A Class 4-like Chromosomal Integron Found in Aeromonas sp. Genomospecies paramedia Isolated from Human Feces
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strains Isolation and Characterization
2.2. Species Identification Based on Conventional Multilocus Phylogenetic Analyses (MLPA)
2.3. Genome Sequencing and MLPA
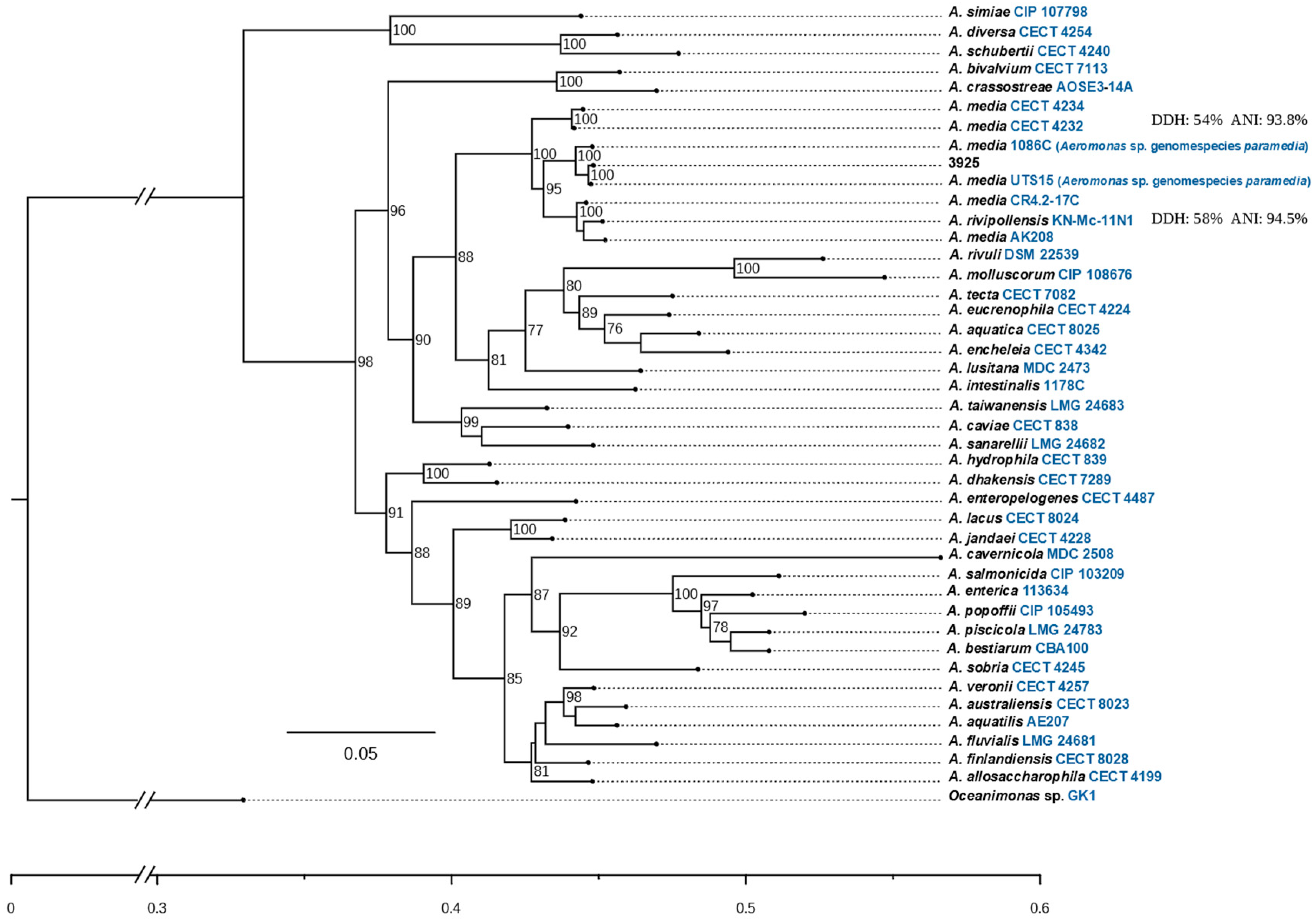
2.4. Phylogenomic Analysis by isDDH and ANI
2.5. Genome Analysis and Comparative Annotations
2.6. Characterization and Functionality of the Integron
2.7. Analysis of intI4-like Integrase Expression by RT-PCR
3. Results
3.1. Species Assignment of Aeromonas sp. 3925
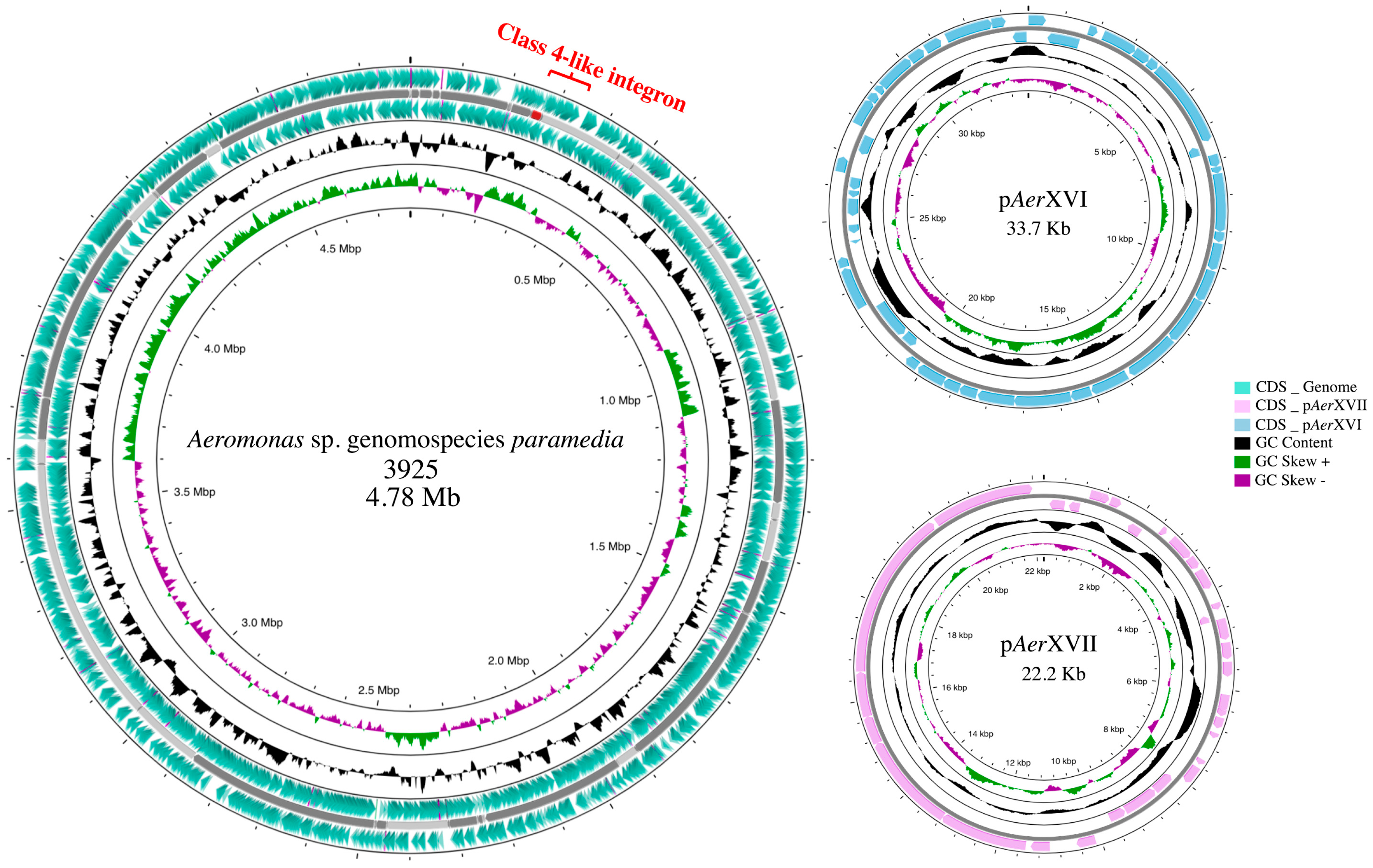
3.2. Genome Analysis
3.3. Plasmid Analysis
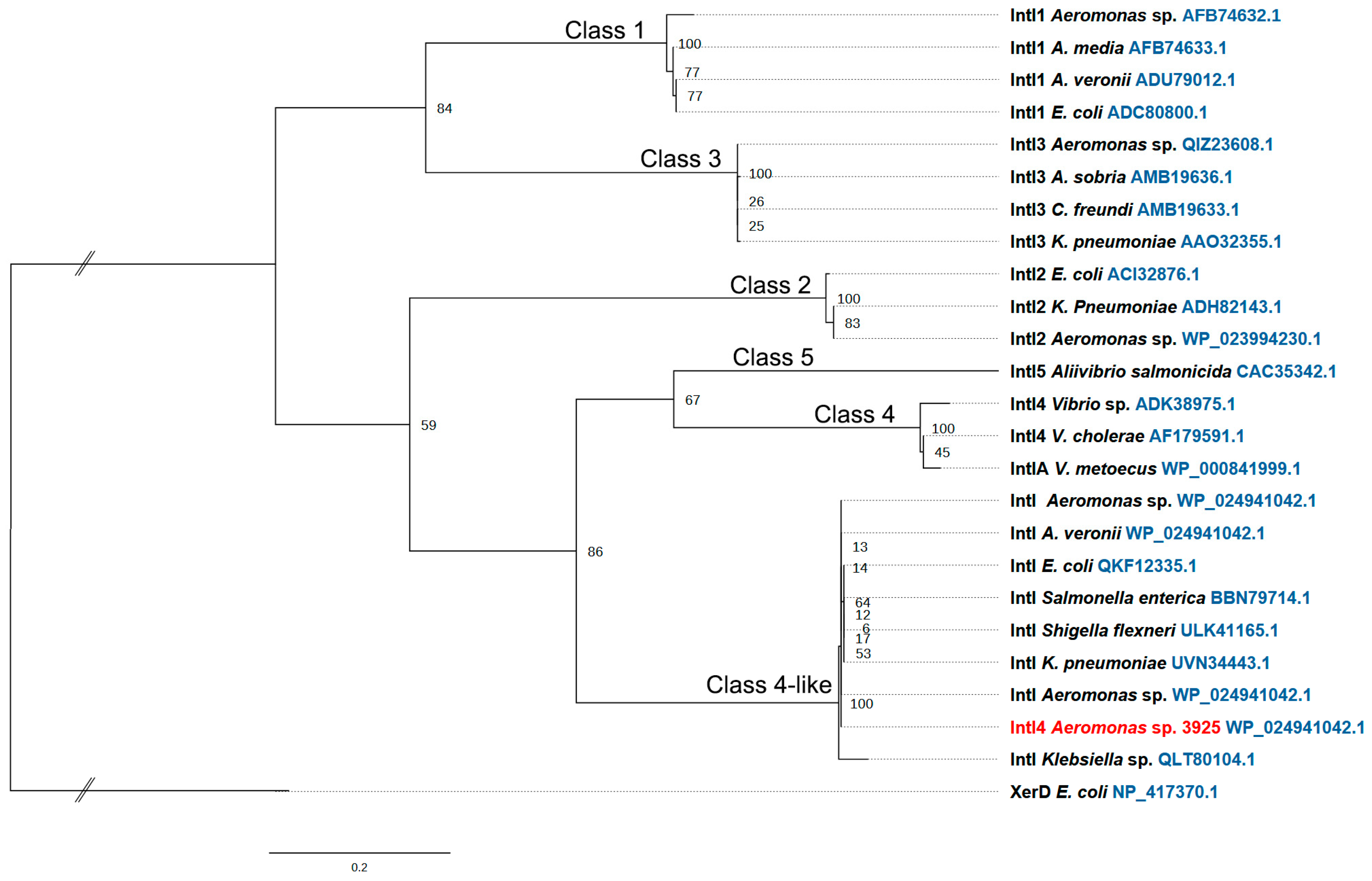
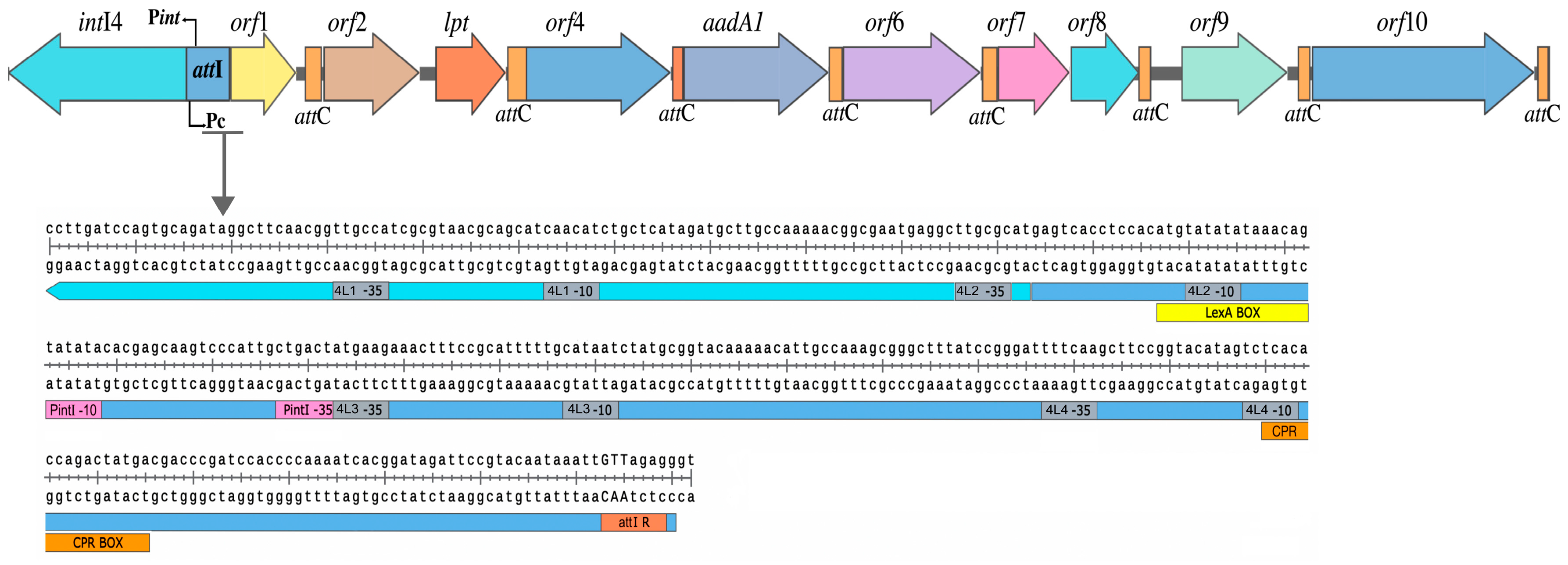
3.4. Integron Characterization
3.5. Cassette Expression
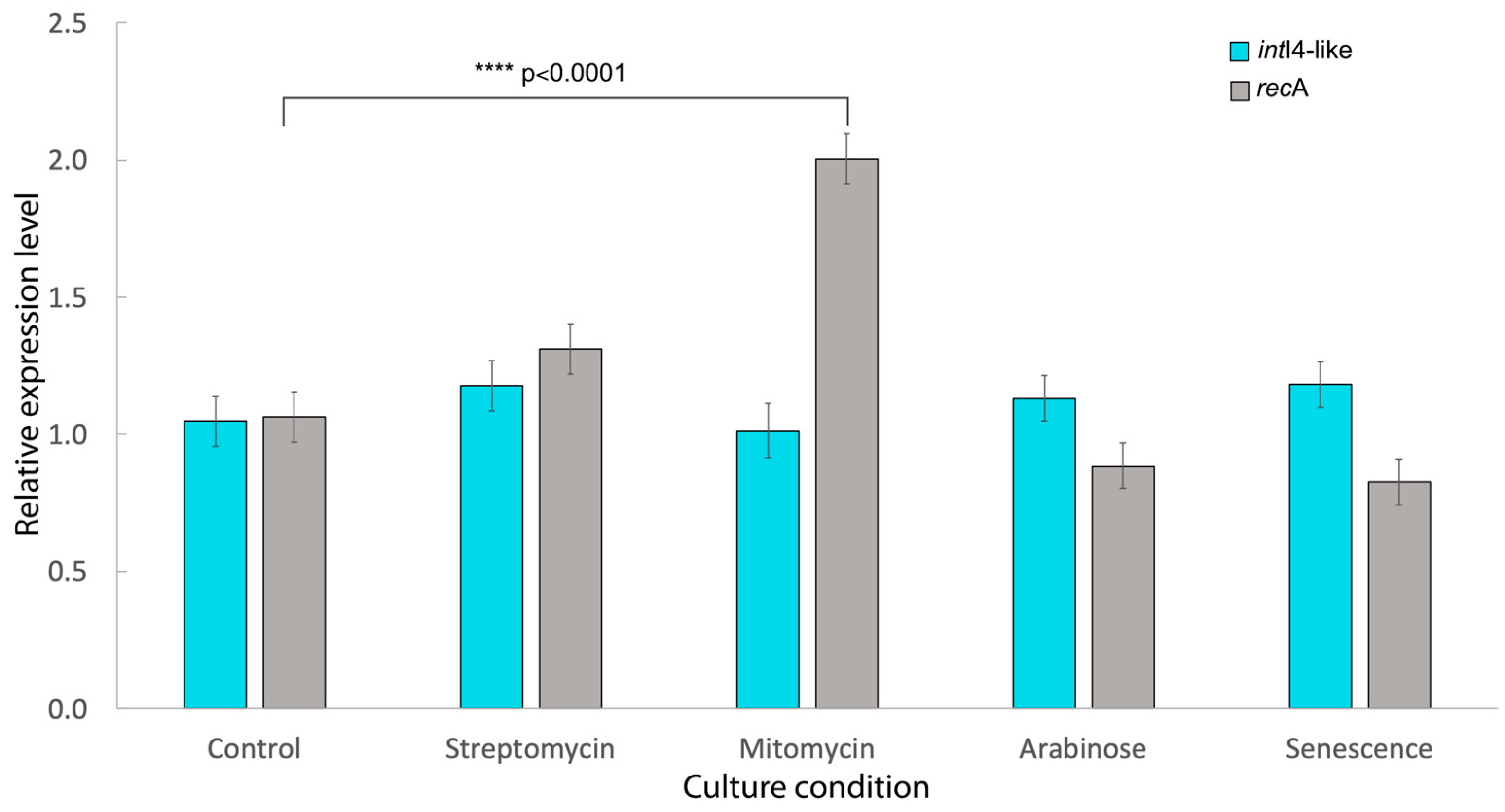
3.6. Expression of Class 4-like Integrase
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fernández-Bravo, A.; Figueras, M.J. An Update on the Genus Aeromonas: Taxonomy, Epidemiology, and Pathogenicity. Microorganisms 2020, 8, 129. [Google Scholar] [CrossRef] [PubMed]
- Figueras, M.J.; Latif-Eugenín, F.; Ballester, F.; Pujol, I.; Tena, D.; Berg, K.; Hossain, M.J.; Beaz-Hidalgo, R.; Liles, M.R. ‘Aeromonas Intestinalis’ and ‘Aeromonas Enterica’ Isolated from Human Faeces, ‘Aeromonas Crassostreae’ from Oyster and ‘Aeromonas Aquatilis’ Isolated from Lake Water Represent Novel Species. New Microbes New Infect. 2017, 15, 74–76. [Google Scholar] [CrossRef] [PubMed]
- Alperi, A.; Figueras, M.J.; Inza, I.; Martínez-Murcia, A.J. Analysis of 16S rRNA Gene Mutations in a Subset of Aeromonas Strains and Their Impact in Species Delineation. Int. Microbiol. 2008, 11, 185–194. [Google Scholar] [PubMed]
- Borrell, N.; Acinas, S.G.; Figueras, M.J.; Martínez-Murcia, A.J. Identification of Aeromonas Clinical Isolates by Restriction Fragment Length Polymorphism of PCR-Amplified 16S rRNA Genes. J. Clin. Microbiol. 1997, 35, 1671–1674. [Google Scholar] [CrossRef] [PubMed]
