Bacteria Associated with Acute Oak Decline: Where Did They Come From? We Know Where They Go
Abstract
:1. Introduction
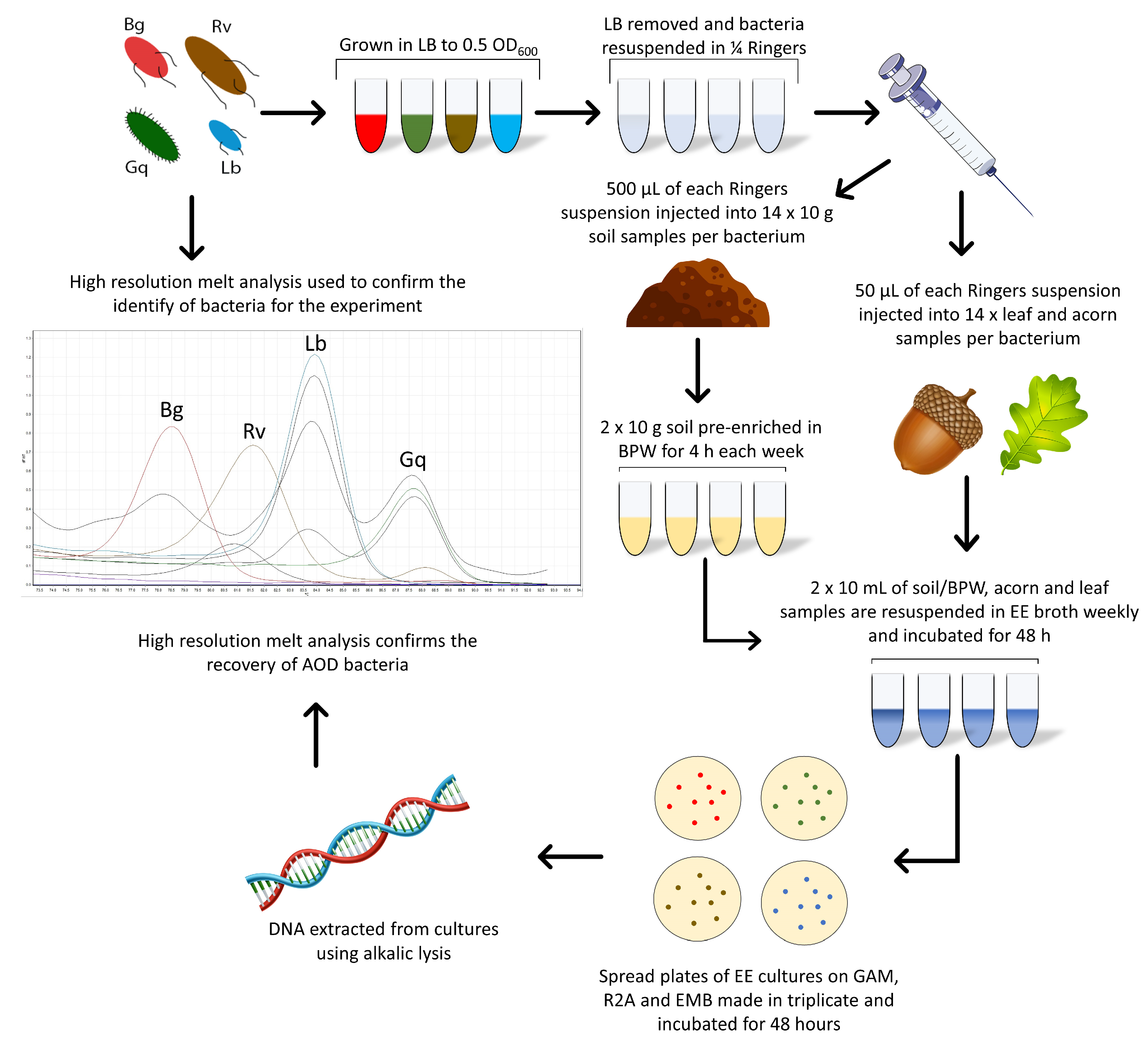
2. Materials and Methods
2.1. Strains Utilised
2.2. Surface Sterilisation of Oak Material
2.3. Spiking of Microcosms with AOD Bacteria
- (a)
- To spike soil: 500 µL of pure washed bacterial culture was pipetted into the middle of a Falcon tube containing 10 g of rhizosphere soil collected from an oak located on the University of the West of England, Frenchay Campus. The soil microcosms were then shaken on the vortex and hand-shaken to ensure the dispersal of bacteria throughout the soil.
