S5. Surface-disinfected maize seeds were sown into pots (bottom, 5 cm, top, 7.5 cm, height, 10.5 cm) containing 200 g of soil [45], placing 3 seeds per pot. The control group (CK) received 10 mL of sterile water. In the experimental group (TG), we applied 10 mL of bacterial solution. Plant height and root length (cm), stem and root fresh weight (g), stem and root dry weight (g), stem thickness (cm), and relative leaf chlorophyll content were measured after 2 weeks [45,46]. The relative content of chlorophyll (SPAD) was measured by a handheld chlorophyll meter (Konica Minolta SPAD-502 Plus, Tokyo, Japan) [47,48].
The GenBank accession numbers for the 16S rRNA gene sequence and draft genome sequences are OR016664 and JAOBQJ000000000, respectively.
RAST data: https://rast.nmpdr.org/seedviewer.cgi?page=Organism&organism=2977121.4 (accessed on 14 November 2023); antiSMASH data: https://antismash.secondarymetabolites.org/upload/bacteria-0f8777f2-48fc-4834-ac26-cad8c79c868e/index.html (accessed on 20 October 2023).
3. Results
3.1. Phylogenetic Analysis
The 16S rRNA gene sequence obtained from the analysis of the genomic DNA of strain Q11T was found to comprise 1524 bp (accession number OR016664). After comparing and querying the gene sequence of this strain in EzBioCloud and Blast, it was observed that the similarity between this strain and strains of species of Flexivirga was the highest. The strain had high sequence similarity with F. aerilata ID2601ST (99.24%), F. caeni BO-16T (97.64%), F. endophytica KCTC 39536T (97.30%), F. alba ST13T (97.09%), F. oryzae R1T (97.02%), and F. lutea KCTC 39625T (96.31%). Phylogenetic analysis based on the NJ, MP, and ML methods revealed that strain Q11T clustered together with members of the genus Flexivirga in the phylogenetic tree (Figure 1, Figures S1 and S2). This indicated that strain Q11T belongs to the genus Flexivirga.
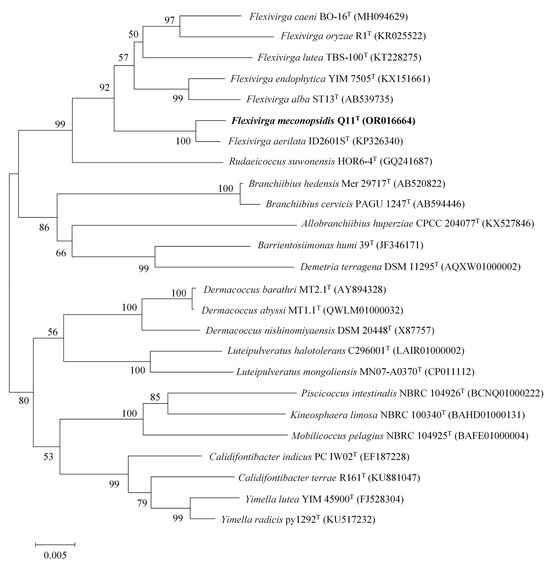
Figure 1.
Neighbor-joining phylogenetic tree of the 16S rRNA gene sequence showing the relationship between strain Q11T and strains of species of the genus Flexivirga. The number of bootstrap replications was 1000. Shown in parentheses are the accession numbers of the 16S rRNA sequences in the NCBI database. Bar, 0.005 nucleotide substitutions per position.
3.2. Genome Sequence Analysis
The genome size of strain Q11T was found to be 4.28 Mbp. The genome appeared to contain 10 contigs with an N50 value of 750,316 coding sequences. We found that the G + C content of the DNA was 68.47 mol%, the protein-coding genes were 4144, and the average gene length was 959.06, and there were 50 tRNAs and 4 rRNAs. The ANI and dDDH values of strain Q11 T and strains F. aerilata ID2601ST, F. caeni BO-16T, F. Endophytica KCTC 39536T, and F. oryzae R1T were in the 77.24–88.82% and 21.00–36.20% ranges (Table S1), respectively, and were lower than the threshold values of these spccies [49].
The RAST annotation of the strain Q11T genome showed 281 subsystems, with a coverage rate of 24%. These subsystems mainly include amino acids and derivatives, carbohydrates, and protein metabolism (Figure S3). The genome of strain Q11T was annotated using Proksee. Antibiotic resistance gene predictions indicated that there were two antibiotic resistance gene clusters. These clusters include the vanY gene in the vanB cluster [50] and the vanW gene in the vanI cluster [51]. Both the vanW and the vanY genes contribute to the gene cluster encoding glycopeptide antibiotics with a resistance mechanism associated with antibiotic target alteration [52,53]. The results of CRISPRCasFinder Annotation analysis showed that strain Q11T had five CRISPR arrays and no CAS array (Figure 2).

Figure 2.
Annotation of the genome of strain Q11T through Proksee. CARD: comprehensive antibiotic resistance database, CRISPER: CRISPR arrays, ORF: open reading frames, CDS: coding sequence.
The biosynthesis gene clusters were analyzed on the antiSMASH webpage, and the results are shown in Table 1. In Region 1, we found four types of protein synthesis gene clusters. The first was the terpene gene cluster, which is involved in the first step of the production of terpenoid compounds, which can be processed into perfumes, plant hormones, drugs, etc. [54]. It showed 38% similarity to the hopene biosynthetic gene cluster, which is involved in the synthesis of bioactive hopanoids that contribute to bacterial stress resistance [55]. The second included the non-alpha poly-amino acid (NAPAA) and RIPP-like biosynthetic gene clusters, showing 100% similarity to known gene clusters for the biosynthesis ε-poly-L-lysine, which mediates resistance to pathogenic bacteria [56]. The third was the proteusin gene cluster, which encodes ribosomally synthesized and post-translationally modified peptide (RiPP) natural products [57] and has a similarity of 14% to the aborycin biosynthetic gene cluster, which is involved in aborycin production [58]. The fourth was the betalactone gene cluster, which is involved in the production of natural β-lactone [59]. In Region 2, three types of protein synthesis gene clusters were identified. The first was also a terpene gene cluster. The second was the phenazine gene cluster, showing 30% similarity with the 5-acetyl-5,10-dihydrophenazine-1-carboxylic acid biosynthetic gene cluster; phenazine mediates resistance to pathogenic bacteria [60]. The third was the NI–siderophore gene cluster, showing 75% similarity to the desferrioxamine E biosynthetic gene cluster [61]. In region 3, there were two types of protein synthesis gene clusters. One was the linaridin gene cluster, showing 33% similarity to the 5-dimethylallylindole-3-acetonitrile gene cluster involved in a new pathway of tryptophan metabolism [62], probably requiring the biosynthesis of isoprenyl indole derivatives. The other was the ectoine gene cluster, showing 8% similarity to the kosinostatin biosynthetic gene cluster, kosinostatin being an antitumor antibiotic [63]. In region 6, only the ectoine gene cluster was predicted. Previous research [64] indicated that both kosinostatin and ectoine could protect cells from the pressure caused by the external environment.