- Figueras, M.J.; Beaz-Hidalgo, R.; Senderovich, Y.; Laviad, S.; Halpern, M. Re-Identification of Aeromonas Isolates from Chironomid Egg Masses as the Potential Pathogenic Bacteria Aeromonas Aquariorum: Aeromonas Aquarirum in Chironomid Eggs. Environ. Microbiol. Rep. 2011, 3, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Latif-Eugenín, F.; Beaz-Hidalgo, R.; Figueras, M.J. Evaluation of Different Conditions and Culture Media for the Recovery of Aeromonas spp. from Water and Shellfish Samples. J. Appl. Microbiol. 2016, 121, 883–891. [Google Scholar] [CrossRef]
- Navarro, A.; Martínez-Murcia, A. Phylogenetic Analyses of the Genus Aeromonas Based on Housekeeping Gene Sequencing and Its Influence on Systematics. J. Appl. Microbiol. 2018, 125, 622–631. [Google Scholar] [CrossRef]
- Martinez-Murcia, A.J.; Monera, A.; Saavedra, M.J.; Oncina, R.; Lopez-Alvarez, M.; Lara, E.; Figueras, M.J. Multilocus Phylogenetic Analysis of the Genus Aeromonas. Syst. Appl. Microbiol. 2011, 34, 189–199. [Google Scholar] [CrossRef]
- Roger, F.; Marchandin, H.; Jumas-Bilak, E.; Kodjo, A.; the colBVH study group; Lamy, B. Multilocus Genetics to Reconstruct Aeromonad Evolution. BMC Microbiol. 2012, 12, 62. [Google Scholar] [CrossRef]
- de Melo, B.S.T.; Mendes-Marques, C.L.; Campos, T.D.L.; Almeida, A.M.P.D.; Leal, N.C.; Xavier, D.E. High-Resolution Genome-Wide Analysis Is Essential for the Identification of Ambiguous Aeromonas Strains. FEMS Microbiol. Lett. 2019, 366, fnz245. [Google Scholar] [CrossRef]
- Piotrowska, M.; Popowska, M. Insight into the Mobilome of Aeromonas Strains. Front. Microbiol. 2015, 6, 494. [Google Scholar] [CrossRef] [PubMed]
- Richard, E.; Darracq, B.; Loot, C.; Mazel, D. Unbridled Integrons: A Matter of Host Factors. Cells 2022, 11, 925. [Google Scholar] [CrossRef]
- Escudero, J.A.; Loot, C.; Nivina, A.; Mazel, D. The Integron: Adaptation On Demand. Microbiol. Spectr. 2015, 3, MDNA3-0019-2014. [Google Scholar] [CrossRef] [PubMed]
- Abella, J.; Bielen, A.; Huang, L.; Delmont, T.O.; Vujaklija, D.; Duran, R.; Cagnon, C. Integron Diversity in Marine Environments. Environ. Sci. Pollut. Res. 2015, 22, 15360–15369. [Google Scholar] [CrossRef]
- Antelo, V.; Giménez, M.; Azziz, G.; Valdespino-Castillo, P.; Falcón, L.I.; Ruberto, L.A.M.; Mac Cormack, W.P.; Mazel, D.; Batista, S. Metagenomic Strategies Identify Diverse Integron-integrase and Antibiotic Resistance Genes in the Antarctic Environment. Microbiol. 2021, 10, e1219. [Google Scholar] [CrossRef] [PubMed]
