Microbial-Based Products to Control Soil-Borne Pathogens: Methods to Improve Efficacy and to Assess Impacts on Microbiome
Abstract
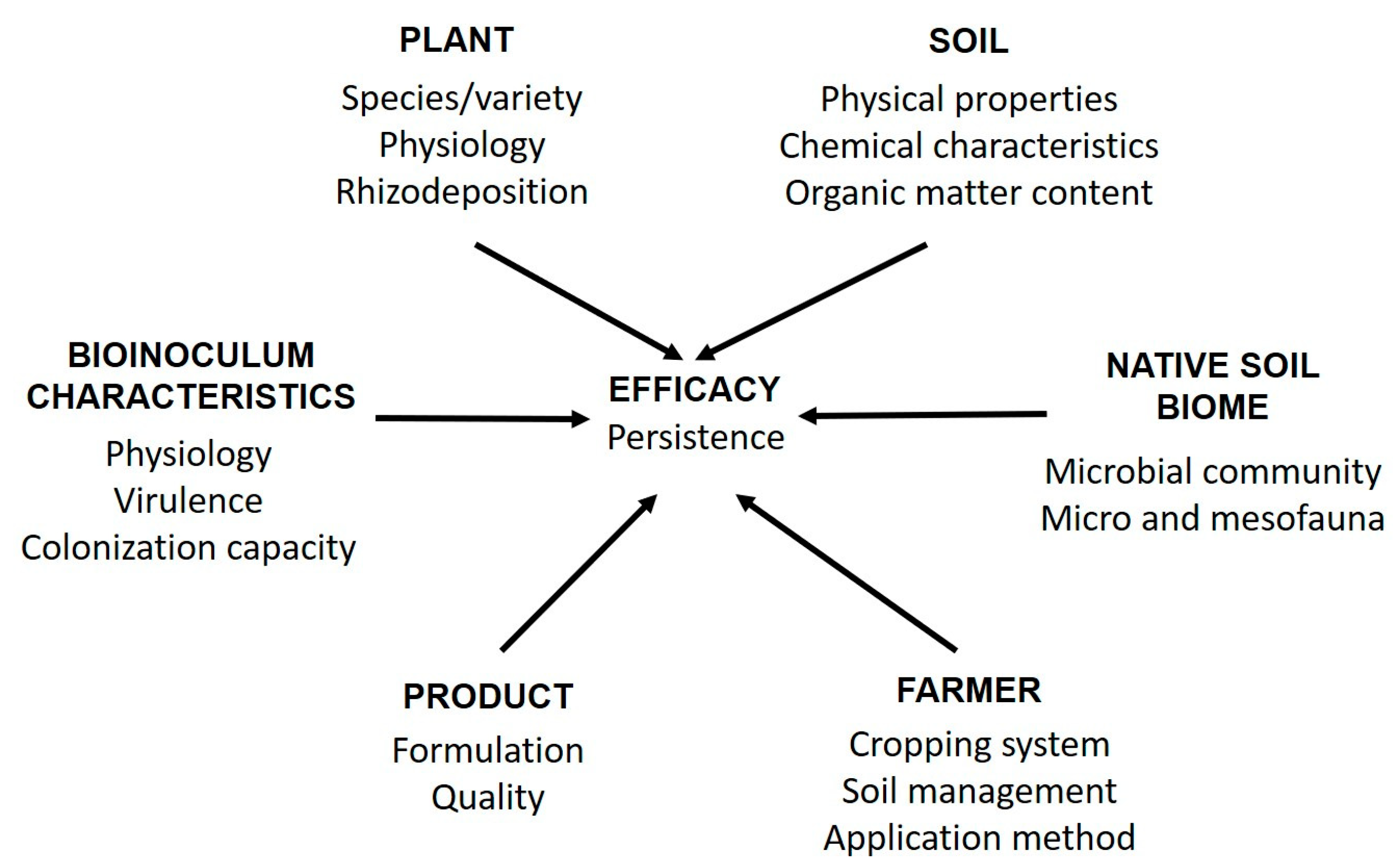
1. Introduction
2. Improving Production and Formulation of Bioinocula
3. Methods for the Study of Bioinocula Interactions with Soil Microbiome to Increase Their Efficacy and Marketing
4. Methods for Evaluating the Effect on the Soil Microbiome to Improve Microbial-Based Product Exploitation and Environmental Impact Assessment
5. Probiotics and Prebiotics to Control Soil-Borne Pathogens
| Crop/Pathogen System | Microbial Inoculant | Method of Application | Observed Effects | Impact on the Soil Native Microorganisms | Reference |
|---|---|---|---|---|---|
| Lettuce/Fusarium oxysporum f. sp. Lactucae |
| Preventative application in nursery and at planting in naturally or artificially infested soil. | Fusarium wilt severity reduction, in all cases and over the years, from 50% to 70%, compared to the untreated control. | The microbiome was not affected by the treatments, and no significant differences were observed when comparing the soil microbial community with that of the untreated control. | [137] |
| Preventative application in nursery and at planting in naturally or artificially infested soil. | Fusarium wilt reduction by as much as 69%. | Relevant impact of the treatments on ammonia-oxidizing bacteria and on the archaea that harbor the amoA gene. Significant negative correlations between Bacillus subtilis, Trichoderma, and Pseudomonas sp. abundances and wilt severity. No negative impact on the indigenous microbial communities. | [139] | |
| Zucchini/Phytophthora capsici |
| Mixed with the potting soil at different concentrations (1–10–20% v/v) in controlled greenhouse pot trials. | Trichoderma-enriched compost administered at 10% v/v reduced P. capsici by 50%, but when applied at 20% did not significantly suppress the pathogen. | Differences in population composition at genera level and in relative abundance according to the mycobiota sequencing. PCA analysis clustered the treated soils separately from the untreated ones. | [144] |
| Zucchini/Phytophthora capsici |
| Three soil applications with BCAs to the plug trays between sowing and transplanting. The microbial-enriched compost was mixed at 20% v/v in the tray and immediately before sowing. | All the treatments reduced disease severity by as much as 50%. | Alphaproteobacteria enrichment and, in particular, a more relative abundance of Bradyrhizobium, Mesorhizobium and Hypomicrobium, suggesting their involvement in disease suppression. No modification of the mycobiota, but. all the treatments reduced pathogen abundance. | [138] |
| Tomato/F. oxysporum f. sp. lycopersici |
| Four soil treatments with bioinoculants to plug tray between sowing and transplanting in a commercial nursery. Microbial enriched compost was applied twice: first mixed with the substrate at sowing, and then mixed with the soil one week before planting | The treatments reduced Fusarium wilt severity by as much as 50%. | No negative effects of the bioinoculants were observed on non-target microbial communities. Decreased F. lycopersici abundance in the soil with a greater abundance of the inoculated microbials and an accumulation of transcripts encoding PR genes. | [138] |
| Tomato/Fusarium oxysporum f. sp. lycopersici |
