Lacticaseibacillus rhamnosus GG Versus Placebo for Eradication of Vancomycin-Resistant Enterococcus faecium in Intestinal Carriers: A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protocol
2.2. Search Strategy
2.3. Data Collection and Analysis
2.4. Inclusion/Exclusion of Studies
2.5. Quality Assessment
2.6. Statistical Analyses
3. Results
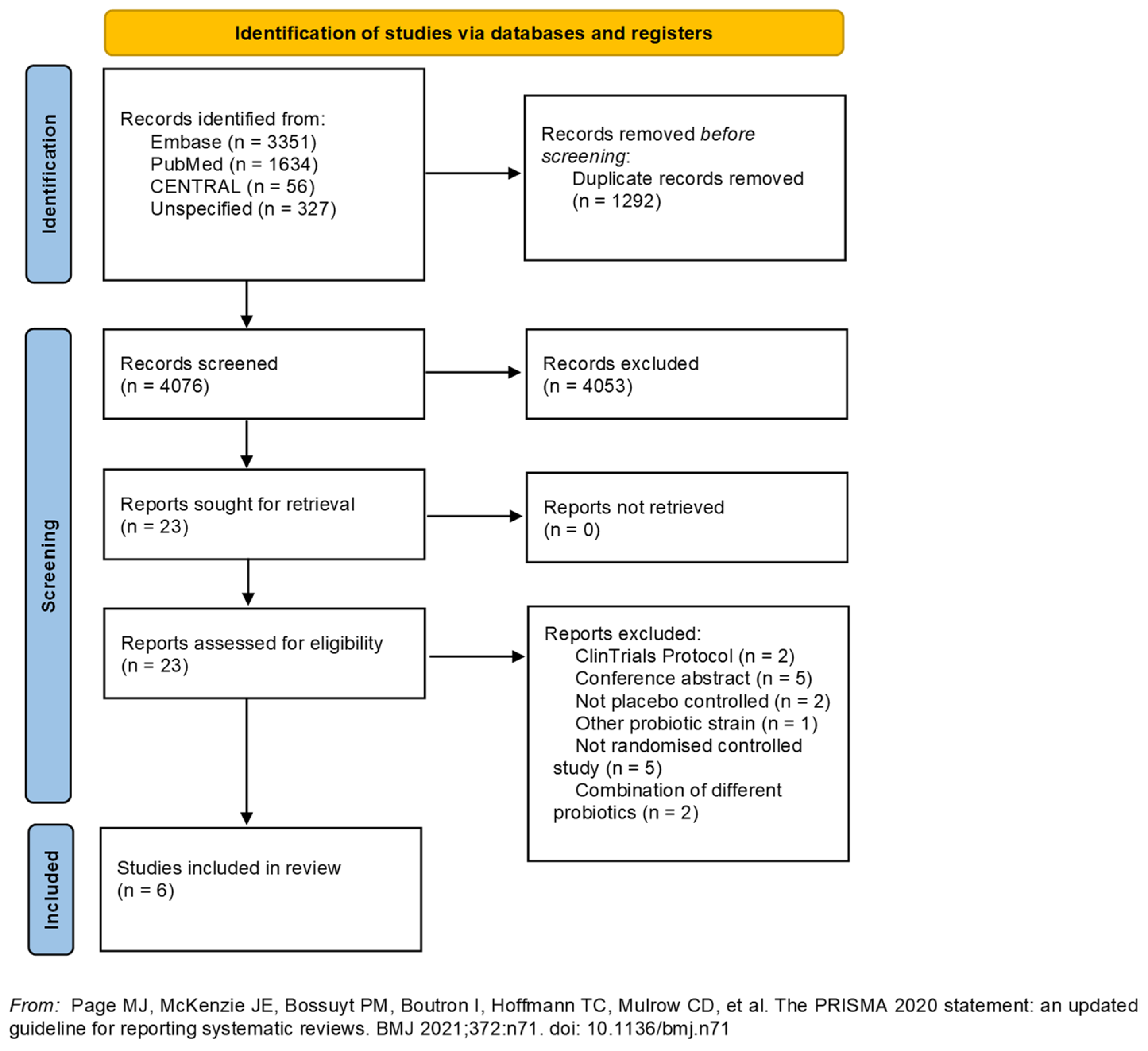
3.1. Study Design and Selection
3.2. Description of the Studies
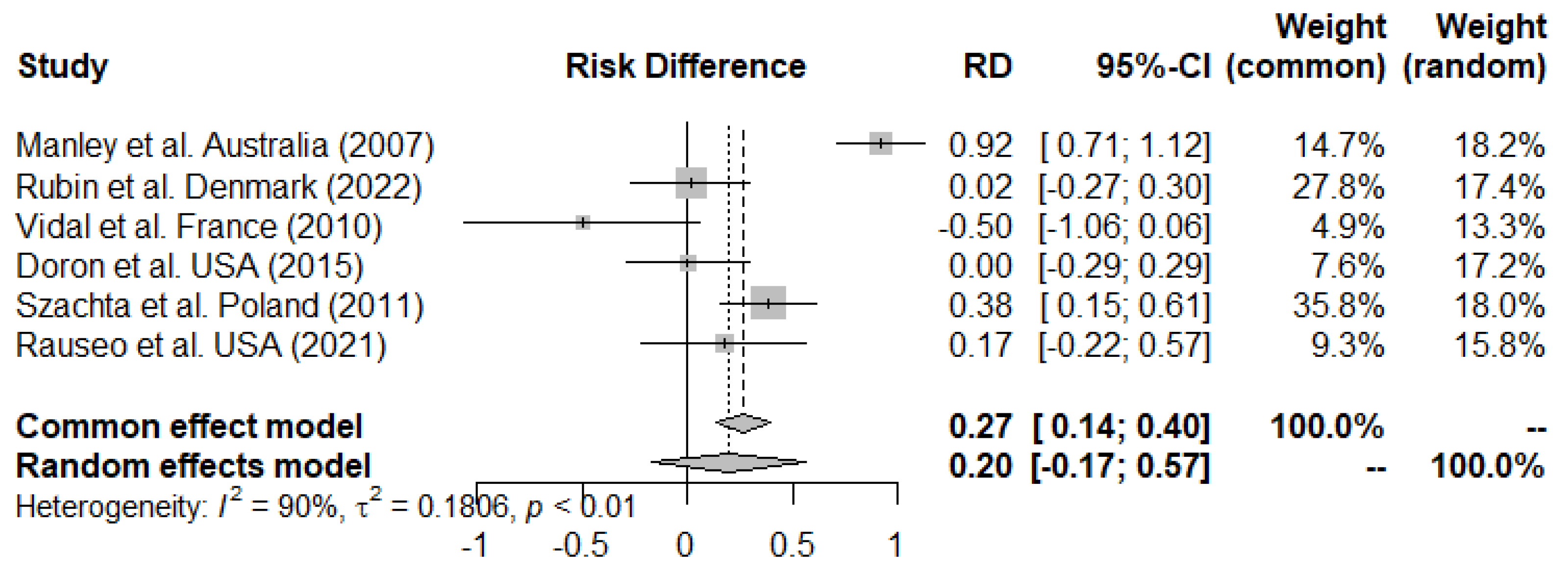
3.3. Primary Outcome
Meta-Analysis and Efficacy of LGG on Eradication of VREfm from the GI-Tract
3.4. Secondary Outcomes
3.4.1. Health-Related Change in the Quality of Life
3.4.2. LGG Colonization at the End of Intervention
3.4.3. Tolerability and Safety
3.4.4. Heading
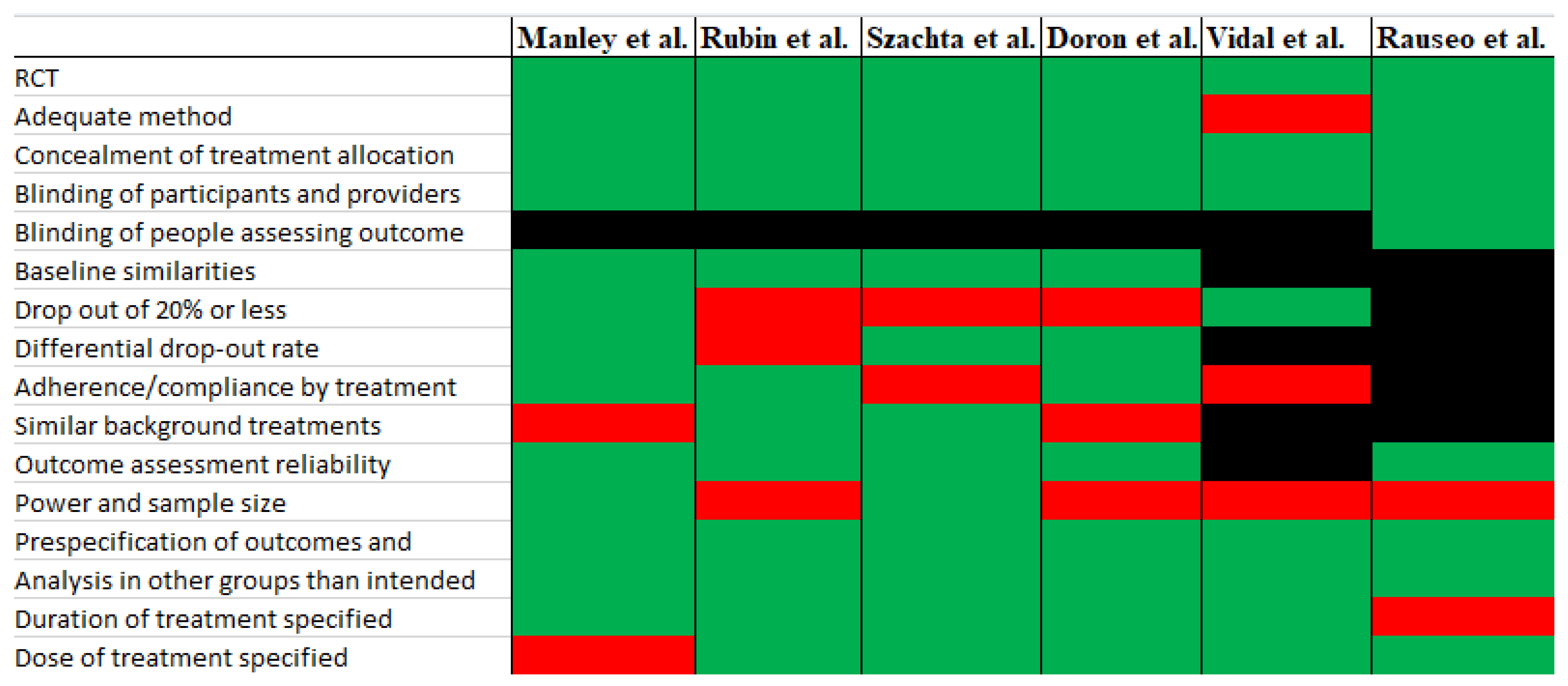
3.5. Risk of Bias
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Majumder, M.A.A.; Singh, K.; Hilaire, M.G.-S.; Rahman, S.; Sa, B.; Haque, M. Tackling Antimicrobial Resistance by promoting Antimicrobial stewardship in Medical and Allied Health Professional Curricula. Expert Rev. Anti-Infect. Ther. 2020, 18, 1245–1258. [Google Scholar] [CrossRef] [PubMed]
- CDC. Antibiotic Resistance Threats in the United States, 2019; U.S. Department of Health and Human Services, Center for Disease control and Prevention: Atlanta, GA, USA, 2019. [Google Scholar]
- Högberg, L.D.; Weist, K.; Suetens, C.; Griskeviciene, J.; Monnet, D.; Heuer, O. Annual Epidemiological Report 2014. Antimicrobial Resistance and Healthcare-Associated Infections; ECDC: Solna, Sweden, 2014; p. 28. [Google Scholar]
- Arias, C.A.; Murray, B.E. The rise of the Enterococcus: Beyond vancomycin resistance. Nat. Rev. Microbiol. 2012, 10, 266–278. [Google Scholar] [CrossRef] [PubMed]
- Kristich, C.J.; Rice, L.B.; Arias, C.A. Enterococcal Infection—Treatment and Antibiotic Resistance; Massachusetts Eye and Ear Infirmary: Boston, MA, USA, 2014. [Google Scholar]
- Cassini, A.; Högberg, L.D.; Plachouras, D.; Quattrocchi, A.; Hoxha, A.; Simonsen, G.S.; Colomb-Cotinat, M.; Kretzschmar, M.E.; Devleesschauwer, B.; Cecchini, M.; et al. Attributable deaths and disability-adjusted life-years caused by infections with antibiotic-resistant bacteria in the EU and the European Economic Area in 2015: A population-level modelling analysis. Lancet Infect. Dis. 2019, 19, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Billington, E.; Phang, S.; Gregson, D.; Pitout, J.; Ross, T.; Church, D.; Laupland, K.; Parkins, M. Incidence, risk factors, and outcomes for Enterococcus spp. blood stream infections: A population-based study. Int. J. Infect. Dis. 2014, 26, 76–82. [Google Scholar] [CrossRef]