- Cambray, G.; Guerout, A.-M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef] [PubMed]
- Gillings, M.R. Class 1 Integrons as Invasive Species. Curr. Opin. Microbiol. 2017, 38, 10–15. [Google Scholar] [CrossRef]
- Biskri, L.; Bouvier, M.; Guérout, A.-M.; Boisnard, S.; Mazel, D. Comparative Study of Class 1 Integron and Vibrio Cholerae Superintegron Integrase Activities. J. Bacteriol. 2005, 187, 1740–1750. [Google Scholar] [CrossRef]
- Dhanapala, P.M.; Kalupahana, R.S.; Kalupahana, A.W.; Wijesekera, D.P.H.; Kottawatta, S.A.; Jayasekera, N.K.; Silva-Fletcher, A.; Jagoda, S.S.S.D.S. Characterization and Antimicrobial Resistance of Environmental and Clinical Aeromonas Species Isolated from Fresh Water Ornamental Fish and Associated Farming Environment in Sri Lanka. Microorganisms 2021, 9, 2106. [Google Scholar] [CrossRef]
- Hossain, S.; De Silva, B.C.J.; Wickramanayake, M.V.K.S.; Dahanayake, P.S.; Wimalasena, S.H.M.P.; Heo, G.-J. Incidence of Antimicrobial Resistance Genes and Class 1 Integron Gene Cassettes in Multidrug-Resistant Motile Aeromonas sp. Isolated from Ornamental Guppy (Poecilia Reticulata). Lett. Appl. Microbiol. 2019, 69, 2–10. [Google Scholar] [CrossRef]
- Igbinosa, I.H.; Okoh, A.I. Antibiotic Susceptibility Profile of Aeromonas Species Isolated from Wastewater Treatment Plant. Sci. World J. 2012, 2012, 764563. [Google Scholar] [CrossRef] [PubMed]
- Otero-Olarra, J.E.; Curiel-Quesada, E.; Baltazar-Cruz, J.; Aguilera-Arreola, M.G.; Pérez-Valdespino, A. Low Cassette Variability in Class 2 and Class 1 Integrons of Aeromonas spp. Isolated from Environmental Samples. Microb. Drug Resist. 2020, 26, 794–801. [Google Scholar] [CrossRef] [PubMed]
- Simo Tchuinte, P.L.; Stalder, T.; Venditti, S.; Ngandjio, A.; Dagot, C.; Ploy, M.-C.; Barraud, O. Characterisation of Class 3 Integrons with Oxacillinase Gene Cassettes in Hospital Sewage and Sludge Samples from France and Luxembourg. Int. J. Antimicrob. Agents 2016, 48, 431–434. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Valdespino, A.; Fernández-Rendón, E.; Curiel-Quesada, E. Detection and Characterization of Class 1 Integrons in Aeromonas spp. Isolated from Human Diarrheic Stool in Mexico: Class 1 Integrons in Clinical Aeromonas. J. Basic Microbiol. 2009, 49, 572–578. [Google Scholar] [CrossRef]