- (b)
- To spike leaves: hypodermic needles were used to inject a total of 20 µL of pure washed bacterial culture into the petiole and midrib of Quercus robur leaves at three and four different points on the leaf, respectively.
- (c)
- To spike acorns: a hypodermic needle was used to break through the pericarp and testa of Q. robur acorns at four symmetrical points around the centre to inject 5 µL at each point for a total of 20 µL.
2.4. Enterobacteriaceae Enrichment of Bacteria from Microcosms
2.5. Sample Collection
2.6. HRM Identification of AOD-Associated Bacteria
2.7. Confirmation of Presence of Oak Roots in Rhizosphere Samples
3. Results
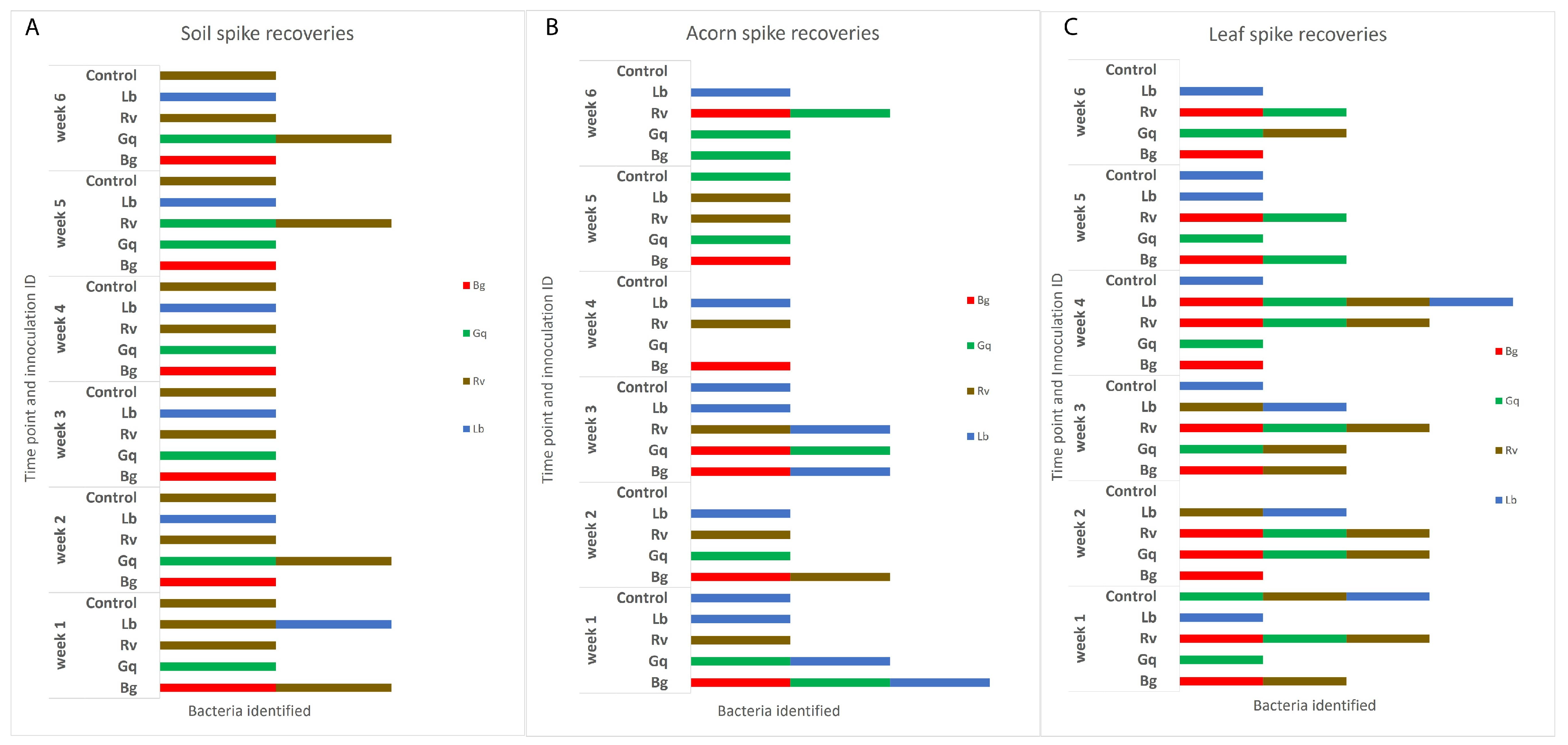
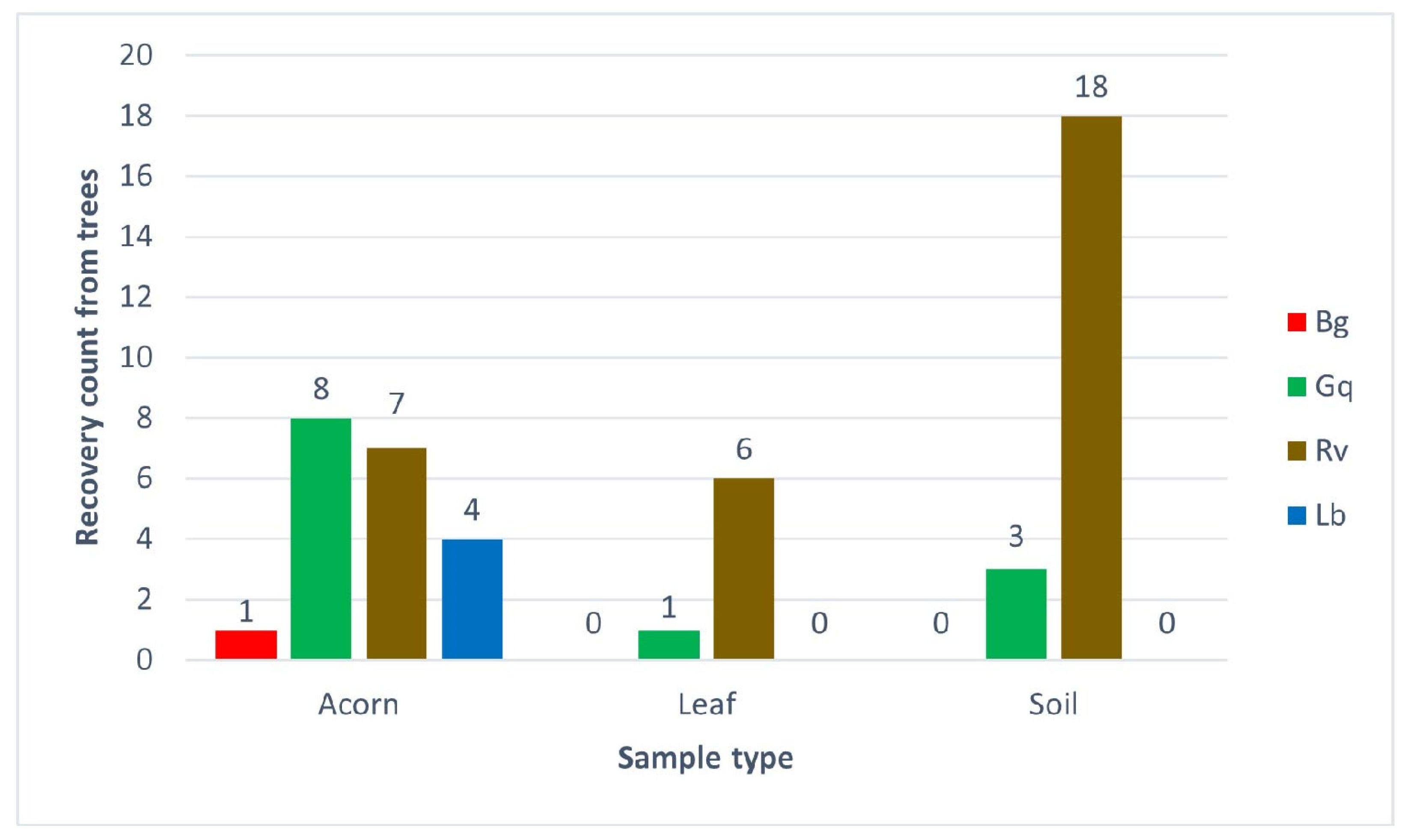