- Salgado, C.D. The risk of developing a vancomycin-resistant Enterococcus bloodstream infection for colonized patients. Am. J. Infect. Control 2008, 36, S175.e5–S175.e8. [Google Scholar] [CrossRef]
- Leavis, H.L.; Bonten, M.J.; Willems, R.J. Identification of high-risk enterococcal clonal complexes: Global dispersion and antibiotic resistance. Curr. Opin. Microbiol. 2006, 9, 454–460. [Google Scholar] [CrossRef]
- Werner, G.; Coque, T.M.; Hammerum, A.M.; Hope, R.; Hryniewicz, W.; Johnson, A.; Klare, I.; Kristinsson, K.G.; Leclercq, R.; Lester, C.H.; et al. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. Bull. Eur. Mal. Transm. Eur. Commun. Dis. Bull. 2008, 13, 19046. [Google Scholar] [CrossRef]
- De Been, M.; Van Schaik, W.; Cheng, L.; Corander, J.; Willems, R.J. Recent recombination events in the core genome are associated with adaptive evolution in Enterococcus faecium. Genome Biol. Evol. 2013, 5, 1524–1535. [Google Scholar] [CrossRef]
- Knudsen, M.J.S.; Rubin, I.M.C.; Gisselø, K.; Mollerup, S.; Petersen, A.M.; Pinholt, M.; Westh, H.; Bartels, M.D. The use of core genome multilocus sequence typing to determine the duration of vancomycin-resistant Enterococcus faecium outbreaks. APMIS 2022, 130, 323–329. [Google Scholar] [CrossRef]
- Fournier, S.; Brossier, F.; Fortineau, N.; Gillaizeau, F.; Akpabie, A.; Aubry, A.; Barbut, F.; Chedhomme, F.X.; Kassis-Chikhani, N.; Lucet, J.C.; et al. Long-term control of vancomycin-resistant Enterococcus faecium at the scale of a large multihospital institution: A seven-year experience. Eurosurveillance 2012, 17, 20229. [Google Scholar] [CrossRef]
- Söderblom, T.; Aspevall, O.; Erntell, M.; Hedin, G.; Heimer, D.; Hökeberg, I.; Kidd-Ljunggren, K.; Melhus, Å.; Olsson-Liljequist, B.; Sjögren, I.; et al. Alarming spread of vancomycin resistant enterococci in Sweden since 2007. Eurosurveillance 2010, 15, 19620. [Google Scholar] [CrossRef]
- Cattoir, V.; Leclercq, R. Twenty-five years of shared life with vancomycin-resistant enterococci: Is it time to divorce. J. Antimicrob. Chemother. 2013, 68, 731–742. [Google Scholar] [CrossRef] [PubMed]