- Green, M.R.; Sambrook, J.; Sambrook, J. Molecular Cloning: A Laboratory Manual, 4th ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2012; ISBN 978-1-936113-41-5. [Google Scholar]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef]
- Lefort, V.; Longueville, J.-E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC Results into a Web-Based, Interactive, and Extensible FASTQ Quality Control Tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Rissman, A.I.; Mau, B.; Biehl, B.S.; Darling, A.E.; Glasner, J.D.; Perna, N.T. Reordering Contigs of Draft Genomes Using the Mauve Aligner. Bioinformatics 2009, 25, 2071–2073. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid Annotations Using Subsystems Technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Antipov, D.; Raiko, M.; Lapidus, A.; Pevzner, P.A. Plasmid Detection and Assembly in Genomic and Metagenomic Data Sets. Genome Res. 2019, 29, 961–968. [Google Scholar] [CrossRef]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An Integrated Platform for Visualization and Analysis of High-Throughput Sequence-Based Experimental Data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef]
- Lemoine, F.; Domelevo Entfellner, J.-B.; Wilkinson, E.; Correia, D.; Dávila Felipe, M.; De Oliveira, T.; Gascuel, O. Renewing Felsenstein’s Phylogenetic Bootstrap in the Era of Big Data. Nature 2018, 556, 452–456. [Google Scholar] [CrossRef]
- Colston, S.M.; Fullmer, M.S.; Beka, L.; Lamy, B.; Gogarten, J.P.; Graf, J. Bioinformatic Genome Comparisons for Taxonomic and Phylogenetic Assignments Using Aeromonas as a Test Case. mBio 2014, 5, e02136-14. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA–DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.-P.; Göker, M. Genome Sequence-Based Species Delimitation with Confidence Intervals and Improved Distance Functions. BMC Bioinform. 2013, 14, 60. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; García-Fernández, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Møller Aarestrup, F.; Hasman, H. In Silico Detection and Typing of Plasmids Using PlasmidFinder and Plasmid Multilocus Sequence Typing. Antimicrob Agents Chemother 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Moran, R.A.; Hall, R.M. pBuzz: A Cryptic Rolling-Circle Plasmid from a Commensal Escherichia Coli Has Two Inversely Oriented oriTs and Is Mobilised by a B/O Plasmid. Plasmid 2019, 101, 10–19. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Wei, Y.; Shen, Y.; Li, X.; Zhou, H.; Tai, C.; Deng, Z.; Ou, H.-Y. TADB 2.0: An Updated Database of Bacterial Type II Toxin–Antitoxin Loci. Nucleic Acids Res. 2018, 46, D749–D753. [Google Scholar] [CrossRef] [PubMed]
- Néron, B.; Littner, E.; Haudiquet, M.; Perrin, A.; Cury, J.; Rocha, E. IntegronFinder 2.0: Identification and Analysis of Integrons across Bacteria, with a Focus on Antibiotic Resistance in Klebsiella. Microorganisms 2022, 10, 700. [Google Scholar] [CrossRef]
- Pereira, M.B.; Wallroth, M.; Kristiansson, E.; Axelson-Fisk, M. HattCI: Fast and Accurate attC Site Identification Using Hidden Markov Models. J. Comput. Biol. 2016, 23, 891–902. [Google Scholar] [CrossRef]
- Solovyev, V.; Salamov, A. Automatic Annotation of Microbial Genomes and Metagenomic Sequences Automatic Annotation of Microbial Genomes and Metagenomic Sequences; Nova Biomedical: New York, NY, USA, 2011; ISBN 978-1-61728-731-2. [Google Scholar]
- Klucar, L.; Stano, M.; Hajduk, M. phiSITE: Database of Gene Regulation in Bacteriophages. Nucleic Acids Res. 2010, 38, D366–D370. [Google Scholar] [CrossRef]
- Grant, J.R.; Enns, E.; Marinier, E.; Mandal, A.; Herman, E.K.; Chen, C.; Graham, M.; Van Domselaar, G.; Stothard, P. Proksee: In-Depth Characterization and Visualization of Bacterial Genomes. Nucleic Acids Res. 2023, 51, W484–W492. [Google Scholar] [CrossRef]
- Pérez-Valdespino, A.; Lazarini-Martínez, A.; Rivera-González, A.X.; García-Hernández, N.; Curiel-Quesada, E. Dynamics of a Class 1 Integron Located on Plasmid or Chromosome in Two Aeromonas spp. Strains. Front. Microbiol. 2016, 7, 1556. [Google Scholar] [CrossRef]
- Shearer, J.E.S.; Summers, A.O. Intracellular Steady-State Concentration of Integron Recombination Products Varies with Integrase Level and Growth Phase. J. Mol. Biol. 2009, 386, 316–331. [Google Scholar] [CrossRef]
- Weinstein, M.P.; Patel, J.B. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria That Grow Aerobically: M07-A11, 11th ed.; Documents/Clinical and Laboratory Standards Institute; Committee for Clinical Laboratory Standards: Wayne, PA, USA, 2018; ISBN 978-1-56238-836-2. [Google Scholar]
