The Signal Transduction Protein PII Controls the Levels of the Cyanobacterial Protein PipX
Abstract
1. Introduction
2. Materials and Methods
2.1. Cyanobacterial Growth Conditions
2.2. Plasmid and Strains Construction
2.3. Protein Extraction and Immunodetection Assays
2.4. Computational Methods
3. Results and Discussion
3.1. PipX Toxicity Is Altered by Point Mutations Targeting Residues at Each of Its Two Domains
3.2. Most but Not All PipX Point Mutations Alter Toxicity and NtcA Coactivation in a Similar Manner
3.3. Not All Loss-of-Function Mutations for Toxicity Decrease PipX Levels in S. elongatus
3.4. Most Mutations Impairing Binding to PII and NtcA Reduce PipX Levels in S. elongatus
3.5. PipX Levels Are Down Regulated by Switching from Dark to Light Conditions
3.6. Regulatory Complexities of the PipX-PII Interaction Network, the Dual Role of PII
3.7. PipX Complexes and Toxicity
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blank, C.E.; Sanchez-Baracaldo, P. Timing of morphological and ecological innovations in the cyanobacteria—A key to understanding the rise in atmospheric oxygen. Geobiology 2010, 8, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-W.; Noh, J.-H.; Choi, D.-H.; Yun, M.; Bhavya, P.S.; Kang, J.-J.; Lee, J.-H.; Kim, K.-W.; Jang, H.-K.; Lee, S.-H. Picocyanobacterial Contribution to the Total Primary Production in the Northwestern Pacific Ocean. Water 2021, 13, 1610. [Google Scholar] [CrossRef]
- Zhang, Y.; Burkhardt, D.H.; Rouskin, S.; Li, G.W.; Weissman, J.S.; Gross, C.A. A Stress Response that Monitors and Regulates mRNA Structure Is Central to Cold Shock Adaptation. Mol. Cell 2018, 70, 274–286.e277. [Google Scholar] [CrossRef]
- Forchhammer, K.; Selim, K.A. Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol. Rev. 2020, 44, 33–53. [Google Scholar] [CrossRef]
- Muro-Pastor, M.I.; Reyes, J.C.; Florencio, F.J. Ammonium assimilation in cyanobacteria. Photosynth. Res. 2005, 83, 135–150. [Google Scholar] [CrossRef]
- Senior, P.J. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: Studies with the continuous-culture technique. J. Bacteriol. 1975, 123, 407–418. [Google Scholar] [CrossRef] [PubMed]
- Huergo, L.F.; Dixon, R. The Emergence of 2-Oxoglutarate as a Master Regulator Metabolite. Microbiol. Mol. Biol. Rev. 2015, 79, 419–435. [Google Scholar] [CrossRef]
- Forchhammer, K.; Selim, K.A.; Huergo, L.F. New views on PII signaling: From nitrogen sensing to global metabolic control. Trends Microbiol. 2022, 30, 722–735. [Google Scholar] [CrossRef]
- Kamberov, E.S.; Atkinson, M.R.; Ninfa, A.J. The Escherichia coli PII signal transduction protein is activated upon binding 2-ketoglutarate and ATP. J. Biol. Chem. 1995, 270, 17797–17807. [Google Scholar] [CrossRef]
- Forchhammer, K.; Hedler, A. Phosphoprotein PII from cyanobacteria—Analysis of functional conservation with the PII signal-transduction protein from Escherichia coli. Eur. J. Biochem. 1997, 244, 869–875. [Google Scholar] [CrossRef]
- Burillo, S.; Luque, I.; Fuentes, I.; Contreras, A. Interactions between the nitrogen signal transduction protein PII and N-acetyl glutamate kinase in organisms that perform oxygenic photosynthesis. J. Bacteriol. 2004, 186, 3346–3354. [Google Scholar] [CrossRef]
- Espinosa, J.; Forchhammer, K.; Burillo, S.; Contreras, A. Interaction network in cyanobacterial nitrogen regulation: PipX, a protein that interacts in a 2-oxoglutarate dependent manner with PII and NtcA. Mol. Microbiol. 2006, 61, 457–469. [Google Scholar] [CrossRef] [PubMed]
- Labella, J.I.; Obrebska, A.; Espinosa, J.; Salinas, P.; Forcada-Nadal, A.; Tremino, L.; Rubio, V.; Contreras, A. Expanding the Cyanobacterial Nitrogen Regulatory Network: The GntR-Like Regulator PlmA Interacts with the PII-PipX Complex. Front. Microbiol. 2016, 7, 1677. [Google Scholar] [CrossRef]