- Ubeda, C.; Taur, Y.; Jenq, R.R.; Equinda, M.J.; Son, T.; Samstein, M.; Viale, A.; Socci, N.D.; van den Brink, M.R.M.; Kamboj, M.; et al. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Investig. 2010, 120, 4332–4341. [Google Scholar] [CrossRef]
- Taur, Y.; Xavier, J.B.; Lipuma, L.; Ubeda, C.; Goldberg, J.; Gobourne, A.; Lee, Y.J.; Dubin, K.A.; Socci, N.D.; Viale, A.; et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clin. Infect. Dis. 2012, 55, 905–914. [Google Scholar] [CrossRef]
- Bhalla, A.; Pultz, N.J.; Ray, A.J.; Hoyen, C.K.; Eckstein, E.C.; Donskey, C.J. Antianaerobic Antibiotic Therapy Promotes Overgrowth of Antibiotic-Resistant, Gram-Negative Bacilli and Vancomycin-Resistant Enterococci in the Stool of Colonized Patients. Infect. Control Hosp. Epidemiol. 2003, 24, 644–649. [Google Scholar] [CrossRef] [PubMed]
- Taur, Y.; Coyte, K.; Schluter, J.; Robilotti, E.; Figueroa, C.; Gjonbalaj, M.; Littmann, E.R.; Ling, L.; Miller, L.; Gyaltshen, Y.; et al. Reconstitution of the gut microbiota of antibiotic-treated patients by autologous fecal microbiota transplant. Sci. Transl. Med. 2018, 10, eaap9489. [Google Scholar] [CrossRef]
- Hayden, M.K.; Bonten, M.J.M.; Blom, D.W.; Lyle, E.A.; van de Vijver, D.A.M.C.; Weinstein, R.A. Reduction in Acquisition of Vancomycin-Resistant Enterococcus after Enforcement of Routine Environmental Cleaning Measures. Clin. Infect. Dis. 2006, 42, 1552–1560. [Google Scholar] [CrossRef] [PubMed]
- Pinholt, M.; Bayliss, S.C.; Gumpert, H.; Worning, P.; Jensen, V.V.S.; Pedersen, M.; Feil, E.J.; Westh, H. WGS of 1058 Enterococcus faecium from Copenhagen, Denmark, reveals rapid clonal expansion of vancomycin-resistant clone ST80 combined with widespread dissemination of a vanA-containing plasmid and acquisition of a heterogeneous accessory ge. J. Antimicrob. Chemother. 2019, 74, 1776–1785. [Google Scholar] [CrossRef]
- Lautenbach, E.; Bilker, W.B.; Brennan, P.J. Enterococcal bacteremia: Risk factors for vancomycin resistance and predictors of mortality. Infect. Control Hosp. Epidemiol. Off. J. Soc. Hosp. Epidemiol. Am. 1999, 20, 318–323. [Google Scholar] [CrossRef]
- Shenoy, E.S.; Paras, M.L.; Noubary, F.; Walensky, R.P.; Hooper, D.C. Natural history of colonization with methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant Enterococcus (VRE): A systematic review. BMC Infect. Dis. 2014, 14, 177. [Google Scholar] [CrossRef]
