The Effect of Probiotics in a Milk Replacer on Leukocyte Differential Counts, Phenotype, and Function in Neonatal Dairy Calves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Blood Collection
2.3. Lung Lavage
2.4. Flow Cytometry
2.5. Gene Expression Analysis
2.6. Statistics
3. Results
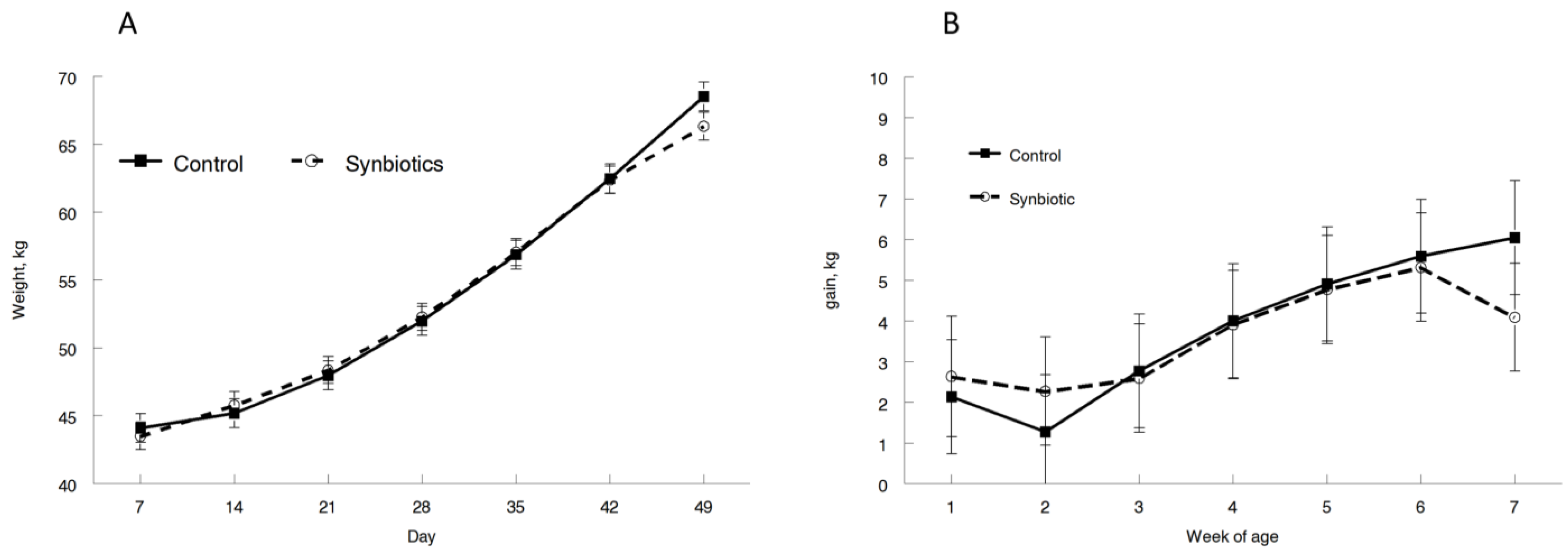
3.1. Performance
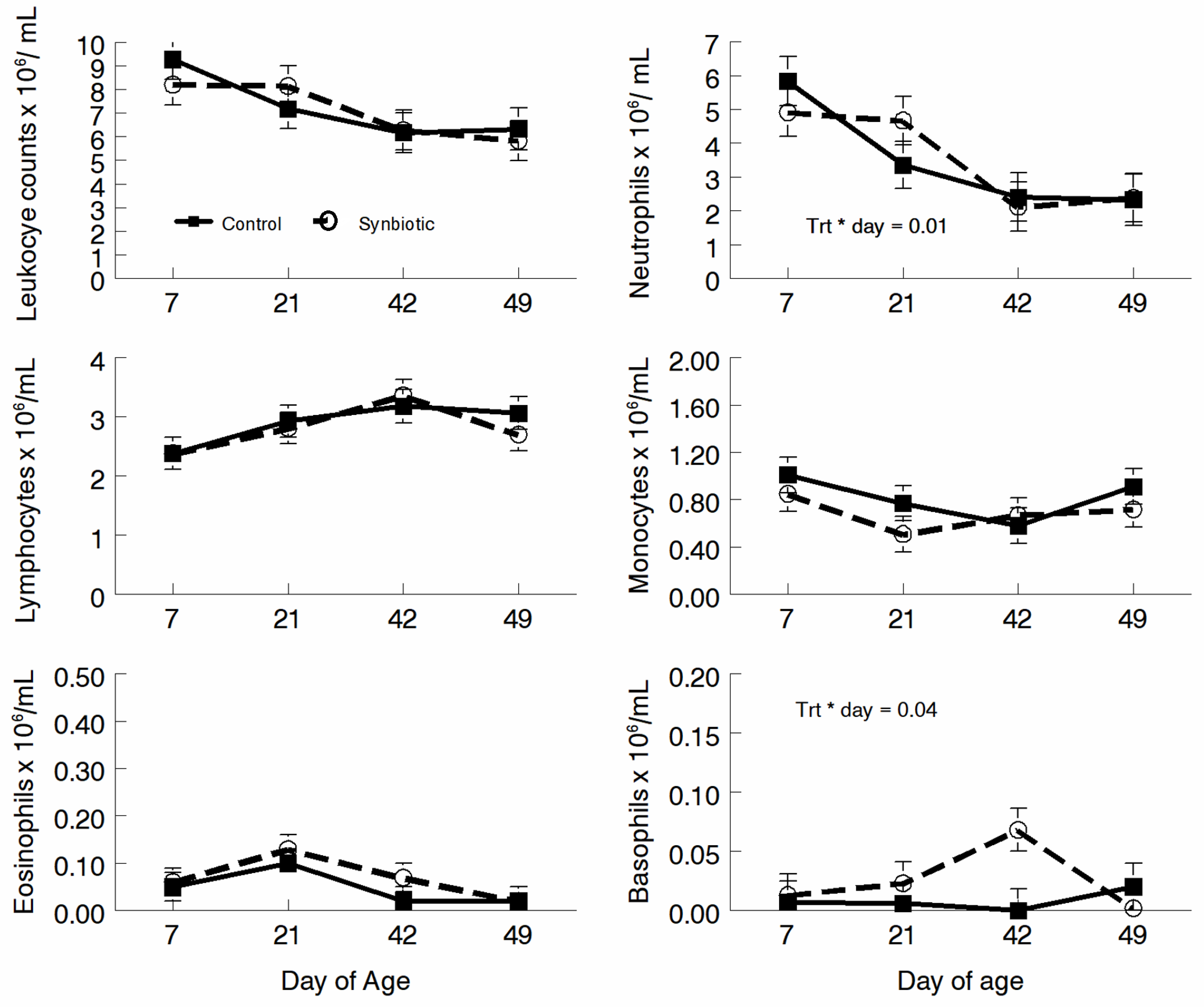
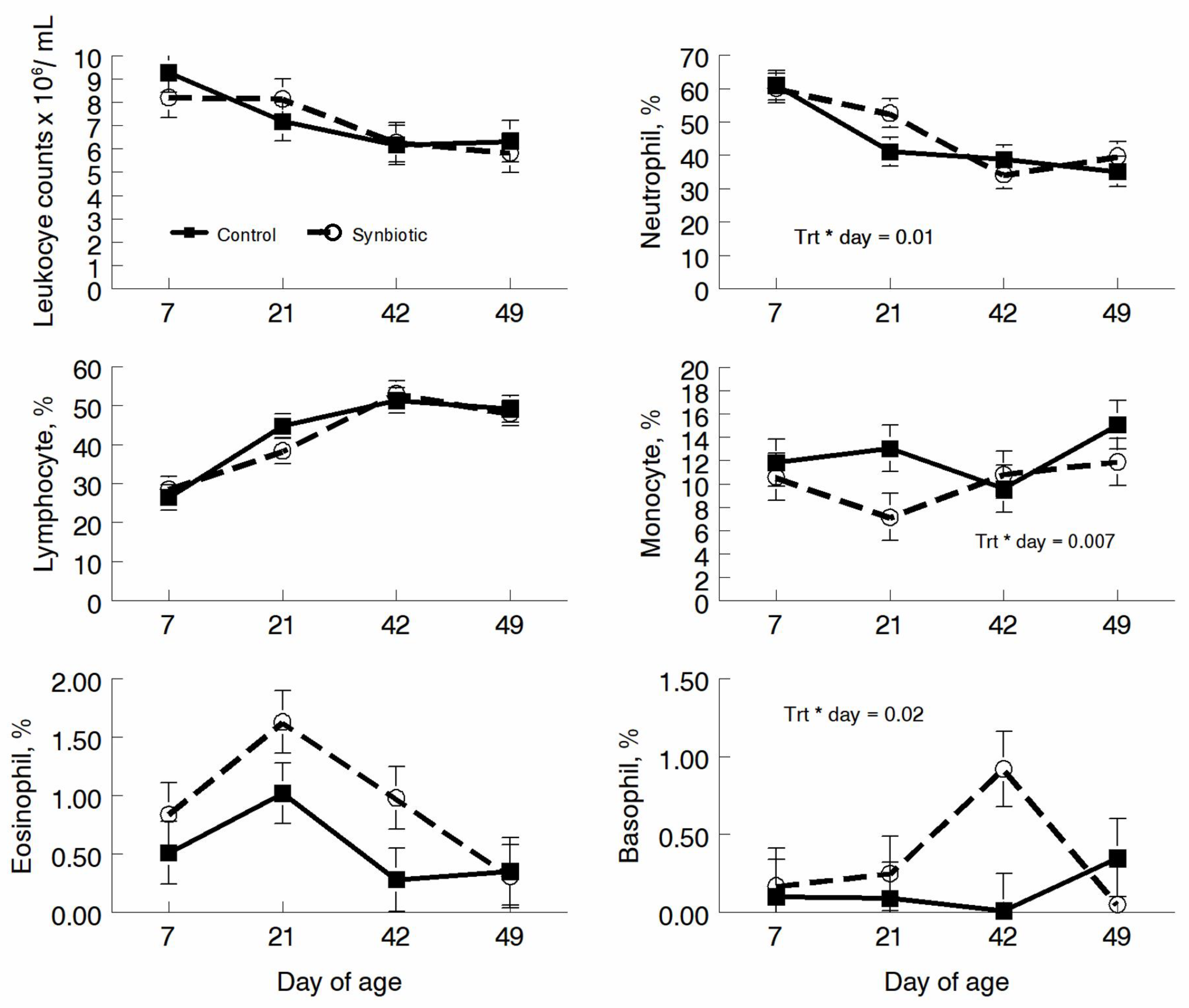
3.2. Hematology
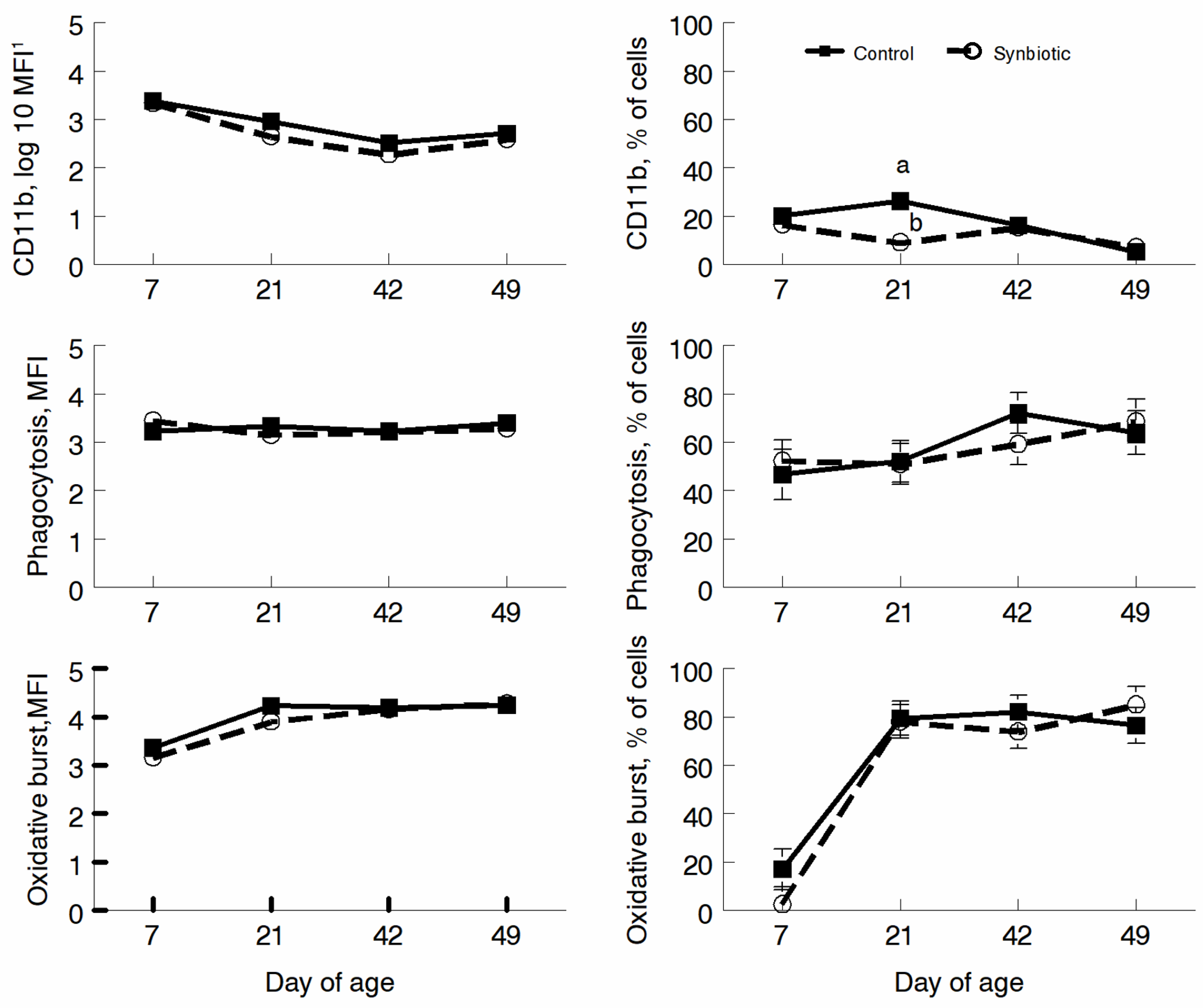
3.3. Flow Cytometry
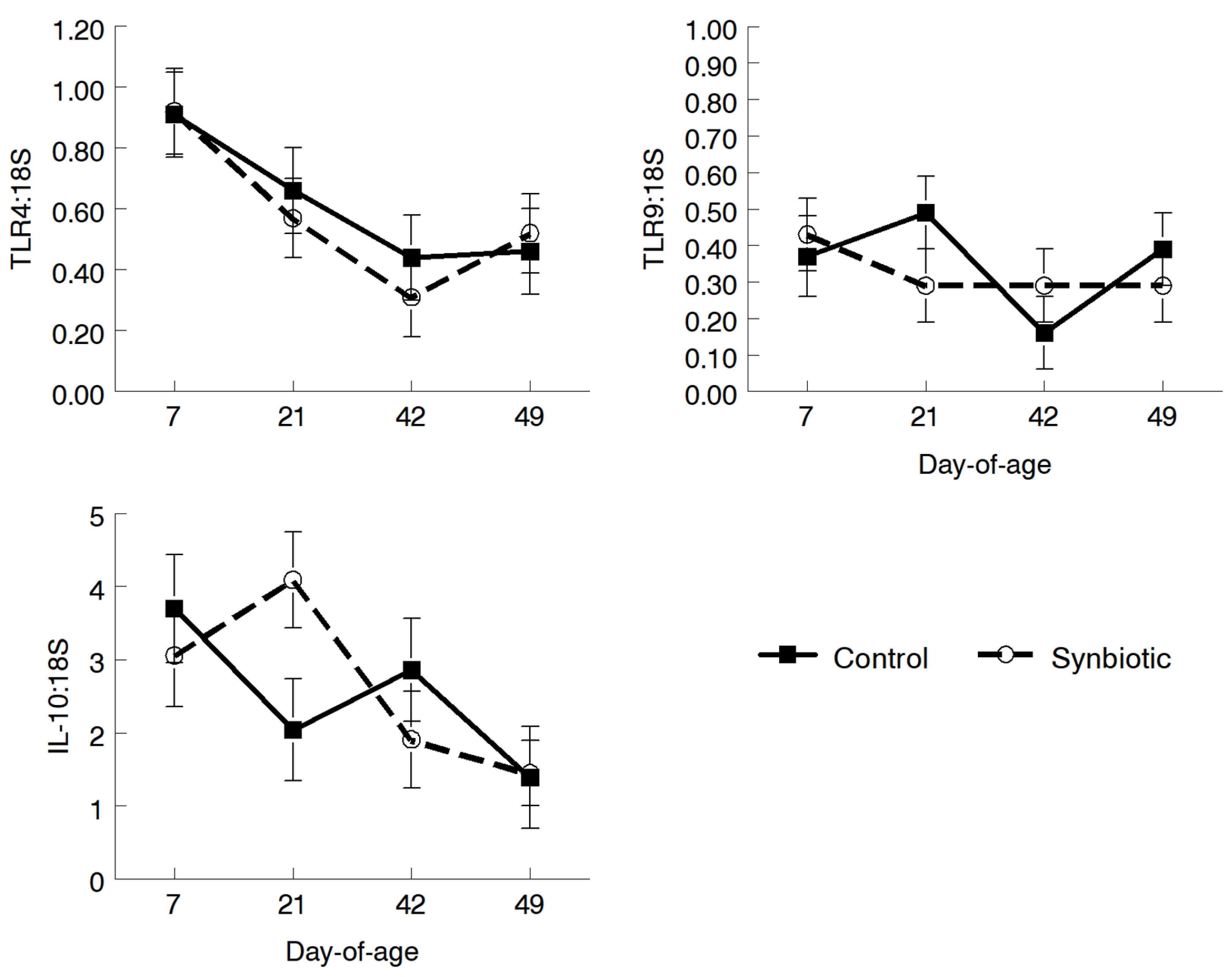
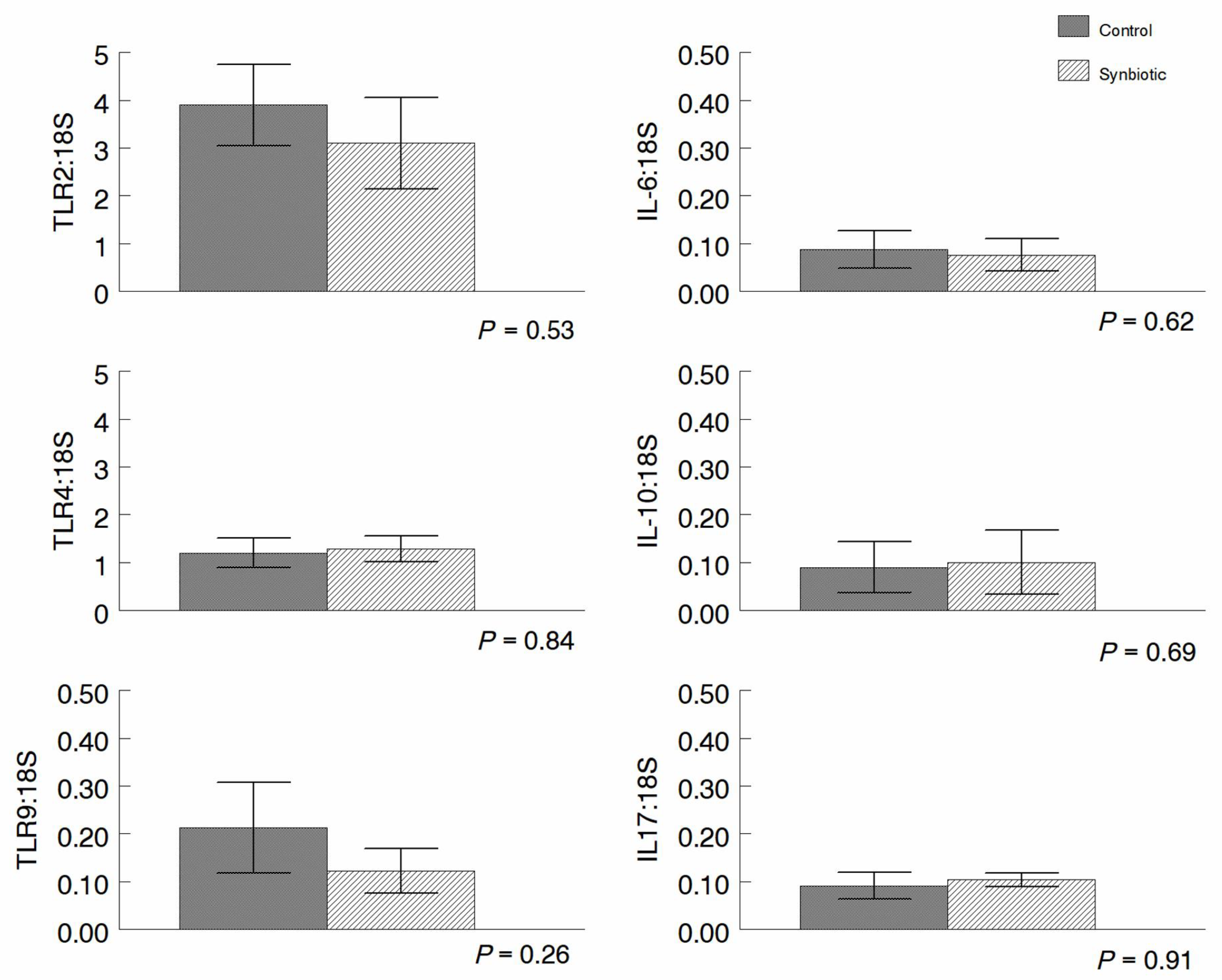
3.4. qRT-PCR Analysis of Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chase, C.C.L. Enteric Immunity Happy Gut, Healthy Animal. Vet. Clin. Food Anim. 2018, 34, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Welch, C.B.; Ryman, V.R.; Pringe, T.D.; Lourenco, J.M. Utilizing the gastrointestinal microbiota to modulate cattle health through the microbiome-gut-organ axes. Microorganisms 2022, 10, 1391. [Google Scholar] [CrossRef] [PubMed]
- Arsene, M.M.J.; Davares, A.K.L.; Andreevna, S.L.; Vladimirovish, E.A.; Carime, B.Z.; Marouf, R.; Khelfi, I. The use of probiotics in animal feeding for safe production and as potential alternatives to antibiotics. Vet. World 2021, 14, 319–328. [Google Scholar] [CrossRef] [PubMed]
- Holman, D.B.; McAllister, G.A.; Topp, E.; Wright, A.G.; Alexander, T.W. The nasopharygeal microbiota of feedlot cattle that develop bovine respiratory disease. Vet. Microbiol. 2015, 180, 90–95. [Google Scholar] [CrossRef]
- Osman, R.; Malmuthuge, N.; Gonzalez-Cano, P.; Griebel, P. Development and Function of the Mucosal Immune System in the Upper Respiratory Tract of Neonatal Calves. Annu. Rev. Anim. Biosci. 2018, 6, 141–155. [Google Scholar] [CrossRef]