- Labella, J.I.; Llop, A.; Contreras, A. The default cyanobacterial linked genome: An interactive platform based on cyanobacterial linkage networks to assist functional genomics. FEBS Lett. 2020, 594, 1661–1674. [Google Scholar] [CrossRef]
- Labella, J.I.; Cantos, R.; Salinas, P.; Espinosa, J.; Contreras, A. Distinctive Features of PipX, a Unique Signaling Protein of Cyanobacteria. Life 2020, 10, 79. [Google Scholar] [CrossRef]
- Forcada-Nadal, A.; Llacer, J.L.; Contreras, A.; Marco-Marin, C.; Rubio, V. The P(II)-NAGK-PipX-NtcA Regulatory Axis of Cyanobacteria: A Tale of Changing Partners, Allosteric Effectors and Non-covalent Interactions. Front. Mol. Biosci. 2018, 5, 91. [Google Scholar] [CrossRef] [PubMed]
- Llacer, J.L.; Espinosa, J.; Castells, M.A.; Contreras, A.; Forchhammer, K.; Rubio, V. Structural basis for the regulation of NtcA-dependent transcription by proteins PipX and PII. Proc. Natl. Acad. Sci. USA 2010, 107, 15397–15402. [Google Scholar] [CrossRef] [PubMed]
- Kyrpides, N.C.; Woese, C.R.; Ouzounis, C.A. KOW: A novel motif linking a bacterial transcription factor with ribosomal proteins. Trends Biochem. Sci. 1996, 21, 425–426. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.X.; Jiang, Y.L.; Xu, B.Y.; Chen, Y.; Zhang, C.C.; Zhou, C.Z. Crystal structure of the cyanobacterial signal transduction protein PII in complex with PipX. J. Mol. Biol. 2010, 402, 552–559. [Google Scholar] [CrossRef]
- Laichoubi, K.B.; Beez, S.; Espinosa, J.; Forchhammer, K.; Contreras, A. The nitrogen interaction network in Synechococcus WH5701, a cyanobacterium with two PipX and two P(II)-like proteins. Microbiology 2011, 157 Pt 4, 1220–1228. [Google Scholar] [CrossRef]
- Espinosa, J.; Castells, M.A.; Laichoubi, K.B.; Contreras, A. Mutations at pipX suppress lethality of PII-deficient mutants of Synechococcus elongatus PCC 7942. J. Bacteriol. 2009, 191, 4863–4869. [Google Scholar] [CrossRef]
- Espinosa, J.; Castells, M.A.; Laichoubi, K.B.; Forchhammer, K.; Contreras, A. Effects of spontaneous mutations in PipX functions and regulatory complexes on the cyanobacterium Synechococcus elongatus strain PCC 7942. Microbiology 2010, 156 Pt 5, 1517–1526. [Google Scholar] [CrossRef][Green Version]
- Laichoubi, K.B.; Espinosa, J.; Castells, M.A.; Contreras, A. Mutational analysis of the cyanobacterial nitrogen regulator PipX. PLoS ONE 2012, 7, e35845. [Google Scholar] [CrossRef]
- Chang, Y.; Takatani, N.; Aichi, M.; Maeda, S.; Omata, T. Evaluation of the effects of P(II) deficiency and the toxicity of PipX on growth characteristics of the P(II)-Less mutant of the cyanobacterium Synechococcus elongatus. Plant Cell Physiol. 2013, 54, 1504–1514. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Takatani, N.; Sonoike, K.; Jimbo, H.; Nishiyama, Y.; Omata, T. Dissection of the Mechanisms of Growth Inhibition Resulting from Loss of the PII Protein in the Cyanobacterium Synechococcus elongatus PCC 7942. Plant Cell Physiol. 2021, 62, 721–731. [Google Scholar] [CrossRef] [PubMed]
- Herrero, A.; Muro-Pastor, A.M.; Flores, E. Nitrogen control in cyanobacteria. J. Bacteriol. 2001, 183, 411–425. [Google Scholar] [CrossRef]
- Esteves-Ferreira, A.A.; Inaba, M.; Fort, A.; Araujo, W.L.; Sulpice, R. Nitrogen metabolism in cyanobacteria: Metabolic and molecular control, growth consequences and biotechnological applications. Crit. Rev. Microbiol. 2018, 44, 541–560. [Google Scholar] [CrossRef] [PubMed]
- Espinosa, J.; Rodriguez-Mateos, F.; Salinas, P.; Lanza, V.F.; Dixon, R.; de la Cruz, F.; Contreras, A. PipX, the coactivator of NtcA, is a global regulator in cyanobacteria. Proc. Natl. Acad. Sci. USA 2014, 111, E2423–E2430. [Google Scholar] [CrossRef]