- Roghmann, M.-C.; Qaiyumi, S.; Schwalbe, R.; Morris, J.G. Natural History of Colonization with Vancomycin-Resistant Enterococcus faecium. Infect. Control Hosp. Epidemiol. 1997, 18, 679–680. [Google Scholar] [CrossRef] [PubMed]
- Rubin, I.M.C.; Mollerup, S.; Broholm, C.; Knudsen, S.B.; Baker, A.; Helms, M.; Holm, M.K.A.; Kallemose, T.; Westh, H.; Knudsen, J.D.; et al. No Effect of Lactobacillus rhamnosus GG on Eradication of Colonization by Vancomycin-Resistant Enterococcus faecium or Microbiome Diversity in Hospitalized Adult Patients. Microbiol. Spectr. 2022, 10, e0234821. [Google Scholar] [CrossRef]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. Expert consensus document: The international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed]
- Salminen, S.; von Wright, A.; Morelli, L.; Marteau, P.; Brassart, D.; de Vos, W.M.; Fondén, R.; Saxelin, M.; Collins, K.; Mogensen, G.; et al. Demonstration of safety of probiotics—A review. Int. J. Food Microbiol. 1998, 44, 93–106. [Google Scholar] [CrossRef]
- Gorbach, S.; Doron, S.; Magro, F. Lactobacillus rhamnosus GG. In The Microbiota in Gastrointestinal Pathophysiology: Implications for Human Health, Prebiotics, Probiotics, and Dysbiosis; Elsevier: Amsterdam, The Netherlands, 2017; pp. 79–88. [Google Scholar] [CrossRef]
- Manley, K.J.; Fraenkel, M.B.; Mayall, B.C.; Power, D.A. Probiotic treatment of vancomycin-resistant enterococci: A randomised controlled trial. Med. J. Aust. 2007, 186, 454–457. [Google Scholar] [CrossRef]
- Szachta, P.; Ignyś, I.; Cichy, W. An evaluation of the ability of the probiotic strain Lactobacillus rhamnosus GG to eliminate the gastrointestinal carrier state of vancomycin-resistant enterococci in colonized children. J. Clin. Gastroenterol. 2011, 45, 872–877. [Google Scholar] [CrossRef]
- Doron, S.; Hibberd, P.L.; Goldin, B.; Thorpe, C.; McDermott, L.; Snydman, D.R. Effect of Lactobacillus rhamnosus GG administration on vancomycin-resistant Enterococcus colonization in adults with comorbidities. Antimicrob. Agents Chemother. 2015, 59, 4593–4599. [Google Scholar] [CrossRef]
- Kim, A.S. Using the good to beat out the bad: Probiotics for eliminating vancomycin-resistant enterococci colonization. J. Clin. Gastroenterol. 2011, 45, 844–845. [Google Scholar] [CrossRef]
