Abstract
Antibiotic-resistant bacteria represent a major public health concern, especially impacting medical care centers and hospitals, thereby challenging the effectiveness of current infection treatment protocols. The emergence and persistence of antimicrobial resistance in the environment have been thoroughly researched, with a focus on the aquatic environment as a potential reservoir of these bacteria in areas with anthropogenic contamination. Having this in mind, this work aims to investigate the water streams of Riguinha and Brito Capelo Street, both of which ultimately flow into Matosinhos Beach in Portugal, to determine the potential presence of fecal contamination. Six water samples were collected and analyzed within twenty-four hours from these two water streams. A phenotypic characterization was performed in various volumes on MacConkey agar with antibiotics. Randomly selected lactose-fermenting gram-negative bacteria underwent antimicrobial susceptibility tests using the agar diffusion method following EUCAST guidelines, covering β-lactam and non-β-lactam antibiotics. The isolates were analyzed through Polymerase Chain Reaction. The findings of this study confirm that both water streams were contaminated by multidrug-resistant bacteria such as Enterobacteriaceae, including Escherichia coli, the KESC group, and Pseudomonas, exhibiting extended-spectrum β-lactamases (ESBL), AmpC β-lactamases, and carbapenemases. These indicate the presence of fecal contamination with relevant antimicrobial-resistant threats.
1. Introduction
Multidrug-resistant Enterobacteriaceae poses a global public health threat due to limited treatment options, which undermine the effectiveness of the current range of antibiotics [1]. Enterobacteriaceae (including Escherichia coli, Klebsiella spp., and Enterobacter spp.) are gram-negative bacteria which inhabit a mammal’s intestinal tract, as well as soil, water, sewage, and food. They are among the most usual human opportunistic pathogens, being responsible for community and healthcare-related diseases like gastroenteritis, pneumonia, urinary tract infections, and meningitis. As antibiotic resistance can also be detected in animals and in the environment, the One Health concept has been implemented by the World Health Organization, emphasizing interconnected health systems (WHO) [2]. These bacteria have acquired resistance to the antibiotics most used and prescribed to treat infections, such as β-lactams (penicillin, cephalosporins, and carbapenems), over the years [3].
The misuse of antibiotics significantly contributes to the emergence of antibiotic resistance. On the one hand, over-consumption of antibiotics not only kills beneficial bacteria in our system, our natural and indispensable microbiota, but also enhances the growth of both antibiotic-resistant commensal and pathogenic microorganisms. On the other hand, the underuse of antibiotics, wherein individuals fail to complete the prescribed treatment, provides an opportunity for resistance to develop. It is imperative to use antibiotics wisely and responsibly to ensure their effectiveness now and future [4].
In terms of resistance mechanisms, extended-spectrum β-lactamases (ESBL) producing Enterobacteriaceae (E-ESBL) are relevant opportunistic pathogens considered a threat to human health. They are responsible for treatment failures in severe infections and silent colonization of the community population’s intestinal tracts, with a particularly notable impact on the elderly. The presence of E-ESBL is not confined to hospital environments; they can be found in several ecological niches, such as in the environment. ESBLs are enzymes responsible for the hydrolysis of oxyimino-β-lactam antibiotics, considered significant therapeutic agents for treating severe human and animal infections. CTX-M, SHV, and TEM are the most prevalent types of these enzymes globally, but numerous other variants exist [5]. Among CTX-M enzymes, CTX-M-1, CTX-M-14, and CTX-M-15 are the most widespread in human-related settings and are frequently associated with E-ESBL reports. This suggests that anthropogenic pressure upon natural environments has a strong influence on antimicrobial resistance emergence, including the dissemination of genes encoding these enzymes. The CTX-M-15 variant is responsible for infectious outbreaks worldwide and is often linked to high-risk clones of Escherichia coli, such as CTX-M-15/ST131 E. coli. This clone is accountable for extraintestinal infections resistant to antibiotics. The increasing frequency of infections caused by E. coli isolates producing ESBLs is a serious concern, as the efficacy of third generation cephalosporins against this pathogen is progressively diminishing [3].
AmpC β-lactamases are widely distributed and can also inactivate β-lactam antibiotics such as cephalosporins. Initially believed to be chromosomal, they have been recognized as plasmid-mediated cephalosporinases as well, since the 1990s. These enzymes contribute to therapeutic failures in community-acquired infections, especially involving E. coli, Klebsiella pneumoniae, and Salmonella enterica. The most disseminated AmpC cephalosporinase globally is CMY-2, associated with Enterobacterales infections in humans [3]. AmpC β-lactamases cannot degrade carbapenems; however, they block their action by binding to them. Plasmid-encoded AmpC CMY-2 is often found in Enterobacteriaceae like E. coli, causing global carbapenem resistance [6].
Carbapenems were considered the antibiotics of choice for treating infections caused by extended-spectrum β-lactamase and AmpC producers [1]. Carbapenem-resistant Enterobacteriaceae (CRE) are currently increasing globally, posing a major threat as carbapenems are last-line antibiotics, and leading to longer hospital stays, increased costs, and mortality. The World Health Organization (WHO) categorizes CRE as critical drug-resistant bacteria, and the Centers for Disease Control (CDC) identifies Klebsiella spp., Escherichia coli, and Enterobacter spp. as prominent emerging concerns in carbapenem resistance [1]. CRE are not only nosocomial and community-acquired pathogens but are also found in food-producing animals and in the environment. Key resistance mechanisms include carbapenemases production (β-lactamase enzymes), altered or mutated porins, enhanced efflux pump-action, and modified penicillin-binding proteins. Carbapenemases, hydrolytic enzymes, come in non-metallo and metallo forms, catalyzing carbapenem hydrolysis using serine or zinc as active catalytic substrates. The genes that encode theses enzymes are often located on plasmids or associated with mobile elements (transposons or integrons), facilitating their spread [7]. Carbapenemases mainly belong to three β-lactamase classes: Ambler A (Klebsiella pneumoniae carbapenemase, e.g., blaKPC), B or metallo-β-lactamases (e.g., blaIMP, blaVIM, blaNDM), and D or oxacillinases (e.g., blaOXA-48), which are carried either on chromosomes or acquired via plasmids [8]. KPC- and NDM-producing bacteria have shown resistance to most β-lactams, fluoroquinolones, and aminoglycosides. As healthcare facilities can function as reservoirs for CRE, KPC-producing Klebsiella pneumoniae has already been associated with hospital outbreaks across Europe. OXA-48-like carbapenemases, when associated with extended-spectrum β-lactamases, can also create high resistance levels [7]. Since carbapenems are not entirely metabolized within the body, some residues found in human excreta may enter hospital sewage. This raises the concern of pathogens in hospital effluent developing resistance to carbapenems due to the selective pressure upon them. It is thought that hospital sewage may act as a reservoir for resistance genes, allowing organisms to acquire resistance through horizontal gene transfer events. Likewise, antibiotic residues discharged into wastewater along with human feces could contribute to the selection of carbapenemases-producing Enterobacteriaceae in water sources [7]. Resistance evolves continuously in the Enterobacteriaceae family [1].