- Adjei-Fremah, S.; Ekwemalor, K.; Asiamah, E.K.; Ismail, H.; Ibrahim, S.; Worku, M. Effect of probiotic supplementation on growth and global gene expression in dairy cows. J. Appl. Anim. Res. 2018, 46, 257–263. [Google Scholar] [CrossRef]
- Wilson, H.L.; Obradovic, M.R. Evidence for a common mucosal immune system in the pig. Mol. Immunol. 2014, 66, 22–34. [Google Scholar] [CrossRef]
- Eicher-Pruiett, S.D.; Morrill, J.L.; Nagaraja, T.G.; Higgins, J.J.; Anderson, N.V.; Reddy, P.G. Response of young dairy calves with lasalocid delivery varied in feed sources. J. Dairy Sci. 1992, 75, 857–862. [Google Scholar] [CrossRef]
- Eicher-Pruiett, S.D.; Morrill, J.L.; Blecha, F.; Higgins, J.J.; Anderson, N.V.; Reddy, P.G. Neutrophil and lymphocyte response to supplementation with vitamins C and E in young calves. J. Dairy Sci. 1992, 75, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Abdelfattah, E.M.; Schutz, M.M.; Lay, D.C., Jr.; Marchant-Forde, J.N.; Eicher, S.D. Effect of group size on behavior, health, production, and welfare of veal calves. J. Anim. Sci. 2013, 91, 5455–5465. [Google Scholar] [CrossRef]
- Lloyd, C.M.; Saglani, S. Early-life respiratory infections and developmental immunity determine lifelong lung health. Nat. Immunol. 2023, 24, 1234–1243. [Google Scholar] [CrossRef]
- Lima, S.T.; Teixeira, A.G.; Higgins, C.H.; Lima, F.S.; Bicalho, R.C. The upper respiratory tract microbiome and its potential role in bovine respiratory disease and otitis media. Sci. Rep. 2016, 6, 029050. [Google Scholar] [CrossRef] [PubMed]
- Bosch, A.A.T.M.; de Steenhuijsen Piters, W.A.A.; van Houten, M.A.; Chu, M.L.J.N.; Biesbroek, G.; Kool, J.; Pernet, P.; de Groot, P.C.M.; Eijkemans, M.J.C.; Keijser, B.J.F.; et al. Maturation of the Infant Respiratory Microbiota, Environmental Drivers, and Health Consequences. A Prospective Cohort Study. Am. J. Respir. Crit. Care Med. 2017, 196, 1582–1590. [Google Scholar] [CrossRef] [PubMed]