- Giner-Lamia, J.; Robles-Rengel, R.; Hernandez-Prieto, M.A.; Muro-Pastor, M.I.; Florencio, F.J.; Futschik, M.E. Identification of the direct regulon of NtcA during early acclimation to nitrogen starvation in the cyanobacterium Synechocystis sp. PCC 6803. Nucleic Acids Res. 2017, 45, 11800–11820. [Google Scholar] [CrossRef]
- Espinosa, J.; Forchhammer, K.; Contreras, A. Role of the Synechococcus PCC 7942 nitrogen regulator protein PipX in NtcA-controlled processes. Microbiology 2007, 153 Pt 3, 711–718. [Google Scholar] [CrossRef] [PubMed]
- Zeth, K.; Fokina, O.; Forchhammer, K. Structural basis and target-specific modulation of ADP sensing by the Synechococcus elongatus PII signaling protein. J. Biol. Chem. 2014, 289, 8960–8972. [Google Scholar] [CrossRef]
- Espinosa, J.; Labella, J.I.; Cantos, R.; Contreras, A. Energy drives the dynamic localization of cyanobacterial nitrogen regulators during diurnal cycles. Environ. Microbiol. 2018, 20, 1240–1252. [Google Scholar] [CrossRef] [PubMed]
- Labella, J.I.; Cantos, R.; Espinosa, J.; Forcada-Nadal, A.; Rubio, V.; Contreras, A. PipY, a Member of the Conserved COG0325 Family of PLP-Binding Proteins, Expands the Cyanobacterial Nitrogen Regulatory Network. Front. Microbiol. 2017, 8, 1244. [Google Scholar] [CrossRef]
- Ito, T. Role of the conserved pyridoxal 5’-phosphate-binding protein YggS/PLPBP in vitamin B6 and amino acid homeostasis. Biosci. Biotechnol. Biochem. 2022, 86, 1183–1191. [Google Scholar] [CrossRef] [PubMed]
- Tremino, L.; Llop, A.; Rubio, V.; Contreras, A. The Conserved Family of the Pyridoxal Phosphate-Binding Protein (PLPBP) and Its Cyanobacterial Paradigm PipY. Life 2022, 12, 1622. [Google Scholar] [CrossRef]
- Cantos, R.; Labella, J.I.; Espinosa, J.; Contreras, A. The nitrogen regulator PipX acts in cis to prevent operon polarity. Environ. Microbiol. Rep. 2019, 11, 495–507. [Google Scholar] [CrossRef] [PubMed]
- Llop, A.; Labella, J.I.; Borisova, M.; Forchhammer, K.; Selim, K.A.; Contreras, A. Pleiotropic effects of PipX, PipY, or RelQ overexpression on growth, cell size, photosynthesis, and polyphosphate accumulation in the cyanobacterium Synechococcus elongatus PCC7942. Front. Microbiol. 2023, 14, 1141775. [Google Scholar] [CrossRef]
- Riediger, M.; Spat, P.; Bilger, R.; Voigt, K.; Macek, B.; Hess, W.R. Analysis of a photosynthetic cyanobacterium rich in internal membrane systems via gradient profiling by sequencing (Grad-seq). Plant Cell 2021, 33, 248–269. [Google Scholar] [CrossRef]
- Jerez, C.; Salinas, P.; Llop, A.; Cantos, R.; Espinosa, J.; Labella, J.I.; Contreras, A. Regulatory Connections Between the Cyanobacterial Factor PipX and the Ribosome Assembly GTPase EngA. Front. Microbiol. 2021, 12, 781760. [Google Scholar] [CrossRef]
- Llop, A.; Bibak, S.; Cantos, R.; Salinas, P.; Contreras, A. The ribosome assembly GTPase EngA is involved in redox signaling in cyanobacteria. Front. Microbiol. 2023, 14, 1242616. [Google Scholar] [CrossRef]
- Rippka, R.; Deruelles, J.; Waterbury, J.B.; Hermand, M.; Stanier, R.Y. Generic Assignments, Strain Histories and Properties of Pure Cultures of Cyanobacteria. J. Gen. Microbiol. 1979, 111, 1–61. [Google Scholar] [CrossRef]
- Golden, S.S.; Sherman, L.A. Optimal conditions for genetic transformation of the cyanobacterium Anacystis nidulans R2. J. Bacteriol. 1984, 158, 36–42. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.R.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor: New York, NY, USA, 1989. [Google Scholar]
- Glover, D.M. (Ed.) DNA Cloning: A Practical Approach; IRL Press: Washington, DC, USA, 1985; p. 109. [Google Scholar]
- Wallace, A.C.; Laskowski, R.A.; Thornton, J.M. LIGPLOT: A program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995, 8, 127–134. [Google Scholar] [CrossRef] [PubMed]
- RStudio: Integrated Development for R. RStudio; RStudio, PBC: Boston, MA, USA, 2020; Available online: http://www.rstudio.com/ (accessed on 23 January 2023).