A standardized international terminology was established to harmonize definitions for various patterns of bacterial resistance. This initiative not only improved the comparability of surveillance data but also facilitated global, regional, and local epidemiological assessments of public health impact. Multidrug-resistant (MDR) is characterized by non-susceptibility to at least one agent in three or more antimicrobial categories. Extensively drug-resistant (XDR) is defined by non-susceptibility to at least one agent in all but two or fewer antimicrobial categories (bacterial isolates susceptible to only one or two categories). Lastly, Pandrug-resistant (PDR) means non-susceptibility to all agents in all antimicrobial categories [9].
Contamination of environmental water, including bathing water, through the release of sewage, fecal discharge, agricultural runoff, and healthcare effluents containing huge amounts of antimicrobial-resistant organisms could seriously impact water quality and jeopardize the health of vulnerable people [4,10]. It is imperative that significant measures be taken to reduce infection rates, especially those requiring antibiotic treatment, by identifying environments contaminated with antibiotic-resistant bacteria [11,12].
Matosinhos Beach is a highly frequented beach in Portugal (Figure 1), drawing visitors not only during the peak summer season but also throughout the winter months. This enduring popularity can be attributed to the presence of numerous surf schools. However, recent concerns have arisen regarding the water quality at this beach, prompting several inspection initiatives and water analysis. Reports have shed light on health issues experienced by surfers who frequent Matosinhos Beach, including gastrointestinal discomfort and skin irritations, which may be related to elevated levels of bacteria, notably intestinal Enterococci and Escherichia coli, detected in the waters [13]. Visitors to this beach are often discouraged from swimming due to bacteriological contamination, and it is suspected that one contributing factor could be the streams that flow into the beach [14,15].
Figure 1.
Matosinhos beach, Matosinhos, Portugal.
The rainwater drainage system of Riguinha is mainly situated within the municipality of Matosinhos, though it also receives contributions from the municipality of Porto, encompassing the region spanning from Praça da Cidade de Salvador to Edifício Transparente. This drainage network is bifurcated into two main branches, corresponding to the hydrographic sub-basins of the Riguinha and Carcavelos streams (as illustrated in Figure 2) [16].
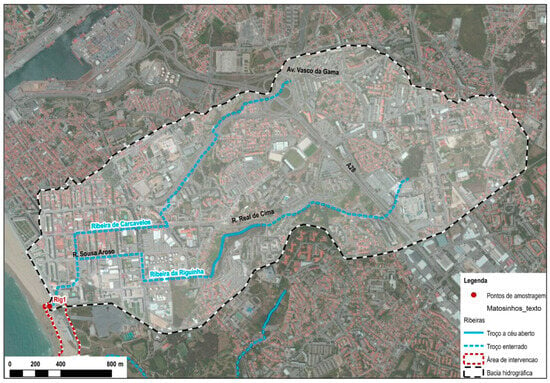
Figure 2.
Route map of Riguinha water stream [16].
The Riguinha stream originates at Fonte das Sete Bicas and runs approximately 3.8 km before merging with the boundary between Matosinhos Beach and International Beach. Meanwhile, the Carcavelos stream, originating near Feira da Senhora da Hora, runs 2.7 km before converging with the Riguinha stream, near the intersection of Sousa Aroso Avenue and D. João I Street (Figure 2) [16]. As both water streams are in a highly urbanized and densely populated area, only a small section of the Riguinha stream is in the open, next to the Parque Real, in Matosinhos. In 2014, water analyses already revealed elevated concentrations of E. coli, suggesting the presence of fecal contamination in this stream. Bathing water quality is one of the most important criteria for the beach to earn a blue flag designation and often requires structural changes to domestic wastewater and rainwater drainage networks [16].
This present study aims to investigate two water streams: the Riguinha natural water stream and the water drainage from Brito Capelo Street, both of which flow into Matosinhos beach. The objective is to assess possible fecal contamination and antimicrobial resistance in selected isolates. For this purpose, several water samples were analyzed, and the authors performed a phenotypic and genotypic characterization, including Polymerase Chain Reaction (PCR) detection of resistance genes for the most relevant bacteria identified.
2. Materials and Methods
2.1. Water Sampling
This research involved the collection of six water samples. Three of these samples were obtained from the Riguinha stream, while the remaining three were taken from Brito Capelo Street’s drainage. To minimize the risk of contamination, all water samples were collected using gloves into a sterile Schott container with a capacity of 1 L, which was opened only at the moment of collection. All samples were collected between October 2021 and April 2022. The first one was on 20 October at 3:35 p.m., the second on 29 November at 7:15 p.m., the third and fourth on 10 February at 10:40 a.m. and 11:25 a.m., respectively, and finally the fifth and sixth were on 11 April at 2:30 p.m.
The samples were coded based on their collection location as follows: the first two samples (MAT1 and MAT2) were obtained from the water drainage system on Brito Capelo Street; the third and fourth (MAT3 and MAT4) from Riguinha stream—specifically, from the mouth of the stream and the open access section, respectively—and the fifth (MAT5) once again from the mouth of Riguinha stream. The last sample (MAT6) was collected from Brito Capelo Street drainage once more. For visual reference, Scheme 1 and Scheme 2 illustrate the precise locations from which theses samples were acquired. All the flow captured by the rainwater drainage system of Riguinha stream and Brito Capelo Street flows into Matosinhos Beach (Scheme 1).

Scheme 1.
Tunnel on the right: final section of Riguinha stream, concrete pipe 4.4 m wide and 2.3 m high. On the left: smaller concrete pipe corresponding to the water drainage of Brito Capelo Street, Matosinhos.

Scheme 2.
Open-air section of Riguinha stream near Parque Real, Matosinhos.
2.2. Culture Medium Preparation
For this study, MacConkey agar, MacConkey agar with antibiotics (ampicillin, cefotaxime, and meropenem), and Mueller–Hinton mediums were used, both sourced from Liofilchem, Italy. MacConkey agar is a selective medium for gram-negative bacteria and differential, whereas Mueller–Hinton is a non-selective and non-differential medium. MacConkey agar medium was prepared following the manufacturer’s instructions. In cases where antibiotics were required, they were added to the medium before distribution, as antibiotics can be degraded by high temperatures. The volume of antibiotic added to each medium was calculated, considering the final concentration needed, the initial concentration of the stock solution, and the final medium volume (400 mL). The final concentrations were 100 μg/mL for ampicillin, 1 μg/mL for cefotaxime, and 0.5 μg/mL for meropenem. After solidifying, the medium was ready for use. The same procedure was followed for preparing the Mueller–Hinton medium.