- Corbett, E.M.; Norby, B.; Halbert, L.W.; Henderson, S.T.; Grooms, D.L.; Manning, S.D.; Kaneene, J.B. Effect of feeding a direct-fed-microbial on total and antimicrobial-resistant fecal coliform counts in preweaned dairy calves. Am. J. Vet. Res. 2015, 76, 780–788. [Google Scholar] [CrossRef]
- Timsit, E.; McMullen, C.; Amat, S.; Alexander, T.W. Respiratory Bacterial Microbiota in Cattle: From development to modulation to enhance respiratory health. Vet. Clin. N. Am. Food Pract. 2020, 36, 297–320. [Google Scholar] [CrossRef] [PubMed]
- Renaud, D.L.; Kelton, D.F.; Weese, J.S.; Noble, C.; Duffield, T.F. Evaluation of a multispecies probiotic as a supportive treatment for diarrhea in dairy calves: A randomized clinical trial. J. Dairy Sci. 2019, 102, 4498–4505. [Google Scholar] [CrossRef] [PubMed]
- Salzar, L.F.L.; Nero, L.A.; Campos-Galvao, M.E.M.; Cortinhas, C.S.; Acedo, T.S.; Tamassia, L.F.M.; Busato, K.C.; Morais, V.C.; Rotta, P.P.; Silva, A.L.; et al. Effect of selected feed additives to improve growth and health of dairy calves. PLoS ONE 2019, 14, e0216066. [Google Scholar] [CrossRef]
- Wang, H.; Yu, Z.; Gao, Z.; Li, Q.; Qui, X.; Wu, F.; Guan, T.; Cao, B.; Su, H. Effects of compound probiotics on growth performance, rumen fermentation, blood parameters, and health status of neonatal Holstein calves. J. Dairy Sci. 2022, 105, 2190–2200. [Google Scholar] [CrossRef]
- de Melo, L.C.; Itavo, L.C.V.; Itavo, C.C.B.F.; Dias, A.M.; Gurgel, A.L.C.; Nonato, L.M.; Arcanjo, A.H.M.; da Sila Zornitta, C.; de Oliviera Monteiro, P.E.; da Silva, A.P. Sequential use of nutritional additives in diets for finishing Nellore steers in confinement. Trop. Anim. Health Prod. 2023, 55, 151. [Google Scholar] [CrossRef]
- Mackey, S.J.; Cooke, R.F.; Colombo, E.A.; Pickett, A.T.; Batista, L.F.D.; Block, E.; Brando, A.P. Supplementing pre- and probiotic ingredients to feedlot steers: Effects on health, growth performance, and physiological responses. Animal 2023, 17, 10070. [Google Scholar] [CrossRef]