Table 1.
The distribution of biosynthetic gene clusters in strain Q11T determined using antiSMASH.
3.3. Physiology and Chemotaxonomy
After 3 days of incubation on BA plates at 30 °C, the strain Q11T colonies had acquired slightly irregular shapes and were light brown, rod-shaped (0.67–0.77 μm in width and 0.77–0.96 μm in length) (Figure S4), and non-motile. The temperature, pH, and NaCl concentration for strain Q11T culture were 25–40 °C, 6.0–8.0, and 0–4.0%, respectively. Characteristic differences between strain Q11T and the reference strains are shown in Table 2.

Table 2.
Differential characteristics of strain Q11T compared to its relative reference strains in the genus Flexivirga. Strains: 1, Q11T; 2, Flexivirga aerilata ID2601ST 3, Flexivirga caeni BO-16T; and 4, Flexivirga endophytica KCTC 39536T. The data of the reference strains are from previous studies. Symbols: +, positive; −, negative; and w, weak reaction.
The cellular fatty acids (>5%) of strain Q11T were summed feature 9 (C16:0 10-methyl and/or iso-C17:1 ω9c) (22.5%), iso-C16:0 (19.7%), iso-C17:0 (10.5%), anteiso-C17:0 (8.7%), C18:0 (7.3%), C18:0 10-methyl (7.0%), and other types (>0.5%), as shown in Table 3. MK-8 (H4) and MK-8 (H6) were the most abundant isoprenoid quinones. The major polar lipids in strain Q11T were diphosphatidylglycerol (DPG), two aminophospholipids (APL1-2), ten phospholipids (PL1-10), two aminolipids (AL1-2), and two glycolipids (GL1-2) (Figure S5).

Table 3.
Cellular fatty acid content of strain Q11T and its relative reference strains in the genus Flexivirga. Strains: 1, Q11T; 2, Flexivirga aerilata ID2601ST 3, Flexivirga caeni BO-16T; and 4, Flexivirga endophytica KCTC 39536T. The data of the reference strains are from previous studies. Values are percentages of total fatty acids. Fatty acids that represented <0.5% in all strains were omitted. TR, trace amount (<0.5%); -, not detected. Summed features are groups of two or more fatty acids that cannot be separated using the MIDI system. Summed feature 3 contains C16:1 ω6c and/or C16:1 ω7c, and summed feature 9 contains C16:0 10-methyl and/or iso-C17:1 ω9c.
3.4. Shotgun Proteomic Analysis
The protein results of strain Q11T are shown in Table S2. It shows that 245 proteins and 794 peptides were identified in the supernatant, and 2401 proteins and 20,413 peptides were identified in the cells (Table S3).
Siderophore-interacting proteins (WP_265447369.1, WP_265443731.1) were detected in the cells [65]. These proteins can interact with other proteins to obtain elemental iron from the environment and also possess their own functions [66,67]. The adenosyl-hopene transferase HpnH (WP_265442571.1) was detected in the cells. Adenosylhopane is a precursor of C35 hopanoids, which can regulate bacterial cell membrane fluidity and permeability [68] and also helps promote beneficial interactions between bacteria and plants [69]. Acetyl-CoA C-acetyltransferase (WP_265443840.1, WP_265444891.1, WP_265445749.1) was detected in both cells and supernatant. It is used to synthesize terpenoids in Platycodon grandifloras, which is known as a traditional Asian herbal medicine and functional food [70]. The enzyme 1-deoxy-D-xylulose-5-phosphate synthase (WP_279671918.1) was detected in the cells; it is involved in the synthesis of terpenes, which are volatile substances with a unique fragrance that attract pollinators and protect against pests and diseases in plants [71]. The gentamicin biosynthesis-related Gfo/Idh/MocA oxidoreductase family (WP_265446697.1) [72] and the cobalamin biosynthesis-related cobaltochelatase subunit CobN (WP_265447325.1) [73] were also detected in the cells. More importantly, the multispecies cold shock protein (WP_171152712.1) was detected in both cells and supernatant. Studies pointed out that this protein can induce frost tolerance in plants in a low-temperature environment without affecting their normal growth and development [74].
3.5. IAA Production and Nitrogen Fixation Ability of Strain Q11T
Strain Q11T could produce IAA. The IAA content per unit volume of fermentation broth was 3.26 mg/L. (Table S4 and Figure S6). Strain Q11T showed the ability of nitrogen fixation, and its nitrogenase activity was 181.80 IU/L (Table S4 and Figure S7). IAA promotes cell division and differentiation, seed germination, and plant growth [75]. Biological nitrogen fixation is an important source of active nitrogen in the ecosystem [76]. It can not only increase soil nutrients but also improve plant habitat [77] and quickly establish a stable community suitable to plant growth [78].
Effects on Maize Growth and Development
Compared with CK, the plant height, root length, fresh weight of stem and root, and dry weight and thickness of stem and root of maize were increased significantly in the experimental group (Figure 3). There were significant differences between TG and CK in root dry weight and stem thickness, plant height, root length, fresh weight of stem and root, and stem dry weight. The experimental treatment did not affect the chlorophyll content of the leaves. The results showed that the inoculation of strain Q11T had a positive effect on the growth and development of maize.