Finally, all the mediums were tested using E. coli ATCC 25922 (ATCC—American Type Culture Collection, Manassas, VA, USA), as a control method. For the mediums to be considered valid, bacterial growth was expected in MacConkey agar but should have been absent in MacConkey agar with antibiotics, as this strain lacks acquired resistance to these antibiotics.
2.3. Water Filtration
To initiate the research, the standard method of membrane filtration was performed on all water samples within 24 h of collection [17]. A sterile cellulose acetate membrane with a pore size of 0.2 μm and a diameter of 47 mm, sourced from the Advantec MFS™ brand (Dublin, CA, USA), was used. This specific pore size allowed the capture of bacteria on top of the membrane, as the bacteria are smaller than the pore size. In the initial exploratory approach, sample volumes of 100 mL and 10 mL were used. However, the bacterial growth was so dense, with no isolated colonies, that it was necessary to modify the approach. As a result, subsequently tested volumes were of 1 mL (with a 1/20 dilution using sterile water) and 100 μL, by spread using a sterile glass Pasteur pipette in L format. Isolated bacterial colonies were obtained only from the last two volumes. The membranes from each filtration volume were placed on both MacConkey agar and MacConkey agar supplemented with ampicillin, cefotaxime, and meropenem. They were then incubated overnight at 37 °C. Scheme 3 depicts the membranes from the 1 mL filtration placed on MacConkey agar, both with and without antibiotics. A noticeable difference in bacterial growth density was observed between MacConkey agar without antibiotics (Scheme 3d) and MacConkey agar with meropenem (Scheme 3a).
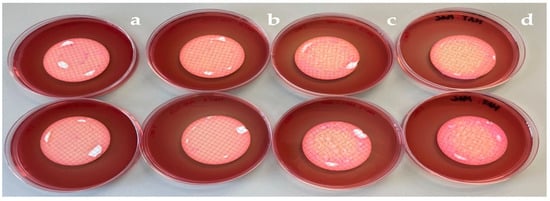
Scheme 3.
Membranes of 1 mL filtration after 24 h incubation at 37 °C placed in MacConkey agar (d), MacConkey agar with meropenem (a), with cefotaxime (b), and ampicillin (c).
2.4. Phenotypic Characterization
The colonies obtained from the filtration membranes were spread in new culture media to achieve isolated and pure colony-forming units (cfu). Mainly lactose-fermenting, gram-negative bacteria were chosen, at random, from MacConkey agar with cefotaxime and meropenem for further phenotypic characterization, as these bacterial isolates are more relevant. Some lactose non-fermenting bacteria were also analyzed, such as Pseudomonas spp., making a total of 38 isolates (16 from Brito Capelo Street and 22 from Riguinha stream).
Antimicrobial susceptibility tests were conducted using the Mueller–Hinton medium for all target isolates, employing the agar diffusion method according to EUCAST [18,19]. This included the assessment of β-lactams, non-β-lactams, and specific antibiotics for Pseudomonas [19].
The placement of antibiotic disks, sourced from Liofilchem, Italy, or OXOID, United Kingdom, was performed with a specific aim to detect synergisms between oxyimino β-lactam antibiotics and the combination of amoxicillin with clavulanic acid that was centrally positioned on the plates. Each disk was applied with a consistent relative distance of 20 mm between them.
The antibiotics used, along with their abbreviations and disk contents (μg), were as follows:
- -
- β-lactam antibiotics: amoxicillin-clavulanic acid (AMC/AUG30), ampicillin (AMP10), aztreonam (ATM30), cefepime (FEP30), cefotaxime (CTX30), cefoxitin (FOX30), ceftaroline (CPT30), ceftazidime (CAZ30), and meropenem (MRP/MEM10).
- -
- Non β-lactam antibiotics: chloramphenicol (C30), ciprofloxacin (CIP5), fosfomycin (FO200), gentamicin (GEN/CN10), nitrofurantoin (F300), tigecycline (TGC15), tetracycline (TE30), tobramycin (TOB10), and trimethoprim-sulfamethoxazole (SXT25).
- -
- Specific antibiotics for Pseudomonas: doripenem (DOR10), aztreonam (ATM30), cefepime (FEP30), levofloxacin (LEV5), ceftazidime (CAZ10), gentamicin (GEN10), meropenem (MRP10), tobramycin (TOB10), netilmicin (NET30), amikacin (AK30), ceftazidime (CAZ30), piperacillin + tazobactam (TZP110), piperacillin (PRL100), and ciprofloxacin (CIP5).
Additionally, a quality control assessment of the antibiogram disks was conducted using a suspension of E. coli ATCC 25922. This was performed to ensure that inhibition zones fell within acceptable ranges. Results were compared to EUCAST quality control (QC) tables [20]. The inhibition zone diameter for each antibiotic was measured with a digital caliper in millimeters and compared with the EUSCAST Breakpoint Tables to categorize the isolates as standard dosing regimen (S), susceptible, increased exposure (I), or resistant (R) [21].
Bacterial isolates were presumptively identified based on their color on Chromogenic UTI Medium from Himedia, Maharashtra, India, which allows the identification of bacteria typically associated with urinary tract infections. Bacteria from the KESC group (Klebsiella spp., Enterobacter spp., Serratia spp., and Citrobacter spp.) exhibited blue-colored colonies, Pseudomonas displayed yellow colonies, and Escherichia coli showed pink/purple colonies.
An oxidase test was performed to confirm that some isolates were Pseudomonas, while the S.I.M test was used to distinguish between E. coli and the KESC group. The oxidase strips used were from Liofilchem, Roseto degli Abruzzi, Italy, and the S.I.M medium was from Microbact, Durham, UK. The oxidase test was employed to determine the presence of the cytochrome c oxidase. This test is useful for differentiating between the families Pseudomonadaceae (oxidase-positive) and Enterobacteriaceae (oxidase-negative). The S.I.M test was used to evaluate whether the isolates could produce hydrogen sulfide, their motility, and their ability to produce indole from tryptophan, with the addition of Ehrlich reagent to the medium.
Regarding the resistance mechanisms, AmpC producers were confirmed via phenotypic tests with inhibitors (cloxacillin), ESBL producers using double-disk synergy test, while carbapenemase producers were detected using Carbapenem Inactivation Method (CIM). To identify AmpC producers, we measured the difference in inhibition zones between cefoxitin (FOX30) and cefoxitin with cloxacillin (FOC230). An isolate was classified as an AmpC producer if the difference was greater than or equal to 3.5 mm. Furthermore, these isolates had to exhibit resistance to CTX, CAZ, AMC, and FOX, while being susceptible to FEP, following the guidelines outlined in EUCAST [22].