- Li, A.; Yang, J.; Zhang, C.; Chi, H.; Zhang, C.; Li, T.; Zhang, J.; Du, P. Lactobacillus acidophilus KLDS 1.0738 inhibits TLR4/NF-kB inflammatory pathway in b-lactoglobulin-induced macrophages via modulating miR-146a. J. Food Biochem. 2021, 45, e13662. [Google Scholar] [CrossRef] [PubMed]
- Lucey, P.M.; Lean, I.J.; Aly, S.S.; Golder, H.M.; Block, E.; Thompson, J.S.; Rossow, H.A. Effects of mannan-oligosaccharide and Bscillus subtilis supplementation to preweaning Holstein dairy heifers on body weight gain, diarrhea, and shedding of fecal pathogens. J. Dairy Sci. 2021, 104, 4290–4302. [Google Scholar] [CrossRef] [PubMed]
- Maamouri, O.; Ben Salem, M. The effect of live yeast Saccharomyces cerevisiae as probiotic supply on growth performance, feed intake, ruminal pH and fermentation in fattening calves. Vet. Med. Sci. 2022, 8, 398–404. [Google Scholar] [CrossRef]
- Colombo, E.A.; Cooke, R.F.; Brandao, A.P.; Wiegand, J.B.; Schubach, K.M.; Sowers, C.A.; Duff, G.C.; Block, E.; Gouvea, V.N. Performance, healthe, and physiological responses of newly received feedlot cattle supplemented with pre- and probiotic ingrediants. Animal 2021, 15, 100214. [Google Scholar] [CrossRef]
- Cull, C.; Singu, V.K.; Cull, B.J.; Lechtenberg, K.F.; Amachawadi, R.G.; Schutz, J.S.; Bryan, K.A. Efficacy of Lactobacillus animalis and Propionibacterium freudenreichii-based feed additives in reducing Salmonella-associated health and performance effects in commercial beef calves. Antibiotics 2022, 11, 1328. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, S.; Fraga, M.; Castells, M.; Colina, R.; Zunino, P. Effect of the administration of Lactobacillus spp. Strains on neonatal diarrhoea, immune parameters and pathogen abundance in pre-weaned calves. Benef. Microbes 2020, 11, 477–488. [Google Scholar] [CrossRef]