- Tytgat, H.L.P.; Douillard, F.P.; Reunanen, J.; Rasinkangas, P.; Hendrickx, A.P.A.; Laine, P.K.; Paulin, L.; Satokari, R.; de Vos, W.M. Lactobacillus rhamnosus GG Outcompetes Enterococcus faecium via Mucus-Binding Pili: Evidence for a Novel and Heterospecific Probiotic Mechanism. Appl. Environ. Microbiol. 2016, 82, 5756–5762. [Google Scholar] [CrossRef]
- Tachedjian, G.; Aldunate, M.; Bradshaw, C.S.; Cone, R.A. The role of lactic acid production by probiotic Lactobacillus species in vaginal health. Res. Microbiol. 2017, 168, 782–792. [Google Scholar] [CrossRef]
- Saxelin, M.; Tynkkynen, S.; Mattila-Sandholm, T.; De Vos, W.M. Probiotic and other functional microbes: From markets to mechanisms. Curr. Opin. Biotechnol. 2005, 16, 204–211. [Google Scholar] [CrossRef]
- Page, M.J.; McKenzie, J.E.; Bossuyt, P.M.; Boutron, I.; Hoffmann, T.C.; Mulrow, C.D.; Shamseer, L.; Tetzlaff, J.M.; Akl, E.A.; Brennan, S.E.; et al. The PRISMA 2020 statement: An updated guideline for reporting systematic reviews. BMJ 2021, 372, 71. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R Core Team 2021 R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 1 September 2023).
- Rauseo, A.M.; Hink, T.; Reske, K.A.; Seiler, S.M.; Bommarito, K.M.; Fraser, V.J.; Burnham, C.-A.D.; Dubberke, E.R.; CDC Prevention Epicenter Program. A randomized controlled trial of Lactobacillus rhamnosus GG on antimicrobial-resistant organism colonization. Infect. Control Hosp. Epidemiol. 2021, 43, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Vidal, M.; Forestier, C.; Charbonnel, N.; Henard, S.; Rabaud, C.; Lesens, O. Probiotics and intestinal colonization by vancomycin-resistant enterococci in mice and humans. J. Clin. Microbiol. 2010, 48, 2595–2598. [Google Scholar] [CrossRef]
- Buyukeren, M.; Yigit, S.; Buyukcam, A.; Kara, A.; Celik, H.T.; Yurdakok, M. A new use of Lactobacillus rhamnosus GG administration in the NICU: Colonized vancomycin-resistant enterococcus eradication in the gastrointestinal system. J. Matern. -Fetal Neonatal Med. 2022, 35, 1192–1198. [Google Scholar] [CrossRef]
- Bäckhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Crouzet, L.; Rigottier-Gois, L.; Serror, P. Potential use of probiotic and commensal bacteria as non-antibiotic strategies against vancomycin-resistant enterococci. FEMS Microbiol. Lett. 2015, 362, fnv012. [Google Scholar] [CrossRef] [PubMed]