ESBL producers were confirmed via the characteristic synergism observed when the antibiotic disks cefotaxime (CTX30) and amoxicillin-clavulanic acid (AMC30) were brought together (20 mm), or/and if the difference between the inhibition zones for cefotaxime (CTX30) and cefotaxime + clavulanic acid (CTL40) was greater than or equal to 3.5. mm. Additionally, the isolates had to exhibit resistance to CTX and CAZ, while remaining susceptible to FOX and AMC [22].
Lastly, carbapenemase producers were identified via the Carbapenem Inactivation Method (CIM), a phenotypic test that detects carbapenemase activity in gram-negative rods within eight hours. It can reliably detect activity encoded by various β-lactamase genes such as blaKPC, blaNDM, blaOXA-48, blaVIM, blaIMP, and blaOXA-23 in species of Enterobacteriaceae (e.g., Klebsiella pneumoniae and Escherichia coli), but also in non-fermenters (e.g., Pseudomonas aeruginosa). In order to perform the CIM, a 10 μL inoculation loop of culture, taken from a Mueller–Hinton, was suspended in 400 μL water on an Eppendorf tube. Subsequently, a susceptibility-testing disk containing 10 μg meropenem was immersed in the suspension and incubated for a minimum of two hours at 37 °C. Afterwards, the disk was removed from the suspension using an inoculation loop, placed on a Mueller–Hinton agar plate inoculated with a susceptible E. coli indicator strain (ATCC 29522), and then incubated overnight at 37 °C. This indicator strain inoculation was performed with a suspension of 0.5 McFarland, spread in Mueller–Hinton agar using a sterile cotton swab. A positive result meant that the bacterial isolate produced carbapenemase, and the meropenem in the susceptibility disk was inactivated, allowing uninhibited growth of the susceptible E. coli ATCC25922, as demonstrated in Scheme 4. A negative one meant that the bacterial isolate did not produce carbapenemases and a clear inhibition zone was observed [23].
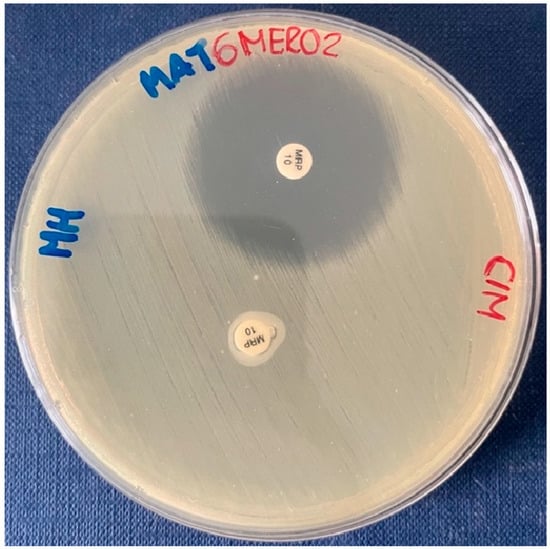
Scheme 4.
CIM positive for MAT6MERO2 isolate, as the meropenem in the susceptibility test disk was inactivated, allowing E. coli ATCC25922 to grow. On the contrary, the antibiotic on the control disk inhibited the growth of this strain.
2.5. Genotypic Detection
The genotypic characterization was only performed on bacterial isolates from samples MAT5 and MAT6 (14 isolates), as representative samples of the bacteria present in the water of Riguinha stream and the water drainage from Brito Capelo Street, respectively. DNA extraction was carried out from all the bacteria of interest using the alkaline lysis method. The purpose of this procedure was to isolate DNA for further analysis.
Procedure:
- 20 mL of alkaline lysis buffer, consisting of 0.05 M NaOH and 0.25% sodium dodecyl sulfate (SDS), was added to a 0.2 mL Eppendorf tube to suspend 2 to 3 bacterial colonies in the solution.
- The bacterial suspension was then incubated in a thermal cycler at 99 °C for 15 min. During this incubation, the detergent (SDS) disrupted cell membranes, allowing the alkaline environment to denature both chromosomal and plasmid DNA.
- Following incubation, 180 mL of Tris-EDTA (TE 1x) buffer solution was gently added, and the solution was homogenized before centrifuging at 12,000 rpm for 15 min.
- The resulting supernatant, approximately 150 mL, was carefully removed and transferred to a new Eppendorf tube for storage. With the addition of a neutralization buffer, the DNA was renatured and a white precipitate that consisted of SDS, lipids, and proteins was formed.
The extracted DNA was stored at 4 °C for a few months (up to six months), but it could also be kept at −20 °C for long-term preservation if necessary. MAT5 and MAT6 isolates were screened via Polymerase Chain Reaction. A multiplex PCR for detection of the most frequent acquired carbapenemase genes, including blaNDM, blaOXA-48, and blaKPC, was performed. All CIM-positive bacterial isolates, as well as those exhibiting resistance to meropenem or reduced susceptibility to this antibiotic, were tested.
In this process, 2 μL of total DNA were subjected to multiplex PCR in a 25 μL reaction mixture. The mix for detecting the mentioned genes included SuperHot Master Mix Taq Polymerase, from BIORON (concentration 2x), along with each primer (forward and reverse) and sterile ultra-pure water. For positive controls, we used standard controls obtained from DNA extraction of well-characterized strains carrying these resistance genes. One control was of blaKPC and the other of blaOXA (OXA-48, TEM, SHV). For negative control, water was used instead of DNA.
Amplification was carried out with the following thermal cycling conditions: 10 min at 94 °C (bacteria lysis and release of DNA); 36 cycles of amplification consisting of 30s at 94 °C (denaturation), 40 s at 52 °C (primer-specific annealing), and 50 s at 72 °C with 5 min at 72 °C for the final extension [24]. Table 1 shows the primers and their sequences (5′–3′) of sense primer (F) and antisense primer (R) that were used in this PCR to amplify specific fragments of the resistance genes and the product size (bp).

Table 1.
Genes that confer resistance to carbapenems that were tested, nucleotide sequence (5′–3′) of sense (F) and antisense (R) primers used, and PCR product size for each primer (bp).
A second Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases was performed. This assay allowed the authors to detect and distinguish alleles-encoding CTX-M enzymes from all five phylogenetic groups. Specific primers for alleles-encoding enzymes belonging to groups 1, 2, 8, 9, and 25 were used. The tested blaCTX-M genes groups, primer pairs, and predicted amplicon sizes are presented in Table 2. Fragments of alleles-encoding enzymes of groups 8 (666 bp) and 25 (327 bp) were amplified using two specific forward primers and a shared reverse primer. Once more, 2 μL of total DNA were subjected to multiplex PCR in a 25 μL reaction mixture. The only variation in the detection mix was the set of primers used [25].

Table 2.
Groups of CTX-M genes that were tested, nucleotide sequence (5′–3′) of sense (F) and antisense (R) primers used, and PCR product size for each primer (bp).