- Gandhar, J.S.; De, U.K.; Kala, A.; Malik, Y.S.; Yadav, S.; Paul, B.R.; Dixit, S.K.; Sircir, S.; Chaudry, P.; Patra, M.K.; et al. Efficacy of microencapsulated probiotic as admunct therapy on resolustion of diarrhea, copper-zinc homeostasis, immunoglobulins, and inflammatory markers in serum of spntaneous rotovirus-infected diarrhoetic calves. Probiotics Antimicrob. Proteins 2021, 14, 1054–1066. [Google Scholar] [CrossRef]
- Kayasaki, F.; Okagawa, T.; Konnai, S.; Kohara, J.; Sajiki, Y.; Watari, K.; Ganbaatar, O.; Goto, S.; Nakamura, H.; Shimakura, H.; et al. Direct evidence of the prevantative effect of milk repacer-based probiotic feeding in calves against sever diarrhea. Vet. Microbiol. 2021, 254, 108976. [Google Scholar] [CrossRef]
- Liu, B.; Wang, C.; Huasau, S.; Han, A.; Zhang, J.; He, L.; Aorigele, C. Compound probiotics improve the diarrhea rate and intestinal microbiota of newborn calves. Animals 2022, 12, 322. [Google Scholar] [CrossRef]
- Urkawa, M.; Zhuang, T.; Sato, H.; Takanashi, S.; Yoshimura, K.; Endo, Y.; Katsura, T.; Umino, T.; Tanaka, K.; Watanabe, H.; et al. Prevention of mastitis in multiparous dairy cows with a previous history of mastitis by oral feeding with probiotic Bacillus subtilis. Anim. Sci. J. 2022, 93, e13764. [Google Scholar] [CrossRef]
- Ikehata, M.; Konnai, S.; Okgawa, T.; Abe, K.; Honma, M.; Kitamura, T.; Maekawa, N.; Suzuki, Y.; Murata, S.; Ohashi, K. In vitro evaluation of Lactiplantibacillus plantrum HOKKAIDO strain, effective lactic acid bacteria for calf diarrhea. Front. Vet. Sci. 2023, 10, 1145445. [Google Scholar] [CrossRef]
- Cattaneo, L.; Lopreiato, V.; Piccioli-Cappelli, F.; Trevisi, E.; Minuti, A. Effect of supplementing live Sacchromyces cerevisiae on performance, rumen function, and metabolism during the transition period in Holstein dairy cows. J. Dairy Sci. 2023, 106, 4353–4365. [Google Scholar] [CrossRef] [PubMed]