- Kovach, M.E.; Elzer, P.H.; Steven Hill, D.; Robertson, G.T.; Farris, M.A.; Roop, R.M.; Peterson, K.M. Four New Derivatives of the Broad-Host-Range Cloning Vector pBBR1MCS, Carrying Different Antibiotic-Resistance Cassettes. Gene 1995, 166, 175–176. [Google Scholar] [CrossRef]
- Yuan, J.S.; Wang, D.; Stewart, C.N. Statistical Methods for Efficiency Adjusted Real-Time PCR Quantification. Biotechnol. J. 2008, 3, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Cambray, G.; Sanchez-Alberola, N.; Campoy, S.; Guerin, É.; Da Re, S.; González-Zorn, B.; Ploy, M.-C.; Barbé, J.; Mazel, D.; Erill, I. Prevalence of SOS-Mediated Control of Integron Integrase Expression as an Adaptive Trait of Chromosomal and Mobile Integrons. Mobile DNA 2011, 2, 6. [Google Scholar] [CrossRef] [PubMed]
- Kaushik, V.; Tiwari, M.; Tiwari, V. Interaction of RecA Mediated SOS Response with Bacterial Persistence, Biofilm Formation, and Host Response. Int. J. Biol. Macromol. 2022, 217, 931–943. [Google Scholar] [CrossRef]
- Talagrand-Reboul, E.; Roger, F.; Kimper, J.-L.; Colston, S.M.; Graf, J.; Latif-Eugenín, F.; Figueras, M.J.; Petit, F.; Marchandin, H.; Jumas-Bilak, E.; et al. Delineation of Taxonomic Species within Complex of Species: Aeromonas Media and Related Species as a Test Case. Front. Microbiol. 2017, 8, 621. [Google Scholar] [CrossRef]
- Pérez-García, D.; Larios-Serrato, V.; Rojas-Rios, R.; Otero-Olarra, J.E.; Mendoza-Sanchez, I.; Curiel-Quesada, E.; Pérez-Valdespino, A. Frequency and Diversity of Small Plasmids in Mesophilic Aeromonas Isolates from Fish, Water and Sediment. Plasmid 2021, 118, 102607. [Google Scholar] [CrossRef]
- Jin, Y.; Zhou, J.; Zhou, J.; Hu, M.; Zhang, Q.; Kong, N.; Ren, H.; Liang, L.; Yue, J. Genome-Based Classification of Burkholderia Cepacia Complex Provides New Insight into Its Taxonomic Status. Biol. Direct. 2020, 15, 6. [Google Scholar] [CrossRef] [PubMed]
- Paul, B.; Dixit, G.; Murali, T.S.; Satyamoorthy, K. Genome-Based Taxonomic Classification. Genome 2019, 62, 45–52. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; Da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed Minimal Standards for the Use of Genome Data for the Taxonomy of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Sandoval-Quintana, E.; Lauga, B.; Cagnon, C. Environmental Integrons: The Dark Side of the Integron World. Trends Microbiol. 2023, 31, 432–434. [Google Scholar] [CrossRef]