- De Regt, M.J.A.; Willems, R.J.L.; Hené, R.J.; Siersema, P.D.; Verhaar, H.J.J.; Hopmans, T.E.M.; Bonten, M.J.M. Effects of probiotics on acquisition and spread of multiresistant enterococci. Antimicrob. Agents Chemother. 2010, 54, 2801–2805. [Google Scholar] [CrossRef]
- Borgmann, S.; Rieß, B.; Meintrup, D.; Klare, I.; Werner, G. Long-lasting decrease of the acquisition of enterococcus faecium and gram-negative bacteria producing extended spectrum beta-lactamase (Esbl) by transient application of probiotics. Int. J. Environ. Res Public Health 2020, 17, 6100. [Google Scholar] [CrossRef] [PubMed]
- Toh, S.-L.; Lee, B.B.; Simpson, J.M.; Rice, S.A.; Kotsiou, G.; Marial, O.; Ryan, S. Effect of probiotics on multi-resistant organism colonisation in persons with spinal cord injury: Secondary outcome of ProSCIUTTU, a randomised placebo-controlled trial. Spinal Cord 2020, 58, 755–767. [Google Scholar] [CrossRef]
- Chanderraj, R.; Brown, C.A.; Hinkle, K.; Falkowski, N.; Ranjan, P.; Dickson, R.P.; Woods, R.J. Gut Microbiota Predict Enterococcus Expansion but Not Vancomycin-Resistant Enterococcus Acquisition. mSphere 2020, 5. [Google Scholar] [CrossRef]
- McDonald, D.; Ackermann, G.; Khailova, L.; Baird, C.; Heyland, D.; Kozar, R.; Lemieux, M.; Derenski, K.; King, J.; Vis-Kampen, C.; et al. Extreme Dysbiosis of the Microbiome in Critical Illness. mSphere 2016, 1. [Google Scholar] [CrossRef]
- Ravi, A.; Halstead, F.D.; Bamford, A.; Casey, A.; Thomson, N.M.; van Schaik, W.; Snelson, C.; Goulden, R.; Foster-Nyarko, E.; Savva, G.M.; et al. Loss of microbial diversity and pathogen domination of the gut microbiota in critically ill patients. Microb. Genom. 2019, 5, e000293. [Google Scholar] [CrossRef]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108, 4554–4561. [Google Scholar] [CrossRef] [PubMed]
- Donskey, C.J.; Hoyen, C.K.; Das, S.M.; Helfand, M.S.; Hecker, M.T. Recurrence of Vancomycin-Resistant Enterococcus Stool Colonization During Antibiotic Therapy. Infect. Control Hosp. Epidemiol. 2002, 23, 436–440. [Google Scholar] [CrossRef] [PubMed]