- Chaumond, E.; Peron, S.; Daniel, N.; Le Gouar, Y.; Guedon, E.; Williams, D.L.; Le Loir, Y.; Jan, G.; Berkova, N. Development of innate immune memory by non-immune cells during Staphylococcus aureus infection dpends on reactive oxygen species. Front. Immunol. 2023, 14, 1138539. [Google Scholar] [CrossRef] [PubMed]
- Kober, A.K.M.H.; Saha, S.; Islan, M.A.; Rajoka, M.S.R.; Fukuyama, K.; Aso, H.; Villena, J.; Kitazawa, H. Immunomodulatory effects of probiotics: A novel preventive approach for the control of bovine mastitis. Microorganisms 2022, 10, 2255. [Google Scholar] [CrossRef]
- Li, K.; Yang, M.; Tian, M.; Jia, L.; Du, J.; Wu, Y.; Li, L.; Yuan, L.; Ma, Y. Lactobacillus platarum 17-5 attenuates Excherichia coli-induced inflammatory responses via inhibitin the NF-kB and MAPK signalling pathways in bovine mammary epithelial cells. BMC Vet. Res. 2022, 18, 250. [Google Scholar] [CrossRef] [PubMed]
- Mansilla, F.; Takagai, M.; Garcia-Castillo, V.; Aso, H.; Nader-Macias, M.E.; Vignolo, G.; Kitazawa, H.; Villena, J. Modulation of Toll-like receptor-mediated innate immunity in bovine intestinal epithelial cells by lacti acid bacteria isolated from feedlot cattle. Benef. Microbes. 2020, 11, 269–282. [Google Scholar] [CrossRef]
- Villena, J.; Oliveira, M.L.S.; Ferreira, P.C.D.; Salva, S.; Alvarez, S. Lactic acid bacteria in the prevention of pneumococcal respiratory infection: Future opportunities and challenges. Int. Immunopharmacol. 2011, 11, 1633–1645. [Google Scholar] [CrossRef]
- Villena, J.; Chiba, E.; Tomosada, Y. Orally administered Lactobacillus rhamnosus modulates the respiratory immune response triggered by the viral pathogen-associated molecular pattern poly(I:C). BMC Immunol. 2012, 13, 53. [Google Scholar] [CrossRef]
- Yang, Y.; Jing, Y.; Yang, J.; Yang, Q. Effects of intranasal administration with Bacillus subtilis on immune cells in the nasal mucosa and tonsils of piglets. Exp. Ther. Med. 2018, 15, 5189–5198. [Google Scholar]