Only two positive controls were applied: +24 (B0HH CTX-M-15 (G1)) and +25 (KP18 SHV-2, CTX-M-2 (G2)). Amplification was carried out with the following conditions: initial denaturation at 94 °C for 5 min; 30 cycles of 94 °C for 25 s, 52 °C for 40 s, and 72 °C for 50 s, and a final elongation step at 72 °C for 6 min [25].
Lastly, two Simplex PCR assays were performed to detect blaAmpC genes, in this specific case: bladha-1, bladha-2 and blaCMY. For this PCR, the genes tested, primer pairs (forward and reverse), and predicted amplicon sizes are presented in Table 3. For the detection of bladha-1 and bladha-2 genes, the amplification conditions were as follows: 3 min at 94 °C; 25 amplification cycles consisting of 30 s at 94 °C, annealing at 64 °C 30 s, and 1 min at 72 °C, with 7 min at 72 °C for the final extension [26]. For the detection of blaCMY gene, amplification conditions were as follows: 5 min at 94 °C; 35 amplification cycles consisting of 1 min at 94 °C, 1 min at 49 °C, and 1 min at 72 °C, with 7 min at 72 °C for the final extension [27].

Table 3.
AmpC genes which were tested, nucleotide sequence (5′–3′) of sense (F) and antisense (R) primers used, and PCR product size for each primer (bp).
PCR products were identified through agarose gel electrophoresis, incorporating Sybr® Safe as DNA intercalating dye. The results were observed with a transilluminator and saved using a BIO-RAD Gel DocTM XR+ System, with Image LabTM Software (https://www.bio-rad.com/en-cn/product/image-lab-software?ID=KRE6P5E8Z accessed on 28 September 2023). The agarose gel was prepared by adding 1–2% agarose (GeneOn) in sterile water and heating in the microwave until a clean solution was obtained, followed by the addiction of 3.5 mL of GreenSafe Premium (nzytech). The amplicons’ sizes were compared to a DNA ladder (NZYDNA Ladder VI from nzytech). A loading Buffer II (from BIORON (concentration 6x)) was mixed with every PCR product before initiating this procedure.
For the separation of Multiplex PCR products, carbapenemases and extended-spectrum β-lactamases DNA fragments were analyzed via electrophoresis in a 2% agarose gel at 90 V for 90 min in 1xTAE (40 mmol/L Tris–HCl [pH 8.3], 2 mmol/L acetate, 1 mmol/L EDTA) containing 0.05 mg/L ethidium bromide [24]. On the other hand, for separation of the Simplex PCR products, AmpC DNA fragments were analyzed via 1.5% agarose gel electrophoresis at 120 V with a running time of 30 min in 1XTAE.
3. Results
3.1. Water Filtration
A surprisingly abundant bacterial growth was observed on all membranes of the various filtration volumes performed on each collected water sample. These samples were collected randomly, considering different time frames and climate conditions. Bacteria from three distinct groups (Pseudomonas, KESC, and E. coli) were consistently obtained. The observation of isolated colony-forming units was achieved by either spreading 100 µL on MacConkey agar with meropenem or cefotaxime, or by using 1 mL filtration membranes placed on MacConkey agar with meropenem.
The reduction in bacterial growth became quite evident when comparing larger and smaller volumes of filtration membranes (100 mL–1 mL with dilution and 100 µL). Similar trends were observed when comparing bacterial growth on MacConkey agar and MacConkey agar with antibiotics, particularly meropenem, as illustrated in Scheme 5. While the authors conducted a random selection of lactose-fermenting isolates resistant to CTX and MRP, it is important to note that all isolates observed were analyzed. Thus, our interpretation of the results obtained was primarily qualitative. Lactose-fermenting and non-fermenting bacteria were found, the latter in large numbers, especially in the MAT6 sample.
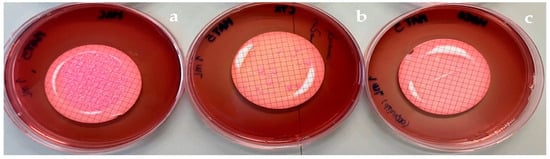
Scheme 5.
1 mL filtration membranes of MAT5 water sample placed in MacConkey agar (a), with cefotaxime (b), and with meropenem (c).
3.2. Bacterial Identification
Among all the bacteria found in the water samples, a total of 38 bacterial isolates were analyzed. Most of these isolates were lactose-fermenting bacteria, which were our main interest; nevertheless, some non-fermenting bacteria were also studied (based on their relevance in terms of resistance). From the water drainage of Brito de Capelo Street (MAT1, MAT2 and MAT6 samples), we analyzed 16 isolates, and from the Riguinha water stream (MAT3 and MAT5 samples from the mouth and MAT4 from the open access section) came 22 isolates.
At first analysis via Chromagar Orientation medium, we presumptively identified five Escherichia coli, six KESC, four Pseudomonas, and one isolate that did not belong to any of these three groups, from the Brito Capelo Street samples. In the case of the Riguinha stream samples, we identified fifteen Escherichia coli, seven KESC, and three Pseudomonas. Two of the Pseudomonas identified were presumptively Pseudomonas aeruginosa, due to their green coloration in the UTI medium.
3.3. Antimicrobial Resistance Phenotype
All the results regarding the inhibition zone diameters obtained for each tested antibiotic, as well as the categorization of the isolates as susceptible (S—green cell), susceptible with increased exposure (I—white cell), or resistant (R—red cell), are presented in Table 4. From the results of the antibiograms, we concluded that the majority of the isolates exhibited resistance to β-lactam antibiotics, including amoxicillin-clavulanic acid (AMC) to fifth-generation cephalosporins such as ceftaroline (CPT). Two isolates out of 38 in total were resistant to all β-lactams antibiotics tested (E. coli—MAT5MERO1 and KESC group—MAT6MERO2).

Table 4.
Compilation of phenotypic characterisation results of isolates from the various water samples, including the presumptive identification, the medium from which it was isolated, resistance mechanism, and the diameters of the zone of inhibition for each antibiotic.
We also observed resistance and reduced susceptibility to carbapenems, particularly meropenem (MRP), in a significant number of isolates. Those classified as “susceptible with increased exposure (I)” already possess carbapenemases and tested positive in the CIM test. In contrast, fewer isolates showed resistance to gentamicin (8 isolates), nitrofurantoin (5 isolates), trimethoprim/sulfamethoxazole (11 isolates), and chloramphenicol (7 isolates). They were 26 out of 38 multidrug-resistant isolates.
Regarding the antibiograms performed with antibiotics specific for Pseudomonas, we measured the inhibition zone diameters for each antibiotic and compared them with the EUCAST Guidelines. However, our primary focus was on Enterobacteriaceae isolates, particularly those resistant to carbapenems due to their clinical significance. In terms of the resistance mechanisms, we identified Enterobacteriaceae bacteria that produce ESBLs, AmpC enzymes, and carbapenemases (Table 4). Carbapenemase-producing isolates exhibited resistance to meropenem or reduced susceptibility, although a few isolates with inhibition zone diameters still considered susceptible tested positive in the CIM test.