- Dinh, A.; Fessi, H.; Duran, C.; Batista, R.; Michelon, H.; Bouchand, F.; Lepeule, R.; Vittecoq, D.; Escaut, L.; Sobhani, I.; et al. Clearance of carbapenem-resistant Enterobacteriaceae vs vancomycin-resistant enterococci carriage after faecal microbiota transplant: A prospective comparative study. J. Hosp. Infect. 2018, 99, 481–486. [Google Scholar] [CrossRef] [PubMed]
| Paper | Study Design | Intervention and Duration | Number of Individuals (LGG:Placebo) | Male:Female Ratio | Mean Age in Years (Range) | Follow-Up | Dose and Formulation of LGG | Number of Cleared Individuals at the End of Intervention | Number of Cleared Individuals at the End of Follow-Up | Adverse Events | Conclusion |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Manley et al. Australia (2007) [29] | Double-blinded, randomized and placebo-controlled trial with two arms. | 1. LGG yoghurt 2. Placebo; 4 weeks | 23 (11:12) | 10:4 | 68 (46–84) | 8 weeks | 100 g of yoghurt, unknown LGG dose | 11/11 in treatment group and 1/12 in placebo group | 8/11 in treatment group and 0/8 in placebo group | Not reported | Significant effect of VRE eradication |
| Vidal et al. France (2010) [39] | Double-blinded, randomized and placebo-controlled pilot trial with two arms. | 1. LGG 2. Placebo; 5 weeks | 8 (6:2) | not reported | 77 (66–92) | 11 weeks or until negative VRE culture | 1 × 109 CFU of LGG daily as capsules | 3/6 in treatment arm and 2/2 in placebo arm | 2/4 in treatment group | Not reported | No effect on VRE clearance |
| Szachta et al. Poland (2011) [30] | Double-blinded, randomized and placebo-controlled trial with two arms. | 1. LGG 2. Placebo; 3 weeks | 61 (32:29) | 40:21 | 2.5 (unknown) | 4 weeks | 3 × 109 CFU of LGG daily as a sachet dissolved in water or milk | 20/32 in treatment arm and 7/29 in placebo group | 10/19 in treatment arm and 9/20 in placebo group | Not reported | Temporary effect of VRE eradication |
| Doron et al. USA (2015) [31] | Double-blinded, randomized and placebo-controlled trial with two arms. | 1. LGG 2. Placebo; 2 weeks | 11 (5:6) | 7:4 | 70 (53–90) | 2 weeks | 2 × 1010 CFU of LGG as capsules daily | 0/5 in treatment arm and 0/6 in placebo arm | 0/4 in treatment arm and 0/5 in placebo arm | No adverse events related to LGG were seen | No effect on VRE clearance |