- Ludwig, I.S.; Broere, F.; Manurung, S.; Lambers, T.T.; van der Zee, R.; van Eden, W. Lactobacillus rhamnosus GG-Derived Soluble Mediators Modulate Adaptive Immune Cells. Front. Immunol. Frontimmun. 2018, 9, 1546. [Google Scholar] [CrossRef]
- Kitazawa, H.; Villena, J. Modulation of respiratory TLR3-anti-viral response by probiotic microorganisms: Lessons learned from Lactobacillus rhamnosus CRL1505. Front. Immunol. 2014, 201. [Google Scholar] [CrossRef]
- Lehtoranta, L.; Pitkäranta, A.; Korpela, R. Probiotics in respiratory virus infections. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 1289–1302. [Google Scholar] [CrossRef]
- Eicher, S.D.; Chitko-McKown, C.G.; Bryan, K.A. Variation in the response of bovine alveolar lavage cells to diverse species of probiotic bacteria. BMC Res. Notes 2020, 13, 159. [Google Scholar] [CrossRef]
- Forsythe, P.; Kunze, W.A. Voices from within: Gut microbes and the CNS. Cell. Mol. Life Sci. 2013, 70, 55–69. [Google Scholar] [CrossRef]
- Marranzino, G.; Villena, J.; Salva, S.; Alvarez, S. Stimulation of macrophages by immunobiotic Lactobacillus Immunogenics: Immunostimustrains: Influence beyond the intestinal tract. Microbiol. Immunol. 2012, 56, 771–781. [Google Scholar] [CrossRef]
- Mack, D.R.; Ahrne, S.; Hyde, L.; Wei, S.; Hollingsworth, M.A. Extracellular MUC3 secretion follows adherence of Lactobacillus strains to intestinal epithelial cells in vitro. Gut 2003, 52, 827–833. [Google Scholar] [CrossRef]
- Popova, M.; Molimard, P.; Courau, S.; Crociani, J.; Dufour, C.; Le Vacon, F.; Carton, T. Beneficial effects of probiotics in upper respiratory tract infections and their mechanical actions to antagonize pathogens. J. Appl. Micro. 2012, 113, 6. [Google Scholar] [CrossRef]
- Lambo, M.T.; Chang, X.; Liu, D. The recent trend in the use of multistrain probiotics in livestock production: An overview. Animals 2021, 11, 2805. [Google Scholar] [CrossRef]