- Deng, Y.; Wu, Y.; Jiang, L.; Tan, A.; Zhang, R.; Luo, L. Multi-Drug Resistance Mediated by Class 1 Integrons in Aeromonas Isolated from Farmed Freshwater Animals. Front. Microbiol. 2016, 7, 935. [Google Scholar] [CrossRef]
- El-Baz, A.M.; Yahya, G.; Mansour, B.; El-Sokkary, M.M.A.; Alshaman, R.; Alattar, A.; El-Ganiny, A.M. The Link between Occurrence of Class I Integron and Acquired Aminoglycoside Resistance in Clinical MRSA Isolates. Antibiotics 2021, 10, 488. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The Conserved Domain Database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Dunin-Horkawicz, S.; Feder, M.; Bujnicki, J.M. Phylogenomic Analysis of the GIY-YIG Nuclease Superfamily. BMC Genom. 2006, 7, 98. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, U.; Roske, Y. Cold-Shock Domains—Abundance, Structure, Properties, and Nucleic-Acid Binding. Cancers 2021, 13, 190. [Google Scholar] [CrossRef]
- Horn, G.; Hofweber, R.; Kremer, W.; Kalbitzer, H.R. Structure and Function of Bacterial Cold Shock Proteins. Cell. Mol. Life Sci. 2007, 64, 1457–1470. [Google Scholar] [CrossRef]
- Collis, C.M.; Hall, R.M. Expression of Antibiotic Resistance Genes in the Integrated Cassettes of Integrons. Antimicrob Agents Chemother 1995, 39, 155–162. [Google Scholar] [CrossRef]
- Lévesque, C.; Brassard, S.; Lapointe, J.; Roy, P.H. Diversity and Relative Strength of Tandem Promoters for the Antibiotic-Resistance Genes of Several Integron. Gene 1994, 142, 49–54. [Google Scholar] [CrossRef]
- Fonseca, É.L.; Vicente, A.C. Integron Functionality and Genome Innovation: An Update on the Subtle and Smart Strategy of Integrase and Gene Cassette Expression Regulation. Microorganisms 2022, 10, 224. [Google Scholar] [CrossRef]
- Da Fonseca, E.L.; Freitas, F.D.S.; Vicente, A.C.P. Pc Promoter from Class 2 Integrons and the Cassette Transcription Pattern It Evokes. J. Antimicrob. Chemother. 2011, 66, 797–801. [Google Scholar] [CrossRef]
- Jové, T.; Da Re, S.; Tabesse, A.; Gassama-Sow, A.; Ploy, M.-C. Gene Expression in Class 2 Integrons Is SOS-Independent and Involves Two Pc Promoters. Front. Microbiol. 2017, 8, 1499. [Google Scholar] [CrossRef]