Regarding the prevalence of these three different resistance mechanisms in the two water samples from the Riguinha stream and Brito Capelo Street, we found that in the first water sample, there were 9 ESBL-producing isolates, 8 carbapenemase-producing isolates, and 2 AmpC-producing isolates out of a total of 22 isolates. In the second sample, we identified 7 AmpC producers, 4 carbapenemase producers, and 2 ESBL producers out of 16 isolates.
In conclusion, it is essential to highlight our findings related to bacterial contamination in the water samples collected at different points along the Riguinha stream and the water drainage system of Brito Capelo Street. Upstream of Riguinha (MAT4), our analysis predominantly revealed the presence of ESBL-producing bacteria. However, downstream at MAT3 and MAT5, we identified carbapenemase-producing bacteria.
3.4. Genotypic Characterization
A 2% agarose gel electrophoresis was performed to separate the products of the Multiplex PCR used for detecting acquired carbapenemase genes, as presented in Figure 3a. Positive controls for blaKPC and blaOXA-48 yielded the expected bands, confirming the specificity of the primers used.
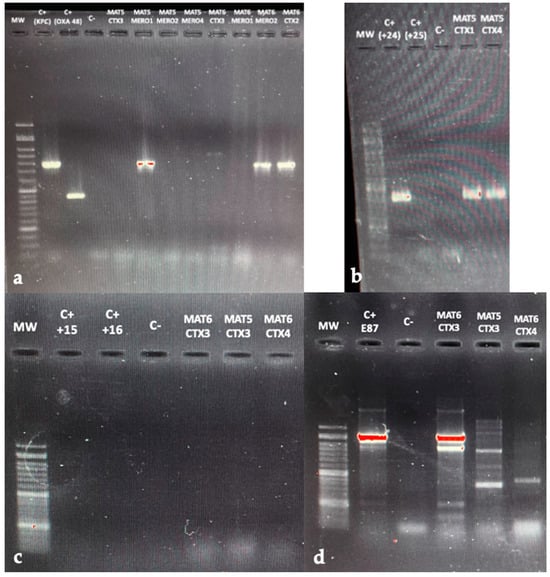
Figure 3.
(a) Agarose gel electrophoresis (2%) used for separation of the different multiplex PCR products, performed to detect carbapenemase genes. (b) Multiplex PCR assay for blaCTX-M alleles from G1, G2, G8, G9, and G25. (c) Agarose gel electrophoresis (1.5%) used for separation of the different multiplex PCR products performed to identify AmpC blaDHA gene. (d) Agarose gel electrophoresis (1.5%) used for separation of the different multiplex PCR products performed to identify AmpC blaCMY gene. Lanes labeled “MW”—correspond to DNA ladder; “C+”—to positive controls used, and “C−“—blank control.
Numerous isolates from the last two water samples were tested, including MAT5CTX3, MAT5MERO1, MAT5MERO2, MAT5MERO4, MAT6CTX3, MAT6MERO1, MAT6MERO2, and MAT6CTX2. All of these isolates were found to be carbapenemase-producers, as confirmed by the CIM test.
Specifically, MAT5MERO1, previously identified as E. coli, MAT6MERO2, and MAT6CTX2, both identified as KESC, carried the blaKPC gene, as evidenced by the amplification of the corresponding gene, resulting in a 798 bp band. In contrast, all other isolates, while confirmed as carbapenemase-producers, tested negative for the blaKPC, blaOXA-48, and blaNDM genes, which are among the most frequently identified carbapenemases genes. Testing for other potential resistance genes, such as blaVIM and blaIMP, was the next step; however, it was not possible due to time constraints. Additionally, there was no band observed in the negative control lane, indicating that gene amplification was not affected by PCR products.
The results of the Multiplex PCR assay for detecting genes-encoding CTX-M extended-spectrum β-lactamases, specifically from groups 1, 2, 8, 9, and 25, are presented in Figure 3b. We conducted tests on two isolates: MAT5CTX1, identified as E. coli, and MAT5CTX4, identified as KESC. These isolates had previously been classified as ESBL-positive. Upon analyzing the agarose gel electrophoresis data, we concluded that these isolates carried the blaCTX-M-group1 gene. This conclusion was based on the presence of a 415 bp band, which matched the positive control 24.
Finally, the results of the two simplex PCR assays performed to detect blaAmpC genes, namely blaDHA and blaCMY, are presented in Figure 3c,d, respectively. The AmpC-positive isolates tested included: MAT6CTX3 (KESC), MAT5CTX3 (E. coli), and MAT6CTX4 (KESC). In the first electrophoresis (Figure 3c), no bands appeared in the positive controls used, as they did not contain the blaDHA gene responsible for the phenotype, and no suitable controls were available in the laboratory. All three isolates tested negative for this gene.
Regarding the second electrophoresis, it was difficult to interpret, since nonspecific bands appeared, which was not expected. It appears to have amplified the positive control’s blaCMY gene correctly, and there was no interference by the PCR reaction products, as the negative control did not display any bands. However, the presence of multiple bands in the isolates introduced uncertainty. To draw definitive conclusions, we will need to repeat the PCR. There is a possibility that the MAT6CTX3 isolate carries the blaCMY gene, since it displayed a band similar to that of the positive control (Figure 3d). Still, we cannot confirm this with absolute certainty.
4. Discussion
In these watercourses we found ESBL −, AmpC −, and carbapenemase-producing Enterobacteriaceae, as well as Pseudomonas, exhibiting multidrug resistance. It is important to note that these bacteria are supposedly of environmental origin, found in two water streams that flow into the beach, which was unexpected. Also, there is a possibility that bacteria may acquire resistance mechanisms through the aquatic environment, which could lead to their spread and subsequent further transmission [10]. Some of these bacteria demonstrated reduced susceptibility and resistance to meropenem. The decreased susceptibility is also concerning, as bacteria classified as “susceptible with increased exposure (I)” already possess carbapenemases, which confer some level of resistance.
Carbapenem resistance rates rose alarmingly in Portugal, especially among Klebsiella pneumoniae, between 2014 and 2017. In several healthcare settings in the north of Portugal, the ST147 K. pneumoniae clone presenting K-type K64 has been one of those responsible for KPC-3 spread. Also, it was responsible for the first large outbreak of carbapenemase-producing Enterobacteriaceae in a hospital in the north of Portugal in 2015. Beyond that, clones, including E. coli ST131 and carbapenemases, predominantly KPC-3 but also OXA-48 and VIM, were identified three years after the onset of carbapenemases spreading in this hospital [28].