- Barreto, M.O.; Soust, M.; Moore, R.J.; Olchowy, T.W.J.; Alawneh, J.I. Systemic review and meta-analysis of probiotic use on inflammatory biomarkers and disease prevention in cattle. Prev. Vet. Med. 2021, 194, 105433. [Google Scholar] [CrossRef]


| Antigen (Ab) | Flurorphore | Source |
|---|---|---|
| CO | 0 | --- |
| CD4 | FITC | BioRad 1 |
| CD8 | PE | BioRad 1 |
| CD14 | RPE | BioRad 1 |
| CD18 | FITC | WSU 2, BioRad goat anti-mouse secondary Ab 1 |
| CD205 | RPE | BioRad 1 |
| CD3 | RPE | BioRad 1 |
| CD11b | FITC | BioRad 1 |
| Phagocytosis | E. coli bioparticles pHrodo Red + OPSONIN | Invitrogen 3 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eicher, S.D.; Kritchevsky, J.E.; Bryan, K.A.; Chitko-McKown, C.G. The Effect of Probiotics in a Milk Replacer on Leukocyte Differential Counts, Phenotype, and Function in Neonatal Dairy Calves. Microorganisms 2023, 11, 2620. https://doi.org/10.3390/microorganisms11112620
Eicher SD, Kritchevsky JE, Bryan KA, Chitko-McKown CG. The Effect of Probiotics in a Milk Replacer on Leukocyte Differential Counts, Phenotype, and Function in Neonatal Dairy Calves. Microorganisms. 2023; 11(11):2620. https://doi.org/10.3390/microorganisms11112620
Chicago/Turabian StyleEicher, Susan D., Janice E. Kritchevsky, Keith A. Bryan, and Carol G. Chitko-McKown. 2023. "The Effect of Probiotics in a Milk Replacer on Leukocyte Differential Counts, Phenotype, and Function in Neonatal Dairy Calves" Microorganisms 11, no. 11: 2620. https://doi.org/10.3390/microorganisms11112620
APA StyleEicher, S. D., Kritchevsky, J. E., Bryan, K. A., & Chitko-McKown, C. G. (2023). The Effect of Probiotics in a Milk Replacer on Leukocyte Differential Counts, Phenotype, and Function in Neonatal Dairy Calves. Microorganisms, 11(11), 2620. https://doi.org/10.3390/microorganisms11112620