3.1. Survival and Isolation of AOD Bacteria from Oak-Related Niches
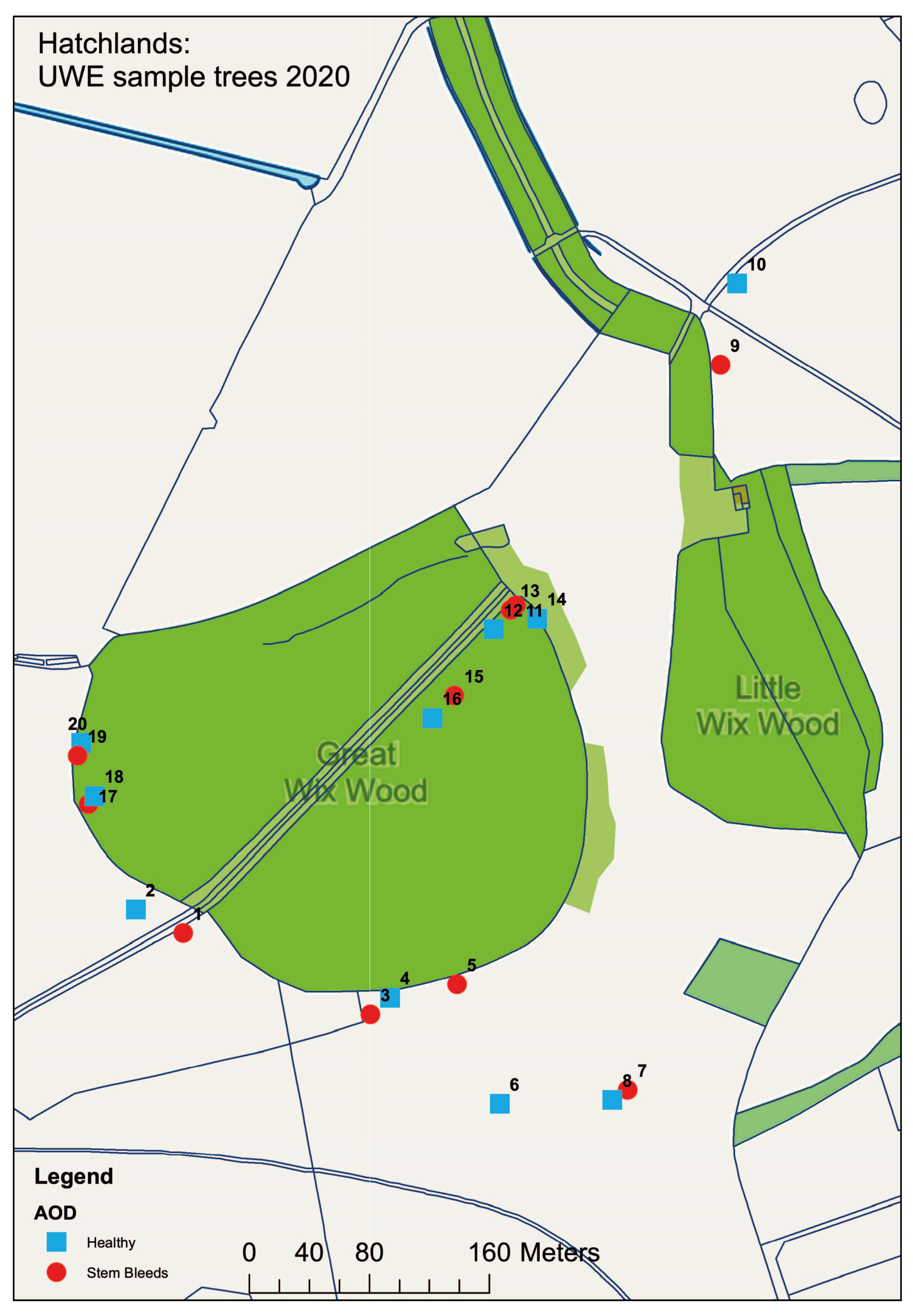
3.2. AOD Confirmation of Symptomatic Oak
3.3. LAMP Confirmation of Oak Roots
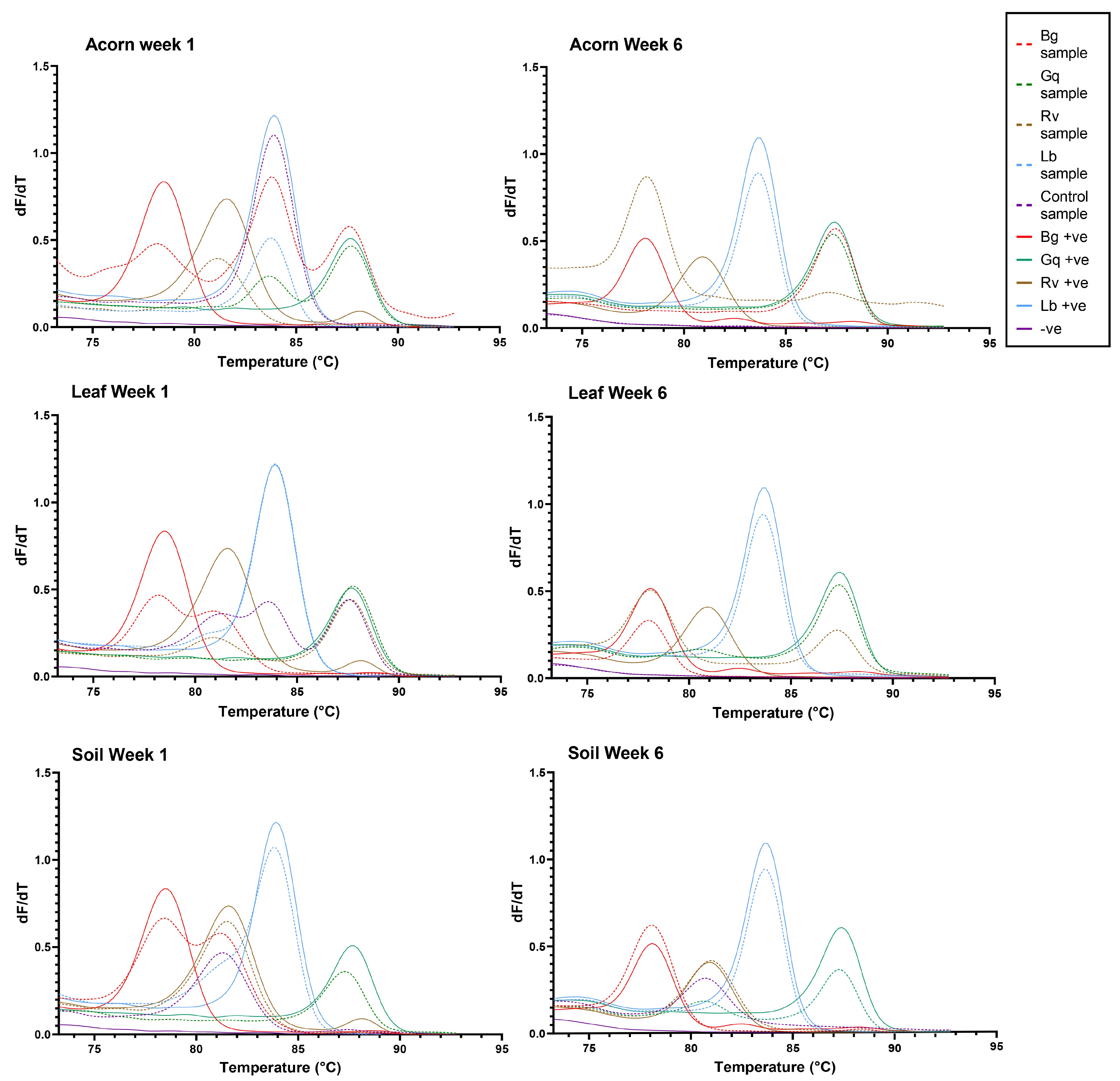
3.4. HRM Analysis of Bacteria from Hatchlands Park Samples
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ruffner, B.; Schneider, S.; Meyer, J.; Queloz, V.; Rigling, D. First report of acute oak decline disease of native and non-native oaks in Switzerland. New Dis. Rep. 2020, 41, 18. [Google Scholar] [CrossRef]
- Crampton, B.; Brady, C.; Denman, S. Bacterial Tree Disease Fact Sheets—Acute oak decline. 2022. Available online: https://bacterialplantdiseases.uk/new-bacterial-tree-disease-factsheets/ (accessed on 11 October 2023).