- Krin, E.; Cambray, G.; Mazel, D. The Superintegron Integrase and the Cassette Promoters Are Co-Regulated in Vibrio Cholerae. PLoS ONE 2014, 9, e91194. [Google Scholar] [CrossRef] [PubMed]
- Frumerie, C.; Ducos-Galand, M.; Gopaul, D.N.; Mazel, D. The Relaxed Requirements of the Integron Cleavage Site Allow Predictable Changes in Integron Target Specificity. Nucleic Acids Res. 2010, 38, 559–569. [Google Scholar] [CrossRef] [PubMed]
- Guerin, É.; Cambray, G.; Sanchez-Alberola, N.; Campoy, S.; Erill, I.; Da Re, S.; Gonzalez-Zorn, B.; Barbé, J.; Ploy, M.-C.; Mazel, D. The SOS Response Controls Integron Recombination. Science 2009, 324, 1034. [Google Scholar] [CrossRef] [PubMed]
- Baharoglu, Z.; Krin, E.; Mazel, D. Connecting Environment and Genome Plasticity in the Characterization of Transformation-Induced SOS Regulation and Carbon Catabolite Control of the Vibrio Cholerae Integron Integrase. J. Bacteriol. 2012, 194, 1659–1667. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baltazar-Cruz, J.; Rojas-Rios, R.; Larios-Serrato, V.; Mendoza-Sanchez, I.; Curiel-Quesada, E.; Pérez-Valdespino, A. A Class 4-like Chromosomal Integron Found in Aeromonas sp. Genomospecies paramedia Isolated from Human Feces. Microorganisms 2023, 11, 2548. https://doi.org/10.3390/microorganisms11102548
Baltazar-Cruz J, Rojas-Rios R, Larios-Serrato V, Mendoza-Sanchez I, Curiel-Quesada E, Pérez-Valdespino A. A Class 4-like Chromosomal Integron Found in Aeromonas sp. Genomospecies paramedia Isolated from Human Feces. Microorganisms. 2023; 11(10):2548. https://doi.org/10.3390/microorganisms11102548
Chicago/Turabian StyleBaltazar-Cruz, Jesús, Rogelio Rojas-Rios, Violeta Larios-Serrato, Itza Mendoza-Sanchez, Everardo Curiel-Quesada, and Abigail Pérez-Valdespino. 2023. "A Class 4-like Chromosomal Integron Found in Aeromonas sp. Genomospecies paramedia Isolated from Human Feces" Microorganisms 11, no. 10: 2548. https://doi.org/10.3390/microorganisms11102548
APA StyleBaltazar-Cruz, J., Rojas-Rios, R., Larios-Serrato, V., Mendoza-Sanchez, I., Curiel-Quesada, E., & Pérez-Valdespino, A. (2023). A Class 4-like Chromosomal Integron Found in Aeromonas sp. Genomospecies paramedia Isolated from Human Feces. Microorganisms, 11(10), 2548. https://doi.org/10.3390/microorganisms11102548