In contrast, fewer isolates showed resistance to gentamicin (CN), nitrofurantoin (F), trimethoprim/sulfamethoxazole (SXT), and chloramphenicol, indicating that these antibiotics are still effective against infections caused by these bacteria. Applying the definitions for MDR (multidrug-resistant), XDR (extensively drug-resistant), and PDR (pandrug-resistant) as defined by Magiorakos [9], we identified several MDR bacteria in the water samples. For example, MAT3MERO6, a KESC isolate, exhibited resistance to meropenem, tetracycline, and tobramycin.
In addition, we highlight the fact that higher contamination levels were observed at the mouth of the river. This discrepancy suggests a potential contamination source from the open section of the Riguinha stream, possibly due to accidental wastewater discharge or contamination from human and animal feces. Unfortunately, we were unable to pinpoint the exact source and location of contamination. Further research is required to address this critical knowledge gap.
Similarly, the water drainage system of Brito Capelo Street may also be susceptible to accidental contamination. It should be emphasized that rainwater from such systems is typically discharged into the environment without treatment or control measures. This situation poses a significant risk to public health and calls for immediate attention and action. Two isolates belonged to the KESC group, and one E. coli carried the blaKPC gene. This finding aligns with the prevalence and widespread distribution of the KPC carbapenemase globally. Despite its initial discovery in Klebsiella pneumoniae isolates, clinical isolates of KPC-producing Escherichia coli and certain species of Klebsiella spp. and Enterobacter spp. have also been identified [6].
In conclusion, relevant acquired genes commonly found in clinical isolates such as blaKPC, and particularly in Enterobacteriaceae, were detected in bacteria from these water samples. A similar study on the occurrence of CRE in a Portuguese river also identified multidrug-resistant isolates, including K. pneumoniae carrying the KPC carbapenemase-encoding gene, specifically blaKPC-3 [29].
Regarding the MAT5CTX1 isolate, it is likely to contain CTX-M-15, as E. coli strains carrying CTX-Ms are known to be widespread ESBL-producing bacteria, with CTX-M-15 being the most commonly detected resistance gene. However, within the scope of our testing conditions, we can only confirm the presence of the blaCTX-M-group1 gene [30]. In a related investigation focusing on wastewater and river water in Tunisia, ESBL-producing Escherichia coli and Klebsiella pneumoniae were also isolated. Most of these isolates exhibited multidrug resistance and were found to produce CTX-M-15 [31].
In the context of future research, it will be imperative to expand the scope by analyzing a more extensive range of water samples collected from both streams and during different seasons throughout the year. This broader analysis aims to comprehensively understand the impact on water quality. Furthermore, analyzing a larger number of bacterial isolates and assessing their resistance, along with identifying the responsible genes, will provide a more accurate characterization of contamination levels and associated threats.
5. Conclusions
The findings of this study allowed us to confirm the presence of antibiotic-resistant bacterial contamination in both the Riguinha stream and the rainwater drainage system of Brito Capelo Street in Matosinhos. Surprisingly, from very small volumes of water samples, we were able to detect great contamination with multidrug-resistant bacteria, including ESBL-producing, AmpC-producing, and carbapenemase-producing Enterobacteriaceae. It is highly likely that these waters have been contaminated by wastewater or fecal discharges. However, we were unable to pinpoint for certain the source and exact location where the contamination occurred. Therefore, there is a need for further research in this area. The results of this study also highlight the importance of assessing water quality, as environmental water can act as reservoirs for these bacteria, enabling their dissemination and posing a significant threat to public health. Consequently, the information retrieved from this investigation could be used to inform bathing water management and policy in Matosinhos beach. Finally, looking to the future, carbapenem resistance in Enterobacteriaceae is expected to continue to increase substantially, limiting more and more the therapeutic options and jeopardizing public health. Hence, it is critical to control water quality and stop the spread of these bacteria.
Author Contributions
Conceptualization, H.F.; methodology, H.F. and J.D.P.; validation, H.F., M.A.P. and J.D.P.; formal analysis, H.F.; investigation, H.F. and M.A.P.; resources, H.F.; data curation, H.F.; writing—original draft preparation, M.A.P.; writing—review and editing, H.F.; visualization, H.F.; supervision, H.F.; project administration, H.F.; funding acquisition, H.F. All authors have read and agreed to the published version of the manuscript.
Funding
This work is financed by national funds from FCT-Fundação para a Ciência e a Tecnologia, I.P., in the scope of the project UIDP/04378/2020 and UIDB/04378/2020 of the Research Unit on Applied Molecular Biosciences-UCIBIO and the project LA/P/0140/2020 of the Associate Laboratory Institute for Health and Bioeconomy-i4HB.
Data Availability Statement
All relevant data are included in this manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Tilahun, M.; Kassa, Y.; Gedefie, A.; Ashagire, M. Emerging Carbapenem-Resistant Enterobacteriaceae Infection, Its Epidemiology and Novel Treatment Options: A Review. Infect. Drug Resist. 2021, 14, 4363–4374. [Google Scholar] [CrossRef] [PubMed]
- Diallo, O.O.; Baron, S.A.; Abat, C.; Colson, P.; Chaudet, H.; Rolain, J.M. Antibiotic resistance surveillance systems: A review. J. Glob. Antimicrob. Resist. 2020, 23, 430–438. [Google Scholar] [CrossRef] [PubMed]
- Palmeira, J.D.; Cunha, M.V.; Carvalho, J.; Ferreira, H.; Fonseca, C.; Torres, R.T. Emergence and Spread of Cephalosporinases in Wildlife: A Review. Animals 2021, 11, 1765. [Google Scholar] [CrossRef]
- Mishra, M.; Arukha, A.P.; Patel, A.K.; Behera, N.; Mohanta, T.K.; Yadav, D. Multi-Drug Resistant Coliform: Water Sanitary Standards and Health Hazards. Front. Pharmacol. 2018, 9, 311. [Google Scholar] [CrossRef]
- Dantas Palmeira, J.; Ferreira, H.M.N. Extended-spectrum beta-lactamase (ESBL)-producing Enterobacteriaceae in cattle production—A threat around the world. Heliyon 2020, 6, e03206. [Google Scholar] [CrossRef] [PubMed]
- Suay-García, B.; Pérez-Gracia, M.T. Present and Future of Carbapenem-resistant Enterobacteriaceae (CRE) Infections. Antibiotics 2019, 8, 122. [Google Scholar] [CrossRef]
- Taggar, G.; Attiq Rheman, M.; Boerlin, P.; Diarra, M.S. Molecular Epidemiology of Carbapenemases in Enterobacterales from Humans, Animals, Food and the Environment. Antibiotics 2020, 9, 693. [Google Scholar] [CrossRef]
- Bassetti, M.; Peghin, M. How to manage KPC infections. Ther. Adv. Infect. Dis. 2020, 7, 2049936120912049. [Google Scholar] [CrossRef]
- Magiorakos, A.-P.; Srinivasan, A.; Carey, R.B.; Carmeli, Y.; Falagas, M.E.; Giske, C.G.; Harbarth, S.; Hindler, J.F.; Kahlmeter, G.; Olsson-Liljequist, B.; et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: An international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 2012, 18, 268–281. [Google Scholar] [CrossRef]
- Farrell, M.L.; Joyce, A.; Duane, S.; Fitzhenry, K.; Hooban, B.; Burke, L.P.; Morris, D. Evaluating the potential for exposure to organisms of public health concern in naturally occurring bathing waters in Europe: A scoping review. Water Res. 2021, 206, 117711. [Google Scholar] [CrossRef]
- Holcomb, D.A.; Stewart, J.R. Microbial Indicators of Fecal Pollution: Recent Progress and Challenges in Assessing Water Quality. Curr. Environ. Health Rep. 2020, 7, 311–324. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Hur, H.-G.; Sadowsky, M.J.; Byappanahalli, M.N.; Yan, T.; Ishii, S. Environmental Escherichia coli: Ecology and public health implications—A review. J. Appl. Microbiol. 2017, 123, 570–581. [Google Scholar] [CrossRef] [PubMed]
- PortoCanal. Água da Praia de Matosinhos Contaminada: MEOBeachCam. Available online: https://beachcam.meo.pt/newsroom/2018/01/agua-na-praia-de-matosinhos-em-risco/ (accessed on 6 February 2022).