- Forcada-Nadal, A.; Palomino-Schatzlein, M.; Neira, J.L.; Pineda-Lucena, A.; Rubio, V. The PipX Protein, When Not Bound to Its Targets, Has Its Signaling C-Terminal Helix in a Flexed Conformation. Biochemistry 2017, 56, 3211–3224. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, A.C.; Benevento, M.; Lehmann, R.; van Breukelen, B.; Post, H.; Giansanti, P.; Maarten Altelaar, A.F.; Axmann, I.M.; Heck, A.J. Daily rhythms in the cyanobacterium synechococcus elongatus probed by high-resolution mass spectrometry-based proteomics reveals a small defined set of cyclic proteins. Mol. Cell Proteom. 2014, 13, 2042–2055. [Google Scholar] [CrossRef]
- Rust, M.J.; Golden, S.S.; O’Shea, E.K. Light-driven changes in energy metabolism directly entrain the cyanobacterial circadian oscillator. Science 2011, 331, 220–223. [Google Scholar] [CrossRef]
- Fadi Aldehni, M.; Sauer, J.; Spielhaupter, C.; Schmid, R.; Forchhammer, K. Signal transduction protein P(II) is required for NtcA-regulated gene expression during nitrogen deprivation in the cyanobacterium Synechococcus elongatus strain PCC 7942. J. Bacteriol. 2003, 185, 2582–2591. [Google Scholar] [CrossRef]
- Collier, J.L.; Grossman, A.R. A small polypeptide triggers complete degradation of light-harvesting phycobiliproteins in nutrient-deprived cyanobacteria. EMBO J. 1994, 13, 1039–1047. [Google Scholar] [CrossRef]
- Salinas, P.; Ruiz, D.; Cantos, R.; Lopez-Redondo, M.L.; Marina, A.; Contreras, A. The regulatory factor SipA provides a link between NblS and NblR signal transduction pathways in the cyanobacterium Synechococcus sp. PCC 7942. Mol. Microbiol. 2007, 66, 1607–1619. [Google Scholar] [CrossRef]
- Groben, R.; Kaloudas, D.; Raines, C.A.; Offmann, B.; Maberly, S.C.; Gontero, B. Comparative sequence analysis of CP12, a small protein involved in the formation of a Calvin cycle complex in photosynthetic organisms. Photosynth. Res. 2010, 103, 183–194. [Google Scholar] [CrossRef]