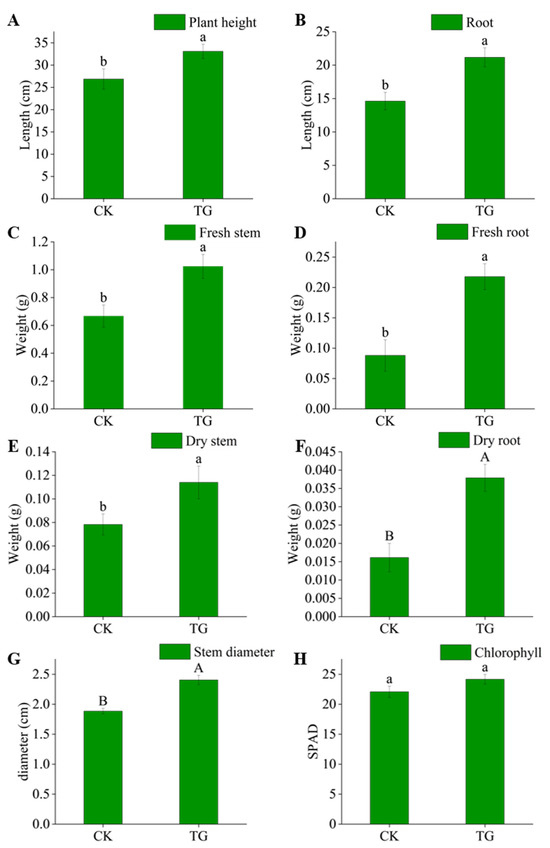
Figure 3.
Effect of strain Q11T on the growth of maize. Plant height (A), root length (B), fresh stem weight (C), fresh root weight (D), dry stem weight (E), dry root weight (F), stem diameter (G), and chlorophyll content (H). Different letters indicate significant differences between treatments (a, b for p < 0.05, A, B for p < 0.01).
4. Description of Flexivirga meconopsidis sp. nov.
Flexivirga meconopsidis (me.co.nop’si.dis. N.L. gen. n. Meconopsidis, of the Plant Meconopsis integrifolia)
The strain is characterized by aerobic, Gram-positive, rod-shaped, non-motile, irregular coccoid cells, 0.67–0.77 μm in width and 0.77–0.96 μm in length. The most suitable conditions for strain growth were 30 °C, pH 7.0, and 1% NaCl concentration. The cells were found to be oxidase-negative and catalase-positive. They showed ability to hydrolyze Tween 60. The API ZYM strip analysis showed positive results for alkaline phosphatase 8, lipase (C14), leucine arylamidase, trypsin, acid phosphatase, naphthol-AS-BI-phosphohydrolase, α-galactosidase, β-galactosidase, α-glucosidase, β-glucosidase, N-acetyl-β-glucosaminidase, α-mannosidase, and β-fucosidase, and negative results for other enzymes. The results for gelatinase, β-galactosidase, glucose, mannose, N-acetyl-glucosamine, maltose, gluconate, malic acid, and citrate of strain Q11T were positive, but the results for other indicators of assimilation were negative in the API 20NE test strip. The API 50CH strip test showed positive results for erythritol, D-galactose, D-glucose, D-fructose, D-mannose, potassium gluconate, and 2-ketogluconate, whereas other results of this test were negative. The major fatty acids (>10%) found were summed feature 9 (C16:0 10-methyl and/or iso-C17:1 ω9c), iso-C16:0, and iso-C17:0. The major respiratory quinones were MK-8 (H4) and MK-8 (H6). The main polar lipids were DPG, APL, PL, AL, and GL.
The strain was designated Q11T (=GDMCC 1.3002T=JCM 36020T); it was isolated from the seeds of Meconopsis integrifolia. The genomic DNA G + C content of this strain is 68.47 mol%.
5. Discussion
Endophytic microorganisms are often closely related to plant physiological metabolism in a reciprocal manner [10,11]. In this study, we isolated the new species Q11T from Meconopsis integrifolia seeds. Genomic ANI and dDDH analyses, together with physiological and biochemical analyses, confirmed that strain Q11T is a novel species of the genus Flexivirga. Compared with the reported strains of the genus Flexivirga [1,2,3,4,5,6], strain Q11T has the abilities of nitrogen fixation (181.80 IU/L), IAA production (3.26 mg/L in 2 days), and plant growth promotion, this last function being described for the first time for the genus Flexivirga. Previous research showed that endophytes promote plant growth through nitrogen fixation, phosphorus dissolution, and growth hormone production [12,79]. Endophytic bacteria with growth-promoting effects were isolated from plants such as rice [45], maize [80], black pepper [81], Arctium lappa L. [79], and Camellia sinensis [82]. Therefore, the isolated strain Q11T has important application potential in plant growth and agriculture.
Antibiotic resistance genes are the root cause of bacterial drug resistance. Various types of antibiotic resistance genes have been found, which cause environmental pollution [83]. They can be transmitted through bird feces [83]. Glycopeptide resistance genes (the vanY gene in the vanB cluster [50] and the vanW gene in the vanI cluster [51]) were found in strain Q11T isolated from the seeds of Meconopsis integrifolia grown in alpine areas. The secondary-metabolite synthesis gene cluster has significant value in a wide range of fields, such as drug discovery [84] and biological protection [62]. In this study, IAA-related genes (5-dimethylallylindole-3-acetonitrile [62]) and extreme environment tolerance genes (ectoine biosynthetic gene cluster [64]) were predicted in strain Q11T, which were related to the habitat of the host Meconopsis integrifolia.
Proteomics is a method to study the interactions, functions, composition, structures, and cellular activities of proteins. Proteomics results reflects cell function better than those from genomic studies [85]. Proteomics also partially confirmed the genomic predictions that the new strain contains antibiotic resistance proteins (class A beta-lactamase [86], D-alanine ligase family protein [87]), siderophore transport proteins (siderophore-interacting protein [65]), a protein that improves plant cold tolerance (multispecies cold shock protein [74]), and some proteins involved in terpenoid synthesis (adenosyl-hopene transferase HpnH [68], acetyl-CoA C-acetyltransferase [70], and 1-deoxy-D-xylulose-5-phosphate synthase [71]). Microorganisms use iron carriers to provide plants with this metal, which stimulates growth and increases plant resistance to stress [88,89]. It was reported that endophytes obtain nutrients more easily than other microorganisms [90]. Cold shock proteins could help bacteria not only withstand freezing temperatures [90] but also grow in cold environments [91]. They are mainly isolated from bacteria living in extreme environments such as glaciers [92] and cold ecosystems [93]. Therefore, it is speculated that the production of the identified cold shock protein could make the new strain tolerate extreme environments and further ensure the growth and development of the host Meconopsis integrifolia seeds at low temperatures.
Previous studies showed that the growth-promoting characteristics of the known strains are related to many mechanisms such as the presence of iron carriers, IAA production, and nitrogen fixation [12,79]. This study also confirmed through various experiments that the isolated strain Q11T has the ability to promote plant growth.