- Fernandes, C.; Duarte, L.; Naves, P.; Sousa, E.; Cruz, L. First report of Brenneria goodwinii causing acute oak decline on Quercus suber in Portugal. J. Plant Pathol. 2022, 143, 837–838. [Google Scholar] [CrossRef]
- González, A.J.; Ciordia, M. Brenneria goodwinii and Gibbsiella quercinecans isolated from weeping cankers on Quercus robur L. in Spain. Eur. J. Plant Pathol. 2020, 156, 965–969. [Google Scholar] [CrossRef]
- Zalkalns, O.; Celma, L. The distribution of bacteria Gibbsiella quercinecans and Brenneria goodwinii in oak (Quercus robur L.) stands in Latvia. IOP Conf. Ser. Earth Environ. Sci. 2021, 875, 012033. [Google Scholar] [CrossRef]
- Moradi-Amirabad, Y.; Rahimian, H.; Babaeizad, V.; Denman, S. Brenneria spp. and Rahnella victoriana associated with acute oak decline symptoms on oak and hornbeam in Iran. For. Pathol. 2019, 49, e12535. [Google Scholar] [CrossRef]
- Denman, S.; Brown, N.; Vanguelova, E.; Crampton, B. Temperate Oak Declines: Biotic and abiotic predisposition drivers. In Forest Microbiology; Academic Press: Cambridge, MA, USA, 2022; pp. 239–263. [Google Scholar]
- Denman, S.; Doonan, J.; Ransom-Jones, E.; Broberg, M.; Plummer, S.; Kirk, S.; Scarlett, K.; Griffiths, A.R.; Kaczmarek, M.; Forster, J.; et al. Microbiome and infectivity studies reveal complex polyspecies tree disease in Acute Oak Decline. ISME J. 2018, 12, 386–399. [Google Scholar] [CrossRef]
- Brady, C.; Arnold, D.; McDonald, J.; Denman, S. Taxonomy and identification of bacteria associated with acute oak decline. World J. Microbiol. Biotechnol. 2017, 33, 143. [Google Scholar] [CrossRef]
- Doonan, J.; Denman, S.; Pachebat, J.A.; McDonald, J.E. Genomic analysis of bacteria in the Acute Oak Decline pathobiome. Microb. Genom. 2019, 5, e000240. [Google Scholar] [CrossRef]
- Denman, S.; Plummer, S.; Kirk, S.; Peace, A.; McDonald, J.E. Isolation studies reveal a shift in the cultivable microbiome of oak affected with Acute Oak Decline. Syst. Appl. Microbiol. 2016, 39, 484–490. [Google Scholar] [CrossRef]
- Li, A.; He, W. Molecular Aspects of an Emerging Poplar Canker Caused by Lonsdalea populi. Front. Microbiol. 2019, 10, 2496. [Google Scholar] [CrossRef]
- Sapp, M.; Lewis, E.; Moss, S.; Barrett, B.; Kirk, S.; Elphinstone, J.G.; Denman, S. Metabarcoding of bacteria associated with the acute oak decline syndrome in England. Forests 2016, 7, 95. [Google Scholar] [CrossRef]
- Meaden, S.; Metcalf, C.J.E.; Koskella, B. The effects of host age and spatial location on bacterial community composition in the English oak tree (Quercus robur). Environ. Microbiol. Rep. 2016, 8, 649–658. [Google Scholar] [CrossRef]
- Gathercole, L.A.P.; Nocchi, G.; Brown, N.; Coker, T.L.R.; Plumb, W.J.; Stocks, J.J.; Nichols, R.A.; Denman, S.; Buggs, R.J.A. Evidence for the widespread occurrence of bacteria implicated in acute oak decline from incidental genetic sampling. Forests 2021, 12, 1683. [Google Scholar] [CrossRef]
- Santander, R.D.; Català-Senent, J.F.; Figàs-Segura, À.; Biosca, E.G. From the roots to the stem: Unveiling pear root colonization and infection pathways by Erwinia amylovora. FEMS Microbiol. Ecol. 2020, 96, fiaa210. [Google Scholar] [CrossRef]
- An, S.Q.; Potnis, N.; Dow, M.; Vorhölter, F.J.; He, Y.Q.; Becker, A.; Teper, D.; Li, Y.; Wang, N.; Bleris, L.; et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas. FEMS Microbiol. Rev. 2019, 44, 1–32. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2010, 42, 669–678. [Google Scholar] [CrossRef]
- Rahman, M.M.; Flory, E.; Koyro, H.W.; Abideen, Z.; Schikora, A.; Suarez, C.; Schnell, S.; Cardinale, M. Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.). Syst. Appl. Microbiol. 2018, 41, 386–398. [Google Scholar] [CrossRef]
- Compant, S.; Cambon, M.C.; Vacher, C.; Mitter, B.; Samad, A.; Sessitsch, A. The plant endosphere world—Bacterial life within plants. Environ. Microbiol. 2021, 23, 1812–1829. [Google Scholar] [CrossRef]
- Brady, C.; Allainguillaume, J.; Denman, S.; Arnold, D. Rapid identification of bacteria associated with Acute Oak Decline by high-resolution melt analysis. Lett. Appl. Microbiol. 2016, 63, 89–95. [Google Scholar] [CrossRef]
- Bueno-Gonzalez, V. Towards a Rapid Diagostic Method to Identify Bacteria Associated with AOD. Ph.D. Thesis, University of the West of England, Bristol, UK, 2022. [Google Scholar]
- Pettifor, B.J.; Doonan, J.; Denman, S.; McDonald, J.E. Survival of Brenneria goodwinii and Gibbsiella quercinecans, Associated with Acute Oak Decline, in Rainwater and Forest Soil. Syst. Appl. Microbiol. 2020, 43, 126052. [Google Scholar] [CrossRef]
- Microclimates—Fact Sheet 14, National Meteorological Library and Archive. 2019. Available online: https://www.metoffice.gov.uk/binaries/content/assets/metofficegovuk/pdf/research/library-and-archive/library/publications/factsheets/factsheet_14-microclimates.pdf (accessed on 11 October 2023).