- Storz, G.; Wolf, Y.I.; Ramamurthi, K.S. Small proteins can no longer be ignored. Annu. Rev. Biochem. 2014, 83, 753–777. [Google Scholar] [CrossRef]
- Klahn, S.; Schaal, C.; Georg, J.; Baumgartner, D.; Knippen, G.; Hagemann, M.; Muro-Pastor, A.M.; Hess, W.R. The sRNA NsiR4 is involved in nitrogen assimilation control in cyanobacteria by targeting glutamine synthetase inactivating factor IF7. Proc. Natl. Acad. Sci. USA 2015, 112, E6243–E6252. [Google Scholar] [CrossRef]
- Klahn, S.; Bolay, P.; Wright, P.R.; Atilho, R.M.; Brewer, K.I.; Hagemann, M.; Breaker, R.R.; Hess, W.R. A glutamine riboswitch is a key element for the regulation of glutamine synthetase in cyanobacteria. Nucleic Acids Res. 2018, 46, 10082–10094. [Google Scholar] [CrossRef]
- Baumgartner, D.; Kopf, M.; Klahn, S.; Steglich, C.; Hess, W.R. Small proteins in cyanobacteria provide a paradigm for the functional analysis of the bacterial micro-proteome. BMC Microbiol. 2016, 16, 285. [Google Scholar] [CrossRef]
- Brandenburg, F.; Klahn, S. Small but Smart: On the Diverse Role of Small Proteins in the Regulation of Cyanobacterial Metabolism. Life 2020, 10, 322. [Google Scholar] [CrossRef] [PubMed]
- Krauspe, V.; Fahrner, M.; Spat, P.; Steglich, C.; Frankenberg-Dinkel, N.; Macek, B.; Schilling, O.; Hess, W.R. Discovery of a small protein factor involved in the coordinated degradation of phycobilisomes in cyanobacteria. Proc. Natl. Acad. Sci. USA 2021, 118, e2012277118. [Google Scholar] [CrossRef] [PubMed]
- Merida, A.; Candau, P.; Florencio, F.J. Regulation of glutamine synthetase activity in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803 by the nitrogen source: Effect of ammonium. J. Bacteriol. 1991, 173, 4095–4100. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Florencio, F.J. A novel mechanism of glutamine synthetase inactivation by ammonium in the cyanobacterium Synechocystis sp. PCC 6803. Involvement of an inactivating protein. FEBS Lett. 1995, 367, 45–48. [Google Scholar] [CrossRef] [PubMed]