- RTPnotícias. Praia De Matosinhos Está Desaconselhada a Banhos: RTPnotícias. 2021. Available online: https://www.rtp.pt/noticias/pais/praia-de-matosinhos-esta-desaconselhada-a-banhos_v1330195 (accessed on 6 February 2022).
- PortoCanal. Contaminação Bacteriológica Coloca em Alerta Praias de Matosinhos e Arcos de Valdevez: SAPO. 2021. Available online: https://portocanal.sapo.pt/noticia/269302 (accessed on 7 July 2022).
- Roque, L.A. Plano de Estrutura Para a Frente Marítima Do Porto; Aqualogus/Biodesign/Apload: Maia, Portugal, 2014. [Google Scholar]
- ISO 9308-1; Water Quality—Enumeration of Escherichia coli and Coliform bacteria—Part 1: Membrane Filtration Method for Waters with Low Bacterial Background Flora. ISO: Geneva, Switzerland, 2014.
- Microbiology. ASf. Kirby-Bauer Disk Diffusion Susceptibility Test Protocol. 2009. Available online: https://asm.org/getattachment/2594ce26-bd44-47f6-8287-0657aa9185ad/Kirby-Bauer-Disk-Diffusion-Susceptibility-Test-Protocol-pdf.pdf (accessed on 27 October 2023).
- EUCAST. Antimicrobial Susceptibility Testing. EUCAST Disk Diffusion Method: European Society of Clinical Microbiology and Infectious Diseases. 2022. Available online: https://www.eucast.org/ast_of_bacteria/disk_diffusion_methodology/ (accessed on 27 June 2022).
- EUCAST. Routine and Extended Internal Quality Control for MIC Determination and Disk Diffusion as Recommended by EUCAST: European Committee on Antimicrobial Susceptibility Testing. 2022. Available online: https://www.eucast.org/ast_of_bacteria/quality_control/ (accessed on 28 June 2022).
- EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters European Committee on Antimicrobial Susceptibility Testing. 2022. Available online: https://www.eucast.org/clinical_breakpoints/ (accessed on 28 June 2022).
- EUCAST. Guidelines for Detection of Resistance Mechanisms and Specific Resistances of Clinical and/or Epidemiological Importance: European Society of Clinical Microbiology and Infectious Diseases. 2017. Available online: https://www.eucast.org/resistance_mechanisms/ (accessed on 28 June 2022).
- van der Zwaluw, K.; de Haan, A.; Pluister, G.N.; Bootsma, H.J.; de Neeling, A.J.; Schouls, L.M. The carbapenem inactivation method (CIM), a simple and low-cost alternative for the Carba NP test to assess phenotypic carbapenemase activity in gram-negative rods. PLoS ONE 2015, 10, e0123690. [Google Scholar] [CrossRef]
- Poirel, L.; Walsh, T.R.; Cuvillier, V.; Nordmann, P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn. Microbiol. Infect. Dis. 2011, 70, 119–123. [Google Scholar] [CrossRef] [PubMed]
- Woodford, N.; Fagan, E.J.; Ellington, M.J. Multiplex PCR for rapid detection of genes encoding CTX-M extended-spectrum β-lactamases. J. Antimicrob. Chemother. 2005, 57, 154–155. [Google Scholar] [CrossRef] [PubMed]
- Roche, C.; Boo, T.W.; Walsh, F.; Crowley, B. Detection and molecular characterisation of plasmidic AmpC β-lactamases in Klebsiella pneumoniae isolates from a tertiary-care hospital in Dublin, Ireland. Clin. Microbiol. Infect. 2008, 14, 616–618. [Google Scholar] [CrossRef]
- D’Andrea, M.M.; Nucleo, E.; Luzzaro, F.; Giani, T.; Migliavacca, R.; Vailati, F.; Rossolini, G.M. CMY-16, a novel acquired AmpC-type beta-lactamase of the CMY/LAT lineage in multifocal monophyletic isolates of Proteus mirabilis from northern Italy. Antimicrob. Agents Chemother. 2006, 50, 618–624. [Google Scholar] [CrossRef]
- Guerra, A.M.; Lira, A.; Lameirão, A.; Selaru, A.; Abreu, G.; Lopes, P.; Peixe, L. Multiplicity of Carbapenemase-Producers Three Years after a KPC-3-Producing K. pneumoniae ST147-K64 Hospital Outbreak. Antibiotics 2020, 9, 806. [Google Scholar] [CrossRef]
- Teixeira, P.; Tacão, M.; Pureza, L.; Gonçalves, J.; Silva, A.; Cruz-Schneider, M.P.; Henriques, I. Occurrence of carbapenemase-producing Enterobacteriaceae in a Portuguese river: blaNDM, blaKPC and blaGES among the detected genes. Environ. Pollut. 2020, 260, 113913. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, W.E.; Yang, Q. Clinical Perspective of Antimicrobial Resistance in Bacteria. Infect. Drug Resist. 2022, 15, 735–746. [Google Scholar] [CrossRef]
- Hassen, B.; Abbassi, M.S.; Benlabidi, S.; Ruiz-Ripa, L.; Mama, O.M.; Ibrahim, C.; Hassen, A.; Hammami, S.; Torres, C. Genetic characterization of ESBL-producing Escherichia coli and Klebsiella pneumoniae isolated from wastewater and river water in Tunisia: Predominance of CTX-M-15 and high genetic diversity. Environ. Sci. Pollut. Res. 2020, 27, 44368–44377. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).