- Niemann, S.; Pühler, A.; Tichy, H.V.; Simon, R.; Selbitschka, W. Evaluation of the resolving power of three different DNA fingerprinting methods to discriminate among isolates of a natural Rhizobium meliloti population. J. Appl. Microbiol. 1997, 82, 477–484. [Google Scholar] [CrossRef] [PubMed]
- Corry, J.E.L.; Curtis, G.D.W.; Baird, R.M. Enterobacteriaceae enrichment (EE) broth. Prog. Ind. Microbiol. 2003, 37, 462–464. [Google Scholar] [CrossRef]
- Maddock, D.; Kile, H.; Denman, S.; Arnold, D.; Brady, C. Description of three novel species of Scandinavium: Scandinavium hiltneri sp. nov., Scandinavium manionii sp. nov. and Scandinavium tedordense sp. nov., isolated from the oak rhizosphere and bleeding cankers of broadleaf hosts. Front. Microbiol. 2022, 13, 3900. [Google Scholar] [CrossRef] [PubMed]
- Aslam, Z.; Yasir, M.; Khaliq, A.; Matsui, K.; Chung, Y.R. Mini Review Too much bacteria still unculturable. Crop Environ. 2010, 1, 59–60. [Google Scholar]
- Vos, M.; Wolf, A.B.; Jennings, S.J.; Kowalchuk, G.A. Micro-scale determinants of bacterial diversity in soil. FEMS Microbiol. Rev. 2013, 37, 936–954. [Google Scholar] [CrossRef] [PubMed]
- Brady, C.; Orsi, M.; Doonan, J.M.; Denman, S.; Arnold, D. Brenneria goodwinii growth in vitro is improved by competitive interactions with other bacterial species associated with Acute Oak Decline. Curr. Res. Microb. Sci. 2022, 3, 100102. [Google Scholar] [CrossRef] [PubMed]
- Bashir, I.; War, A.F.; Rafiq, I.; Reshi, Z.A.; Rashid, I.; Shouche, Y.S. Phyllosphere microbiome: Diversity and functions. Microbiol. Res. 2022, 254, 126888. [Google Scholar] [CrossRef]
- Lu, S.; Pathology, P.; State, M. Drippy Pod of White Lupine: A New Bacterial Disease Caused by a Pathovar of Brenneria quercina. Plant Dis. 2010, 94, 1431. [Google Scholar] [CrossRef]
- Janda, J.M.; Abbott, S.L. The changing face of the family Enterobacteriaceae (Order:“Enterobacterales”): New members, taxonomic issues, geographic expansion, and new diseases and disease syndromes. Clin. Microbiol. Rev. 2021, 34, 1–45. [Google Scholar] [CrossRef]
- Brady, C.L.; Cleenwerck, I.; Denman, S.; Venter, S.N.; Rodríguez-Palenzuela, P.; Coutinho, T.A.; De Vos, P. Proposal to reclassify Brenneria quercina (Hildebrand and Schroth 1967) Hauben et al. 1999 into a new genus, Lonsdalea gen. nov., as Lonsdalea quercina comb. nov., descriptions of Lonsdalea quercina subsp. quercina comb. nov., Lonsdalea quercina subsp. iberica subsp. nov. and Lonsdalea quercina subsp. britannica subsp. nov., emendation of the description of the genus Brenneria, reclassification of Dickeya dieffenbachiae as Dickeya dadantii subsp. dieffenbachiae comb. nov., and emendation of the description of Dickeya dadantii. Int. J. Syst. Evol. Microbiol. 2012, 62, 1592–1602. [Google Scholar] [CrossRef]
- Tian, B.; Zhang, C.; Ye, Y.; Wen, J.; Wu, Y.; Wang, H.; Li, H.; Cai, S.; Cai, W.; Cheng, Z.; et al. Beneficial traits of bacterial endophytes belonging to the core communities of the tomato root microbiome. Agric. Ecosyst. Environ. 2017, 247, 149–156. [Google Scholar] [CrossRef]
- Huvenne, H.; Goeminne, G.; Maes, M.; Messens, E. Identification of quorum sensing signal molecules and oligolignols associated with watermark disease in willow (Salix sp.). J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2008, 872, 83–89. [Google Scholar] [CrossRef]