- Reyes, J.C.; Muro-Pastor, M.I.; Florencio, F.J. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J. Bacteriol. 1997, 179, 2678–2689. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Dominguez, M.; Reyes, J.C.; Florencio, F.J. Glutamine synthetase inactivation by protein-protein interaction. Proc. Natl. Acad. Sci. USA 1999, 96, 7161–7166. [Google Scholar] [CrossRef]
- Watzer, B.; Spat, P.; Neumann, N.; Koch, M.; Sobotka, R.; Macek, B.; Hennrich, O.; Forchhammer, K. The Signal Transduction Protein P(II) Controls Ammonium, Nitrate and Urea Uptake in Cyanobacteria. Front. Microbiol. 2019, 10, 1428. [Google Scholar] [CrossRef] [PubMed]
- Bolay, P.; Rozbeh, R.; Muro-Pastor, M.I.; Timm, S.; Hagemann, M.; Florencio, F.J.; Forchhammer, K.; Klahn, S. The Novel P(II)-Interacting Protein PirA Controls Flux into the Cyanobacterial Ornithine-Ammonia Cycle. mBio 2021, 12, e00229-21. [Google Scholar] [CrossRef] [PubMed]
- Orthwein, T.; Scholl, J.; Spat, P.; Lucius, S.; Koch, M.; Macek, B.; Hagemann, M.; Forchhammer, K. The novel P(II)-interactor PirC identifies phosphoglycerate mutase as key control point of carbon storage metabolism in cyanobacteria. Proc. Natl. Acad. Sci. USA 2021, 118, e2019988118. [Google Scholar] [CrossRef] [PubMed]
- Bharat, A.; Brown, E.D. Phenotypic investigations of the depletion of EngA in Escherichia coli are consistent with a role in ribosome biogenesis. FEMS Microbiol. Lett. 2014, 353, 26–32. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Majumdar, S.; Acharya, A.; Tomar, S.K.; Prakash, B. Disrupting domain-domain interactions is indispensable for EngA-ribosome interactions. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Gaal, T.; Bartlett, M.S.; Ross, W.; Turnbough, C.L., Jr.; Gourse, R.L. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 1997, 278, 2092–2097. [Google Scholar] [CrossRef]