6. Conclusions
In conclusion, this study showed that strain Q11T isolated from the seeds of Meconopsis integrifolia is a novel species of the genus Flexivirga. Genomic analysis showed that it contains antibiotic resistance genes, IAA production-related genes, and extreme environment tolerance genes. The proteomic results showed that the strain produces antibiotic resistance proteins, iron transport proteins, a plant cold tolerance protein, and terpenoid synthesis proteins. The results of the functional experiments showed that the strain has the ability to produce IAA, fix nitrogen, and promote plant growth. This study is also the first to report that species of the Flexivirga genus have the function of promoting plant growth and provides some insights for improving plant growth and development.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11122899/s1, Table S1: Average nucleotide identity (ANI) and digital DNA–DNA hybridization (dDDH) values between the genome sequence of strain Q11T and those of closely related strains. Table S2: Statistics of the protein identification results. Table S3: The worksheet Supernatant_Proteome.id.anno.al. Proteins and peptides identified in the supernatant of strain Q11T; the worksheet Cells_Proteome.id.anno.all.inte: proteins and peptides identified in the cell of strain Q11T. Table S4: Data of IAA production and nitrogenase activity in strain Q11T. Table S5: Physicochemical properties of the soil. Figure S1: Maximum-likelihood phylogenetic tree of strain Q11T and its relatives based on the comparison of the 16S rRNA gene sequences. Genbank accession numbers are shown in parentheses. Figure S2: Maximum-parsimony phylogenetic tree of strain Q11T and its relatives based on the comparison of the 16S rRNA gene sequences. Genbank accession numbers are shown in parentheses. Figure S3: Different types of subsystems in the Q11T genome. The data were obtained from the RAST server. Figure S4: Transmission electron micrograph of cells of strain Q11T. Bar, 1.0 μm. The cells were incubated on BA at 30 °C for 3 days. Figure S5: The polar lipids of strain Q11T. Polar lipids in the strain, separated by two-dimensional TLC, which were detected by spraying with molybdatophosphoric acid. a, Molybdophosphoric acid was used to reveal the total polar lipids; b, ninhydrin was used for aminolipids; c, ammonium molybdate was used for phospholipids; and d, a-naphthol was used for glycolipids. DPG, diphosphatidylglycerol; PL (1–2), unidentified phospholipids; APL (1–2), unidentified aminophospholipid; AL (1–10), unidentified aminolipids; GL (1–2), unidentified glycolipids. Figure S6: The indole acetic acid (IAA) standard curve. Figure S7: Standard curve of nitrogenase activity.
Author Contributions
Conceptualization: Y.K. and L.Z.; formal analysis and investigation: Y.K.; research experiments: Y.K., L.Z. and Y.W.; writing—original draft: Y.K. and L.Z.; methodology, Y.Z. and X.J.; writing—review and editing: W.Z. and Z.R.; supervision: W.Z., Z.R. and Q.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (3211101206). This research was also supported by the Agricultural Science and Technology Innovation Program (No. CAAS-ZDRW202202 and CAAS-ZDRW202308) and the National Key Research and Development Program of China (2023YFE0104900). This research was also supported by the National Natural Science Foundation of China (No. 32160002).
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Acknowledgments
Thanks to Liu Chang of Shandong University for helping to proofread the language of this article. We thank the editors and the reviewers for their helpful feedback, which has improved this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Anzai, K.; Sugiyama, T.; Sukisaki, M.; Sakiyama, Y.; Otoguro, M.; Ando, K. Flexivirga Alba Gen. Nov., sp. Nov., an Actinobacterial Taxon in the Family Dermacoccaceae. J. Antibiot. 2011, 64, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Liu, B.B.; Yang, W.; Song, P.F.; Chen, W.; Salam, N.; Duan, Y.Q.; Li, Q.Q.; Li, W.J. Flexivirga endophytica sp. Nov., an Endophytic Actinobacterium Isolated from a Leaf of Sweet Basil. Int. J. Syst. Evol. Microbiol. 2016, 66, 3388–3392. [Google Scholar] [CrossRef]
- Kang, W.; Hyun, D.W.; Kim, P.S.; Shin, N.R.; Kim, H.S.; Lee, J.Y.; Tak, E.J.; Roh, J.R.; Park, S.D.; Shim, H.E.; et al. Flexivirga lutea sp. Nov., Isolated from the Faeces of a Crested Ibis, Nipponia Nippon, and Emended Description of the Genus Flexivirga. Int. J. Syst. Evol. Microbiol. 2016, 66, 3594–3599. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.W.; Kim, H.R.; Lee, H.J.; Jeong, S.E.; Choi, S.H.; Jeon, C.O. Flexivirga oryzae sp. Nov., Isolated from Soil of a Rice Paddy, and Emended Description of the Genus Flexivirga Anzai et al. 2012. Int. J. Syst. Evol. Microbiol. 2017, 67, 479–484. [Google Scholar] [CrossRef] [PubMed]
- Keum, D.H.; Lee, Y.J.; Lee, S.Y.; Im, W.T. Flexivirga caeni sp. Nov., Isolated from Activated Sludge. Int. J. Syst. Evol. Microbiol. 2020, 70, 1266–1272. [Google Scholar] [CrossRef]