- Maes, M.; Baeyen, S.; De Croo, H.; De Smet, K.; Steenackers, M. Monitoring of endophytic Brenneria salicis in willow and its relation to watermark disease. Plant Prot. Sci. 2017, 38, 528–530. [Google Scholar] [CrossRef]
- Czajkowski, R.; Pérombelon, M.C.M.; Jafra, S.; Lojkowska, E.; Potrykus, M.; Van Der Wolf, J.M.; Sledz, W. Detection, identification and differentiation of Pectobacterium and Dickeya species causing potato blackleg and tuber soft rot: A review. Ann. Appl. Biol. 2015, 166, 18–38. [Google Scholar] [CrossRef]
- Monteiro, R.A.; Balsanelli, E.; Wassem, R.; Marin, A.M.; Brusamarello-Santos, L.C.C.; Schmidt, M.A.; Tadra-Sfeir, M.Z.; Pankievicz, V.C.S.; Cruz, L.M.; Chubatsu, L.S.; et al. Herbaspirillum-plant interactions: Microscopical, histological and molecular aspects. Plant Soil 2012, 356, 175–196. [Google Scholar] [CrossRef]
- Álvarez-Loayza, P.; White, J.F.; Torres, M.S.; Balslev, H.; Kristiansen, T.; Svenning, J.C.; Gil, N. Light converts endosymbiotic fungus to pathogen, influencing seedling survival and niche-space filling of a common tropical tree, Iriartea deltoidea. PLoS ONE 2011, 6, e16386. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef]
| Primer | Sequence (5′->3′) |
|---|---|
| Bgi2F | CGTCAAACTATTTGCTTCCACCCATC |
| Bgi2R | CGGTATGGGTCGGGACATTTG |
| Bgi3F | CATCGCGTCCAGCGTCTG |
| Bgi3R | GCCTATTGCGTGAACGAACTGGATAG |
| Gqi3F | GCATACGCCTGGTACAGCGC |
| Gqi3R | CCTTGGCGGGACAGTCTTGC |
| Rvii1F | GCATCTCGCAGATCGCTGAAAC |
| Rvii1R | TGGAAGCGGCGGCTGAC |
| Lbi2F | GGAATCGCTTTACCGTCGCTATTG |
| Lbi2R | CAAGGTGGTGATGGTGGTCGATC |
| Tree | Health Status | B. goodwinii | G. quercinecans | R. victoriana | L. britannica |
|---|---|---|---|---|---|
| 1 | AOD active bleeds | + | + | - | - |
| 3 | AOD active bleeds | + | + | - | - |
| 5 | AOD active bleeds | N/A | N/A | N/A | N/A |
| 7 | AOD active bleeds | + | - | - | - |
| 9 | AOD active bleeds | N/A | N/A | N/A | N/A |
| 11 | AOD active bleeds | + | - | - | - |
| 13 * | AOD dry bleeds | - | - | - | - |
| 15 * | AOD dry bleeds | - | - | - | - |
| 17 | AOD dry bleeds | + | - | - | - |
| Totals | 5/9 | 2/9 | 0/9 | 0/9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maddock, D.; Brady, C.; Denman, S.; Arnold, D. Bacteria Associated with Acute Oak Decline: Where Did They Come From? We Know Where They Go. Microorganisms 2023, 11, 2789. https://doi.org/10.3390/microorganisms11112789
Maddock D, Brady C, Denman S, Arnold D. Bacteria Associated with Acute Oak Decline: Where Did They Come From? We Know Where They Go. Microorganisms. 2023; 11(11):2789. https://doi.org/10.3390/microorganisms11112789
Chicago/Turabian StyleMaddock, Daniel, Carrie Brady, Sandra Denman, and Dawn Arnold. 2023. "Bacteria Associated with Acute Oak Decline: Where Did They Come From? We Know Where They Go" Microorganisms 11, no. 11: 2789. https://doi.org/10.3390/microorganisms11112789
APA StyleMaddock, D., Brady, C., Denman, S., & Arnold, D. (2023). Bacteria Associated with Acute Oak Decline: Where Did They Come From? We Know Where They Go. Microorganisms, 11(11), 2789. https://doi.org/10.3390/microorganisms11112789