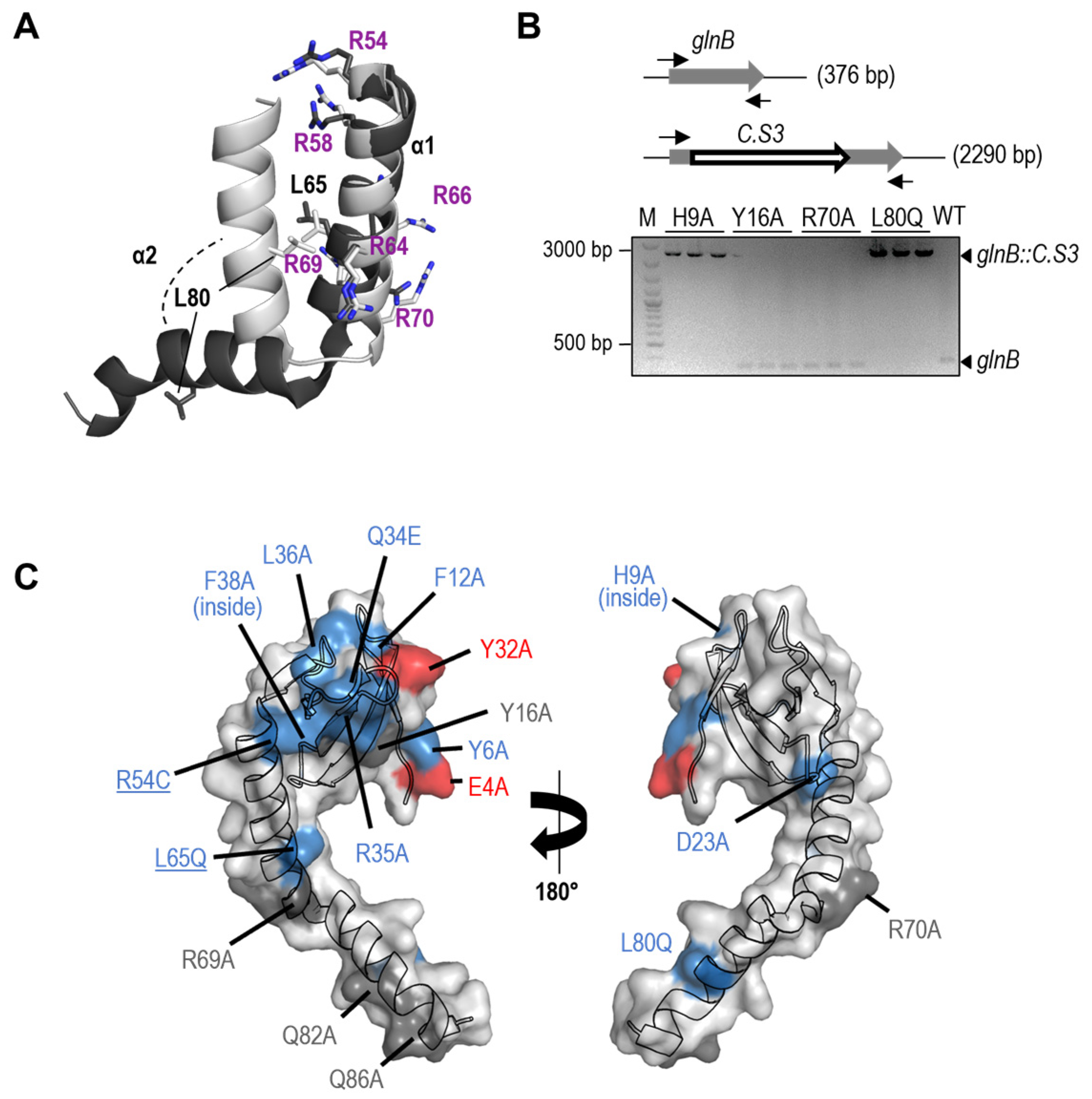
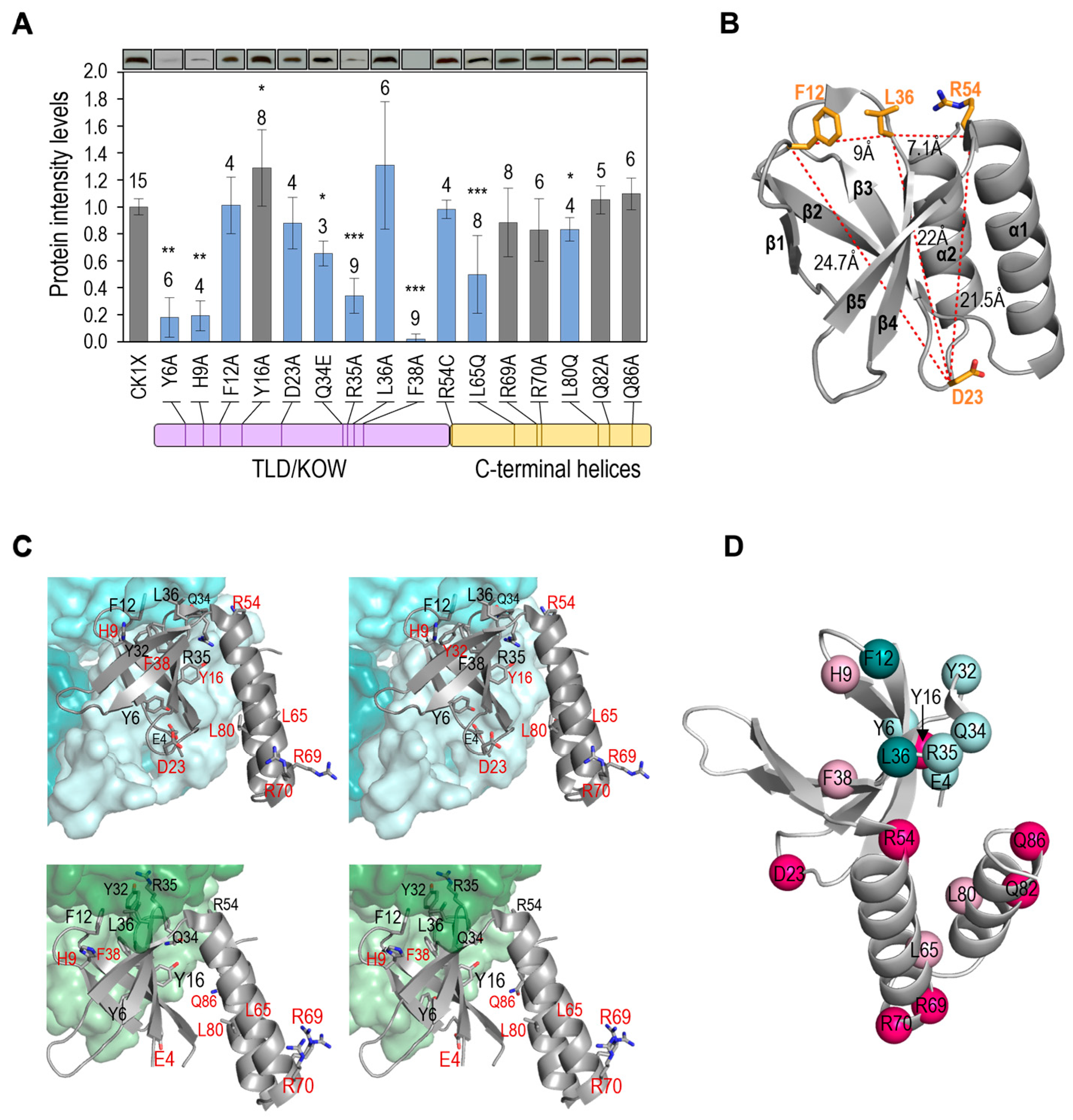
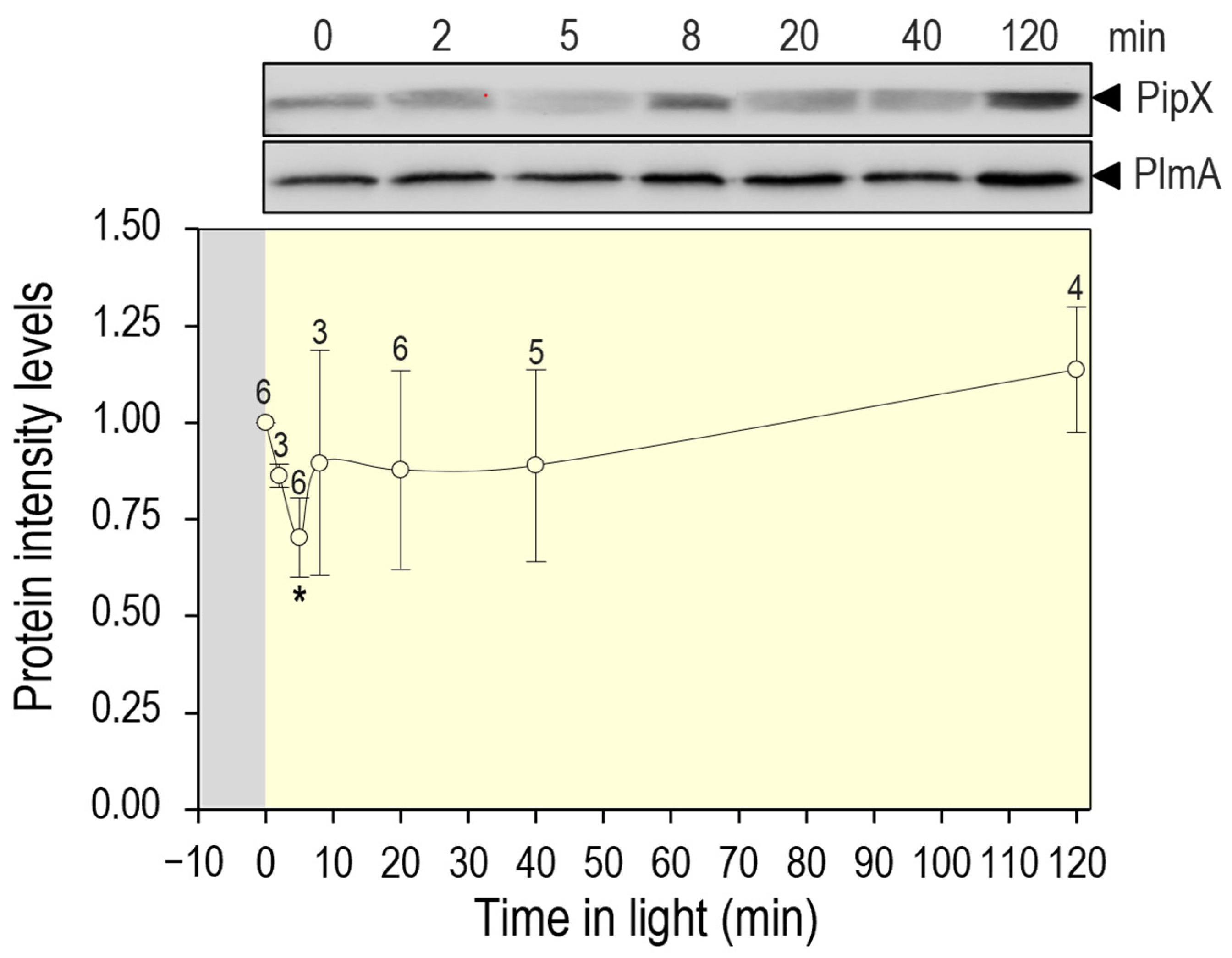
| Strain | Genotype or Relevant Characteristics | Source or Reference |
|---|---|---|
| E. coli DH5α | F− φ80 lacZΔM15 Δ(lacZYA-argF)U169 endA1 recA1 hsdR17(rK− mK+) deoR phoA thi-1 supE44 gyrA96 relA1 λ− | [44] |
| WT | Wild type S. elongatus PCC7942 | Pasteur Culture Collection |
| ∆pipX | pipX::cat, CmR | [33] |
| CK1XY | Φ(C.K1-pipXpipY), KmR | [12] |
| CK1XY6AY | Φ(C.K1-pipXY6ApipY), KmR | [23] |
| CK1XH9AY | Φ(C.K1-pipXH9ApipY), KmR | This work |
| CK1XH9AY-B | Φ(C.K1-pipXH9ApipY)/glnB::CS3, KmR SmR | This work |
| CK1XF12AY | Φ(C.K1-pipXF12ApipY), KmR | [23] |
| CK1XY16AY | Φ(C.K1-pipXY16ApipY), KmR | This work |
| CK1XD23AY | Φ(C.K1-pipXD23ApipY), KmR | [23] |
| CK1XQ34EY | Φ(C.K1-pipXQ34EpipY), KmR | [23] |
| CK1XR35AY | Φ(C.K1-pipXR35ApipY), KmR | [23] |
| CK1XL36AY | Φ(C.K1-pipXL36ApipY), KmR | [23] |
| CK1XF38AY | Φ(C.K1-pipXF38ApipY), KmR | [23] |
| CK1XR54CY | Φ(C.K1-pipXR54CpipY), KmR | [22] |
| CK1XL65QY | Φ(C.K1-pipXL65QpipY), KmR | [22] |
| CK1XR69AY | Φ(C.K1-pipXR69ApipY), KmR | [23] |
| CK1XR70AY | Φ(C.K1-pipXR70ApipY), KmR | This work |
| CK1XL80QY | Φ(C.K1-pipXL80QpipY), KmR | This work |
| CK1XL80QY-B | Φ(C.K1-pipXL80QpipY)/glnB::CS3, KmR SmR | This work |
| CK1XQ82AY | Φ(C.K1-pipXQ82ApipY), KmR | [23] |
| CK1XQ86AY | Φ(C.K1-pipXQ86ApipY), KmR | [23] |
| Plasmid | Genotype or Relevant Characteristics | Source or Reference |
|---|---|---|
| pUAGC126 | pipX replaced with cat, ApR CmR | [33] |
| pUAGC701 | CS3(-) into glnB, ApR SmR | [21] |
| pUAGC410 | [Φ(C.K1-pipXpipY)], ApR KmR | [12] |