- Chaudhary, D.K.; Lee, H.; Dahal, R.H.; Kim, D.H.; Cha, I.T.; Lee, K.; Kim, D.U. Flexivirga aerilata sp. Nov., Isolated from an Automobile Air Conditioning System. Curr. Microbiol. 2021, 78, 796–802. [Google Scholar] [CrossRef]
- Zhou, G.; Chen, Y.; Liu, S.; Yao, X.; Wang, Y. In Vitro and in Vivo Hepatoprotective and Antioxidant Activity of Ethanolic Extract from Meconopsis integrifolia (Maxim.) Franch. J. Ethnopharmacol. 2013, 148, 664–670. [Google Scholar] [CrossRef]
- Wang, Y.; Ma, Q.; Wang, L.; Hu, J.; Xue, H.; Han, D.; Xing, Z.; Ruan, Z. Structure and Function Analysis of Cultivated Meconopsis integrifolia Soil Microbial Community Based on High-Throughput Sequencing and Culturability. Biology 2023, 12, 160. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Hurek, T. Life in Grasses: Diazotrophic Endophytes. Trends Microbiol. 1998, 6, 139–144. [Google Scholar] [CrossRef]
- Bhattacharyya, P.N.; Jha, D.K. Plant Growth-Promoting Rhizobacteria (PGPR): Emergence in Agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef]
- Van Der Heijden, M.G.A.; De Bruin, S.; Luckerhoff, L.; Van Logtestijn, R.S.P.; Schlaeppi, K. A Widespread Plant-Fungal-Bacterial Symbiosis Promotes Plant Biodiversity, Plant Nutrition and Seedling Recruitment. ISME J. 2016, 10, 389. [Google Scholar] [CrossRef]
- Khan, M.S.; Gao, J.; Chen, X.; Zhang, M.; Yang, F.; Du, Y.; Moe, T.S.; Munir, I.; Xue, J.; Zhang, X. Isolation and Characterization of Plant Growth-Promoting Endophytic Bacteria Paenibacillus Polymyxa SK1 from Lilium Lancifolium. BioMed Res. Int. 2020, 2020. [Google Scholar] [CrossRef] [PubMed]
- Abedinzadeh, M.; Etesami, H.; Alikhani, H.A. Characterization of Rhizosphere and Endophytic Bacteria from Roots of Maize (Zea mays L.) Plant Irrigated with Wastewater with Biotechnological Potential in Agriculture. Biotechnol. Rep. 2019, 21, e00305. [Google Scholar] [CrossRef] [PubMed]
- Weilharter, A.; Mitter, B.; Shin, M.V.; Chain, P.S.G.; Nowak, J.; Sessitsch, A. Complete Genome Sequence of the Plant Growth-Promoting Endophyte Burkholderia Phytofirmans Strain PsJN. J. Bacteriol. 2011, 193, 3383–3384. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Li, L.; Chen, Y.; Wang, S.; Xue, L.; Meng, W.; Jiang, J.; Cao, X. Diversity of Endophytic Microbes in Taxus Yunnanensis and Their Potential for Plant Growth Promotion and Taxane Accumulation. Microorganisms 2023, 11, 1645. [Google Scholar] [CrossRef]
- Liu, Y.; Liu, L.; Qiu, F.; Schumann, P.; Shi, Y.; Zou, Y.; Zhang, X.; Song, W. Paenibacillus hunanensis sp. Nov., Isolated from Rice Seeds. Int. J. Syst. Evol. Microbiol. 2010, 60, 1266–1270. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Wang, J.; Ma, Q.; Han, X.; Zhang, W.; Ruan, Z. C Edecea sulfonylureivorans sp. Nov., a Novel Chlorimuron-Ethyldegrading Bacterium Isolated from an Herbicides-Degrading Consortium. Arch. Microbiol. 2023, 205, 21. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef]
- Felsenstein, J. Evolutionary Trees from DNA Sequences: A Maximum Likelihood Approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Miya, M.; Nishida, M. Use of Mitogenomic Information in Teleostean Molecular Phylogenetics: A Tree-Based Exploration under the Maximum-Parsimony Optimality Criterion. Mol. Phylogenet. Evol. 2000, 17, 437–455. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M. A Simple Method for Estimating Evolutionary Rates of Base Substitutions through Comparative Studies of Nucleotide Sequences. J. Mol. Evol. 1980, 16, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Fu, G.Y.; Zhang, C.Y.; Hu, J.; Xu, L.; Wang, R.J.; Su, Y.; Han, S.B.; Yu, X.Y.; Cheng, H.; et al. Isolation and Complete Genome Sequence of Algibacter alginolytica sp. Nov., a Novel Seaweed-Degrading Bacteroidetes Bacterium with Diverse Putative Polysaccharide Utilization Loci. Appl. Environ. Microbiol. 2016, 82, 2975–2987. [Google Scholar] [CrossRef] [PubMed]
- Overbeek, R.; Olson, R.; Pusch, G.D.; Olsen, G.J.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Parrello, B.; Shukla, M.; et al. The SEED and the Rapid Annotation of Microbial Genomes Using Subsystems Technology (RAST). Nucleic Acids Res. 2014, 42, D206–D214. [Google Scholar] [CrossRef] [PubMed]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.Y.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. AntiSMASH 3.0—A Comprehensive Resource for the Genome Mining of Biosynthetic Gene Clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. AntiSMASH 5.0: Updates to the Secondary Metabolite Genome Mining Pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef]
- Ruan, Z.; Wang, Y.; Song, J.; Jiang, S.; Wang, H.; Li, Y.; Zhao, B.; Jiang, R.; Zhao, B. Kurthia huakuii sp. Nov., Isolated from Biogas Slurry, and Emended Description of the Genus Kurthia. Int. J. Syst. Evol. Microbiol. 2014, 64, 518–521. [Google Scholar] [CrossRef]
- Halebian, S.; Harris, B.; Finegold, S.M.; Rolfe, R.D. Rapid Method That Aids in Distinguishing Gram-Positive from Gram-Negative Anaerobic Bacteria. J. Clin. Microbiol. 1981, 13, 444–448. [Google Scholar] [CrossRef]
- Stokes, E.J. A Guide to the Identification of the Genera of Bacteria. J. Clin. Pathol. 1968, 21, 229–230. [Google Scholar] [CrossRef]