| Rauseo et al. USA (2021) [38] | Double-blinded, randomized, and controlled pilot trial with two arms. (VRE clearance = secondary outcome) | 1. LGG 2. Placebo; median duration of intervention was 5.8 and 6.5 days for LGG and placebo respectively | 16 (7:9) | Not reported for the subgroup of VREfm carriers | Not reported for the subgroup of VREfm carriers | Every 3 days after enrollment and at discharge | 2 × 1010 CFU of LGG as capsules or nasogastric administration | 2/7 in treatment arm and 1/9 in placebo arm | Same as end of intervention | No safety concerns and no difference in Bristol stool types between arms | No effect on VRE clearance |
| Rubin et al. Denmark (2022) [25] | Double-blinded, randomized and placebo-controlled trial with two arms. | 1. LGG 2. Placebo; 4 weeks | 48 (21:27) | 18:30 | 75 (64.5–82.5) | 24 weeks | 6 × 1010 CFU of LGG as capsules daily | 12/21 in treatment group and 15/27 in placebo group | 7/8 in treatment group and 9/10 in placebo group | No adverse events related to LGG were seen | No effect on VRE clearance |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rubin, I.M.C.; Knudsen, M.J.S.; Halkjær, S.I.; Ilsby, C.S.; Pinholt, M.; Petersen, A.M. Lacticaseibacillus rhamnosus GG Versus Placebo for Eradication of Vancomycin-Resistant Enterococcus faecium in Intestinal Carriers: A Systematic Review and Meta-Analysis. Microorganisms 2023, 11, 2804. https://doi.org/10.3390/microorganisms11112804
Rubin IMC, Knudsen MJS, Halkjær SI, Ilsby CS, Pinholt M, Petersen AM. Lacticaseibacillus rhamnosus GG Versus Placebo for Eradication of Vancomycin-Resistant Enterococcus faecium in Intestinal Carriers: A Systematic Review and Meta-Analysis. Microorganisms. 2023; 11(11):2804. https://doi.org/10.3390/microorganisms11112804
Chicago/Turabian StyleRubin, Ingrid Maria Cecilia, Maja Johanne Søndergaard Knudsen, Sofie Ingdam Halkjær, Christian Schaadt Ilsby, Mette Pinholt, and Andreas Munk Petersen. 2023. "Lacticaseibacillus rhamnosus GG Versus Placebo for Eradication of Vancomycin-Resistant Enterococcus faecium in Intestinal Carriers: A Systematic Review and Meta-Analysis" Microorganisms 11, no. 11: 2804. https://doi.org/10.3390/microorganisms11112804
APA StyleRubin, I. M. C., Knudsen, M. J. S., Halkjær, S. I., Ilsby, C. S., Pinholt, M., & Petersen, A. M. (2023). Lacticaseibacillus rhamnosus GG Versus Placebo for Eradication of Vancomycin-Resistant Enterococcus faecium in Intestinal Carriers: A Systematic Review and Meta-Analysis. Microorganisms, 11(11), 2804. https://doi.org/10.3390/microorganisms11112804