| pUAGC685 | [Φ(C.K1-pipXY6ApipY)], ApR KmR | [23] |
| pUAGC948 | [Φ(C.K1-pipXH9ApipY)], ApR KmR | This work |
| pUAGC939 | [Φ(C.K1-pipXF12ApipY)], ApR KmR | [23] |
| pUAGC945 | [Φ(C.K1-pipXY16ApipY)], ApR KmR | This wok |
| pUAGC680 | [Φ(C.K1-pipXD23ApipY)], ApR KmR | [23] |
| pUAGC687 | [Φ(C.K1-pipXQ34EpipY)], ApR KmR | [23] |
| pUAGC688 | [Φ(C.K1-pipXR35ApipY)], ApR KmR | [23] |
| pUAGC976 | [Φ(C.K1-pipXL36ApipY)], ApR KmR | [23] |
| pUAGC940 | [Φ(C.K1-pipXF38ApipY)], ApR KmR | [23] |
| pUAGC681 | [Φ(C.K1-pipXR54CpipY)], ApR KmR | [22] |
| pUAGC682 | [Φ(C.K1-pipXL65QpipY)], ApR KmR | [22] |
| pUAGC689 | [Φ(C.K1-pipXR69ApipY)], ApR KmR | [23] |
| pUAGC937 | [Φ(C.K1-pipXR70ApipY)], ApR KmR | This work |
| pUAGC618 | [Φ(C.K1-pipXL80QpipY)], ApR KmR | This work |
| pUAGC619 | [Φ(C.K1-pipXQ82ApipY)], ApR KmR | [23] |
| pUAGC683 | [Φ(C.K1-pipXQ86ApipY)], ApR KmR | [23] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llop, A.; Tremiño, L.; Cantos, R.; Contreras, A. The Signal Transduction Protein PII Controls the Levels of the Cyanobacterial Protein PipX. Microorganisms 2023, 11, 2379. https://doi.org/10.3390/microorganisms11102379
Llop A, Tremiño L, Cantos R, Contreras A. The Signal Transduction Protein PII Controls the Levels of the Cyanobacterial Protein PipX. Microorganisms. 2023; 11(10):2379. https://doi.org/10.3390/microorganisms11102379
Chicago/Turabian StyleLlop, Antonio, Lorena Tremiño, Raquel Cantos, and Asunción Contreras. 2023. "The Signal Transduction Protein PII Controls the Levels of the Cyanobacterial Protein PipX" Microorganisms 11, no. 10: 2379. https://doi.org/10.3390/microorganisms11102379
APA StyleLlop, A., Tremiño, L., Cantos, R., & Contreras, A. (2023). The Signal Transduction Protein PII Controls the Levels of the Cyanobacterial Protein PipX. Microorganisms, 11(10), 2379. https://doi.org/10.3390/microorganisms11102379