- Chen, Y.G.; Cui, X.L.; Pukall, R.; Li, H.M.; Yang, Y.L.; Xu, L.H.; Wen, M.L.; Peng, Q.; Jiang, C.L. Salinicoccus kunmingensis sp. Nov,. a Moderately Halophilic Bacterium Isolated from a Salt Mine in Yunnan, South-West China. Int. J. Syst. Evol. Microbiol. 2007, 57, 2327–2332. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, Y.; Xue, C.X.; Li, B.; Zhou, S.; Liu, L.; Zhang, X.H. Photobacterium chitinilyticum sp. Nov., a Marine Bacterium Isolated from Seawater at the Bottom of the East China Sea. Int. J. Syst. Evol. Microbiol. 2019, 69, 1477–1483. [Google Scholar] [CrossRef] [PubMed]
- Kamlage, B. Methods for General and Molecular Bacteriology. Edited by P. Gerhardt, R.G.E. Murray, W.A. Wood and N. R. Krieg. 791 Pages, Numerous Figures and Tables. American Society for Microbiology, Washington, D.C., 1994. Price: 55.00 £. Food Nahr. 1996, 40, 103. [Google Scholar] [CrossRef]
- Sakamoto, M.; Suzuki, M.; Umeda, M.; Ishikawa, I.; Benno, Y. Reclassification of Bacteroides Forsythus (Tanner et al. 1986) as Tannerella Forsythensis Corrig., Gen. Nov., Comb. Nov. Int. J. Syst. Evol. Microbiol. 2002, 52, 841–849. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.D.; Pirouz, T.; Goodfellow, M.; Minnikin, D.E. Distribution of Menaquinones in Actinomycetes and Corynebacteria. J. Gen. Microbiol. 1977, 100, 221–230. [Google Scholar] [CrossRef]
- Minnikin, D.E.; O’Donnell, A.G.; Goodfellow, M.; Alderson, G.; Athalye, M.; Schaal, A.; Parlett, J.H. An Integrated Procedure for the Extraction of Bacterial Isoprenoid Quinones and Polar Lipids. J. Microbiol. Methods 1984, 2, 233–241. [Google Scholar] [CrossRef]
- Xu, X.W.; Huo, Y.Y.; Wang, C.S.; Oren, A.; Cui, H.L.; Vedler, E.; Wu, M. Pelagibacterium halotolerans Gen. Nov., sp. Nov. and Pelagibacterium luteolum sp. Nov., Novel Members of the Family Hyphomicrobiaceae. Int. J. Syst. Evol. Microbiol. 2011, 61, 1817–1822. [Google Scholar] [CrossRef]
- Kong, D.; Wang, Y.; Li, Q.; Zhou, Y.; Jiang, X.; Xing, Z.; Wang, Z.; Ruan, Z. Chryseobacterium subflavum sp. Nov., Isolated from Soil. Int. J. Syst. Evol. Microbiol. 2022, 72, 005345. [Google Scholar] [CrossRef]
- Omenn, G.S.; Lane, L.; Overall, C.M.; Pineau, C.; Packer, N.H.; Cristea, I.M.; Lindskog, C.; Weintraub, S.T.; Orchard, S.; Roehrl, M.H.A.; et al. The 2022 Report on the Human Proteome from the HUPO Human Proteome Project. J. Proteome Res. 2023, 22, 1024–1042. [Google Scholar] [CrossRef]
- Wilhelm, M.; Schlegl, J.; Hahne, H.; Gholami, A.M.; Lieberenz, M.; Savitski, M.M.; Ziegler, E.; Butzmann, L.; Gessulat, S.; Marx, H.; et al. Mass-Spectrometry-Based Draft of the Human Proteome. Nature 2014, 509, 582–587. [Google Scholar] [CrossRef]
- Hamane, S.; El Yemlahi, A.; Hassani Zerrouk, M.; El Galiou, O.; Laglaoui, A.; Bakkali, M.; Arakrak, A. Promoting the Growth of Sulla flexuosa L. by Endophytic Root Nodule Bacteria Authors and Affiliations. World J. Microbiol. Biotechnol. 2023, 39, 253. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Singh, P.; Sharma, A.; Guo, D.J.; Upadhyay, S.K.; Song, Q.Q.; Verma, K.K.; Li, D.P.; Malviya, M.K.; Song, X.P.; et al. Unraveling Nitrogen Fixing Potential of Endophytic Diazotrophs of Different Saccharum Species for Sustainable Sugarcane Growth. Int. J. Mol. Sci. 2022, 23, 6242. [Google Scholar] [CrossRef] [PubMed]
- Schwyn, B.; Neilands, J.B. Universal Chemical Assay for the Detection and Determination of Siderophores. Anal. Biochem. 1987, 160, 47–56. [Google Scholar] [CrossRef]
- Smith, K.P.; Goodman, R.M. Host Variation for Interactions with Beneficial Plant-Associated Microbes. Annu. Rev. Phytopathol. 1999, 37, 473–491. [Google Scholar] [CrossRef] [PubMed]
- Hernández, I.; Taulé, C.; Pérez-Pérez, R.; Battistoni, F.; Fabiano, E.; Villanueva-Guerrero, A.; Nápoles, M.C.; Herrera, H. Endophytic Seed-Associated Bacteria as Plant Growth Promoters of Cuban Rice (Oryza sativa L.). Microorganisms 2023, 11, 2317. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, H.F.; Fan, J.H.; Li, Y.Y.; Ma, L.J.; Wang, L.L.; Li, X.M. Transcriptome Modulation by Endophyte Drives Rice Seedlings Response to Pb Stress. Ecotoxicol. Environ. Saf. 2023, 254, 114740. [Google Scholar] [CrossRef] [PubMed]
- Netto, A.T.; Campostrini, E.; De Oliveira, J.G.; Bressan-Smith, R.E. Photosynthetic Pigments, Nitrogen, Chlorophyll a Fluorescence and SPAD-502 Readings in Coffee Leaves. Sci. Hortic. 2005, 104, 199–209. [Google Scholar] [CrossRef]
- Kamarianakis, Z.; Panagiotakis, S. Design and Implementation of a Low-Cost Chlorophyll Content Meter. Sensors 2023, 23, 2699. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Courvalin, P. Vancomycin Resistance in Gram-Positive Cocci. Clin. Infect. Dis. 2006, 42 (Suppl. S1), S25–S34. [Google Scholar] [CrossRef]
- Kalan, L.; Ebert, S.; Kelly, T.; Wright, G.D. Noncanonical Vancomycin Resistance Cluster from Desulfitobacterium Hafniense Y51. Antimicrob. Agents Chemother. 2009, 53, 2841–2845. [Google Scholar] [CrossRef]
- Kobayashi, S.; Kuzuyama, T.; Seto, H. Characterization of the FomA and FomB Gene Products from Streptomyces Wedmorensis, Which Confer Fosfomycin Resistance on Escherichia coli. Antimicrob. Agents Chemother. 2000, 44, 647–650. [Google Scholar] [CrossRef]
- Moens, S.; Schloter, M.; Vanderleyden, J. Expression of the Structural Gene, Laf1, Encoding the Flagellin of the Lateral Flagella in Azospirillum Brasilense Sp7. J. Bacteriol. 1996, 178, 5017–5019. [Google Scholar] [CrossRef]
- Redenbach, M.; Kieser, H.M.; Denapaite, D.; Eichner, A.; Cullum, J.; Kinashi, H.; Hopwood, D.A. A Set of Ordered Cosmids and a Detailed Genetic and Physical Map for the 8 Mb Streptomyces Coelicolor A3(2) Chromosome. Mol. Microbiol. 1996, 21, 77–96. [Google Scholar] [CrossRef]
- Belin, B.J.; Busset, N.; Giraud, E.; Molinaro, A.; Silipo, A.; Newman, D.K. Hopanoid Lipids: From Membranes to Plant-Bacteria Interactions. Nat. Rev. Microbiol. 2018, 16, 304–315. [Google Scholar] [CrossRef]
- Purev, E.; Kondo, T.; Takemoto, D.; Niones, J.T.; Ojika, M. Identification of ε-Poly-L-Lysine as an Antimicrobial Product from an Epichloë Endophyte and Isolation of Fungal ε-PL Synthetase Gene. Molecules 2020, 25, 1032. [Google Scholar] [CrossRef]
- Nguyen, N.A.; Cong, Y.; Hurrell, R.C.; Arias, N.; Garg, N.; Puri, A.W.; Schmidt, E.W.; Agarwal, V. A Silent Biosynthetic Gene Cluster from a Methanotrophic Bacterium Potentiates Discovery of a Substrate Promiscuous Proteusin Cyclodehydratase. ACS Chem. Biol. 2022, 17, 1577–1585. [Google Scholar] [CrossRef]
- Shao, M.; Ma, J.; Li, Q.; Ju, J. Identification of the Anti-Infective Aborycin Biosynthetic Gene Cluster from Deep-Sea-Derived Streptomyces sp. SCSIO ZS0098 Enables Production in a Heterologous Host. Mar. Drugs 2019, 17, 127. [Google Scholar] [CrossRef]
- Robinson, S.L.; Christenson, J.K.; Wackett, L.P. Biosynthesis and Chemical Diversity of β-Lactone Natural Products. Nat. Prod. Rep. 2019, 36, 458–475. [Google Scholar] [CrossRef]
- Heine, D.; Martin, K.; Hertweck, C. Genomics-Guided Discovery of Endophenazines from Kitasatospora sp. HKI 714. J. Nat. Prod. 2014, 77, 1083–1087. [Google Scholar] [CrossRef] [PubMed]
- Smits, T.H.M.; Duffy, B. Genomics of Iron Acquisition in the Plant Pathogen Erwinia amylovora: Insights in the Biosynthetic Pathway of the Siderophore Desferrioxamine E. Arch. Microbiol. 2011, 193, 693–699. [Google Scholar] [CrossRef]
- Ozaki, T.; Nishiyama, M.; Kuzuyama, T. Novel Tryptophan Metabolism by a Potential Gene Cluster That Is Widely Distributed among Actinomycetes. J. Biol. Chem. 2013, 288, 9946–9956. [Google Scholar] [CrossRef]
- Ma, H.M.; Zhou, Q.; Tang, Y.M.; Zhang, Z.; Chen, Y.S.; He, H.Y.; Pan, H.X.; Tang, M.C.; Gao, J.F.; Zhao, S.Y.; et al. Unconventional Origin and Hybrid System for Construction of Pyrrolopyrrole Moiety in Kosinostatin Biosynthesis. Chem. Biol. 2013, 20, 796–805. [Google Scholar] [CrossRef]
- Prabhu, J.; Schauwecker, F.; Grammel, N.; Keller, U.; Bernhard, M. Functional Expression of the Ectoine Hydroxylase Gene (ThpD) from Streptomyces chrysomallus in Halomonas elongata. Appl. Environ. Microbiol. 2004, 70, 3130–3132. [Google Scholar] [CrossRef]
- Tu, J.; Lu, F.; Miao, S.; Ni, X.; Jiang, P.; Yu, H.; Xing, L.; Yu, S.; Ding, C.; Hu, Q. The Siderophore-Interacting Protein Is Involved in Iron Acquisition and Virulence of Riemerella Anatipestifer Strain CH3. Vet. Microbiol. 2014, 168, 395–402. [Google Scholar] [CrossRef]
- Li, K.; Chen, W.H.; Bruner, S.D. Structure and Mechanism of the Siderophore-Interacting Protein from the Fuscachelin Gene Cluster of Thermobifida Fusca. Biochemistry 2015, 54, 3989–4000. [Google Scholar] [CrossRef]
- Delepelaire, P. Bacterial ABC Transporters of Iron Containing Compounds. Res. Microbiol. 2019, 170, 345–357. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Kudo, F.; Rohmer, M.; Eguchi, T. Biochemical and Mutational Analysis of Radical S-Adenosyl-L-Methionine Adenosylhopane Synthase HpnH from Zymomonas Mobilis Reveals That the Conserved Residue Cysteine-106 Reduces a Radical Intermediate and Determines the Stereochemistry. Biochemistry 2021, 60, 2865–2874. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Zhang, Y.; Sun, J.; Huang, W.C.; Xue, C.; Mao, X. A Novel Soluble Squalene-Hopene Cyclase and Its Application in Efficient Synthesis of Hopene. Front. Bioeng. Biotechnol. 2020, 8, 526933. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Yu, H.; Li, J.; Dong, N.; Chen, B.; Xu, R.; Wu, J.; Chang, X.; Wang, J.; Peng, H.; et al. Cloning, Expression, and Functional Analysis of the Full-Length CDNA of Acetyl-CoA C-Acetyltransferase (AACT) Genes Related to Terpenoid Synthesis in Platycodon Grandiflorus. Protein Pept. Lett. 2022, 29, 1061–1071. [Google Scholar] [CrossRef]
- Wang, W.; Feng, J.; Wei, L.; Khalil-Ur-Rehman, M.; Nieuwenhuizen, N.J.; Yang, L.; Zheng, H.; Tao, J. Transcriptomics Integrated with Free and Bound Terpenoid Aroma Profiling during “Shine Muscat” (Vitis Labrusca × V. Vinifera) Grape Berry Development Reveals Coordinate Regulation of MEP Pathway and Terpene Synthase Gene Expression. J. Agric. Food Chem. 2021, 69, 1413–1429. [Google Scholar] [CrossRef]
- De Araújo, N.C.; Bury, P.D.S.; Tavares, M.T.; Huang, F.; Parise-Filho, R.; Leadlay, P.; Dias, M.V.B. Crystal Structure of GenD2, an NAD-Dependent Oxidoreductase Involved in the Biosynthesis of Gentamicin. ACS Chem. Biol. 2019, 14, 925–933. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.H.; Yuan, H.; Wang, X.; Dai, H.E.; Zhang, M.; Liu, L. Crystal Structure of the Large Subunit of Cobaltochelatase from Mycobacterium Tuberculosis. Proteins 2021, 89, 462–467. [Google Scholar] [CrossRef]
- Sasaki, K.; Imai, R. Pleiotropic Roles of Cold Shock Domain Proteins in Plants. Front. Plant Sci. 2012, 2, 20693. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Hu, X.; Lu, Q.; Yang, Y.; Linghu, S.; Zhang, X. Study on the Differences in Sludge Toxicity and Microbial Community Structure Caused by Catechol, Resorcinol and Hydroquinone with Metagenomic Analysis. J. Environ. Manag. 2022, 302, 114027. [Google Scholar] [CrossRef]
- Wurzburger, N.; Bellenger, J.P.; Kraepiel, A.M.L.; Hedin, L.O. Molybdenum and Phosphorus Interact to Constrain Asymbiotic Nitrogen Fixation in Tropical Forests. PLoS ONE 2012, 7, e33710. [Google Scholar] [CrossRef]
- Stocker, B.D.; Prentice, I.C.; Cornell, S.E.; Davies-Barnard, T.; Finzi, A.C.; Franklin, O.; Janssens, I.; Larmola, T.; Manzoni, S.; Näsholm, T.; et al. Terrestrial Nitrogen Cycling in Earth System Models Revisited. New Phytol. 2016, 210, 1165–1168. [Google Scholar] [CrossRef]
- Walker Lawrence, R.; Del Moral, R. Lessons from Primary Succession for Restoration of Severely Damaged Habitats. Appl. Veg. Sci. 2009, 12, 55–67. [Google Scholar] [CrossRef]
- Liu, J.Q.; Chen, S.M.; Zhang, C.M.; Xu, M.J.; Xing, K.; Li, C.G.; Li, K.; Zhang, Y.Q.; Qin, S. Abundant and Diverse Endophytic Bacteria Associated with Medicinal Plant Arctium lappa L. and Their Potential for Host Plant Growth Promoting. Antonie Van Leeuwenhoek 2022, 115, 1405–1420. [Google Scholar] [CrossRef]
- Rana, K.L.; Kour, D.; Kaur, T.; Devi, R.; Yadav, A.; Yadav, A.N. Bioprospecting of Endophytic Bacteria from the Indian Himalayas and Their Role in Plant Growth Promotion of Maize (Zea mays L.). J. Appl. Biol. Biotechnol. 2021, 9, 41–50. [Google Scholar] [CrossRef]
- Ashajyothi, M.; Kumar, A.; Sheoran, N.; Ganesan, P.; Gogoi, R.; Subbaiyan, G.K.; Bhattacharya, R. Black Pepper (Piper nigrum L.) Associated Endophytic Pseudomonas Putida BP25 Alters Root Phenotype and Induces Defense in Rice (Oryza sativa L.) against Blast Disease Incited by Magnaporthe Oryzae. Biol. Control 2020, 143, 104181. [Google Scholar] [CrossRef]
- Shan, W.; Zhou, Y.; Liu, H.; Yu, X. Endophytic Actinomycetes from Tea Plants (Camellia sinensis): Isolation, Abundance, Antimicrobial, and Plant-Growth-Promoting Activities. BioMed Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Jian, Z.; Zeng, L.; Xu, T.; Sun, S.; Yan, S.; Yang, L.; Huang, Y.; Jia, J.; Dou, T. Antibiotic Resistance Genes in Bacteria: Occurrence, Spread, and Control. J. Basic Microbiol. 2021, 61, 1049–1070. [Google Scholar] [CrossRef] [PubMed]
- Newman, D.J.; Cragg, G.M. Natural Products as Sources of New Drugs over the Last 25 Years. J. Nat. Prod. 2007, 70, 461–477. [Google Scholar] [CrossRef] [PubMed]
- Al-Amrani, S.; Al-Jabri, Z.; Al-Zaabi, A.; Alshekaili, J.; Al-Khabori, M. Proteomics: Concepts and Applications in Human Medicine. World J. Biol. Chem. 2021, 12, 57. [Google Scholar] [CrossRef] [PubMed]
- Bush, K.; Bradford, P.A. β-Lactams and β-Lactamase Inhibitors: An Overview. Cold Spring Harb. Perspect. Med. 2016, 6, a025247. [Google Scholar] [CrossRef] [PubMed]
- Ambúr, O.H.; Reynolds, P.E.; Arias, C.A. D-Ala:D-Ala Ligase Gene Flanking the VanC Cluster: Evidence for Presence of Three Ligase Genes in Vancomycin-Resistant Enterococcus Gallinarum BM4174. Antimicrob. Agents Chemother. 2002, 46, 95–100. [Google Scholar] [CrossRef]
- Cao, L.; Gao, Y.; Yu, J.; Niu, S.; Zeng, J.; Yao, Q.; Wang, X.; Bu, Z.; Xu, T.; Liu, X.; et al. Streptomyces Hygroscopicus OsiSh-2-Induced Mitigation of Fe Deficiency in Rice Plants. Plant Physiol. Biochem. 2021, 158, 275–283. [Google Scholar] [CrossRef]
- Cui, K.; Xu, T.; Chen, J.; Yang, H.; Liu, X.; Zhuo, R.; Peng, Y.; Tang, W.; Wang, R.; Chen, L.; et al. Siderophores, a Potential Phosphate Solubilizer from the Endophyte streptomyces sp. CoT10, Improved Phosphorus Mobilization for Host Plant Growth and Rhizosphere Modulation. J. Clean. Prod. 2022, 367, 133110. [Google Scholar] [CrossRef]
- Araujo, R.; Kaewkla, O.; Franco, C.M.M. Endophytic Actinobacteria: Beneficial Partners for Sustainable Agriculture. In Endophytes Biology and Biotechnology; Springer: Berlin/Heidelberg, Germany, 2017; pp. 171–191. [Google Scholar] [CrossRef]
- Roberts, M.E.; Inniss, W.E. The Synthesis of Cold Shock Proteins and Cold Acclimation Proteins in the Psychrophilic Bacterium Aquaspirillum Arcticum. Curr. Microbiol. 1992, 25, 275–278. [Google Scholar] [CrossRef]
- Margesin, R.; Zhang, D.C.; Frasson, D.; Brouchkov, A. Glaciimonas frigoris sp. Nov., a Psychrophilic Bacterium Isolated from Ancient Siberian Permafrost Sediment, and Emended Description of the Genus Glaciimonas. Int. J. Syst. Evol. Microbiol. 2016, 66, 744–748. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Margesin, R. Psychrophilic Lifestyles: Mechanisms of Adaptation and Biotechnological Tools. Appl. Microbiol. Biotechnol. 2019, 103, 2857–2871. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).