| Seed coating. | Higher germination rate (22–48%) and lower mean germination time (less than 2.5 days) of tomato seeds than the control; the combination of bioinoculants were more effective than single-isolate treatments. The combined P. fluorescence + T. harzianum, or AMF provided a disease reduction of 67% in the field, compared to the non-treated plants. | The impacts were not evaluated. | [147] |
| Tomato/Ralstonia solanacearum | Fluorescent pseudomonad strains (CHA0, PF5, Q2-87, Q8R1-96, 1M1-96, MVP1-4, F113, and Phl1C2) were combined in different consortium to simulate different levels of strain richness. | Introduced multispecies probiotic consortia of Pseudomonas into a naturally diverse tomato rhizosphere microbiome, 5, 15, 25, and 35 days after the pathogen inoculation in greenhouse experiments. | The survival of introduced Pseudomonas consortia increased with increased diversity. The highest Pseudomonas diversity reduced pathogen density in the rhizosphere and decreased the bacterial wilt incidence. | The most diverse probiotic Pseudomonas communities composed of 8 strains were able to persist at high densities and to compete for resources with the pathogen and the natural bacterial communities. | [157] |
| Tomato and pepper/Verticillium dahliae | AMF combined with humic acids and/or whey | Pre-inoculation in growth chamber | Reduced wilt disease severity and Verticillium dahliae microsclerotia number. | The impacts were not evaluated. | [172] |
| Tomato/Fusarium oxysporum f. sp. lycopersici |
| Applied alone or combined under pot and field conditions. | The combined bioinoculant (F. mosseae + G. fasciculatum + B. velezensis + Bacillus sp.) was the most effective in reducing the Fusarium wilt severity (−77.44%) followed by F. mosseae + B. velezensis + Bacillus sp. or B. velezensis + Bacillus sp. (−66.67%) | The impacts were not evaluated. | [173] |
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Elmerich, C. Historical Perspective: From Bacterization to Endophytes. In Associative and Endophytic Nitrogen-Fixing Bacteria and Cyanobacterial Associations; Springer: Dordrecht, The Netherlands, 2007; pp. 1–20. [Google Scholar] [CrossRef]
- Hartmann, A.; Rothballer, M.; Schmid, M. Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 2008, 312, 7–14. [Google Scholar] [CrossRef]
- Nester, E.W.; Thomashow, L.S.; Metz, M.; Gordon, M. 100 Years of Bacillus thuringiensis: A Critical Scientific Assessment: This report is based on a colloquium, “100 Years of Bacillis thuringiensis, a Paradigm for Producing Transgenic Organisms: A Critical Scientific Assessment”, sponsored by the American Academy of Microbiology and held November 16–18, in Ithaca, New York; American Society for Microbiology: Washington, DC, USA, 2002. [Google Scholar] [CrossRef]
- Smith, R.S. Legume inoculant formulation and application. Can. J. Microbiol. 1992, 38, 485–492. [Google Scholar] [CrossRef]
- Sanahuja, G.; Banakar, R.; Twyman, R.; Capell, T.; Christou, P. Bacillus thuringiensis: A century of research, development and commercial applications. Plant Biotechnol. J. 2011, 9, 283–300. [Google Scholar] [CrossRef] [PubMed]
- Hamdi, Y.A. Application of Nitrogen-Fixing Systems in Soil Improvement and Management; Food and Agriculture Organization: Rome, Italy, 1982; p. 49. [Google Scholar]
- Anonymous. Biopesticides Market Size, Share and Industry Analysis by Product Type, Source, Mode of Application, Crops and Regional Forecast 2018–2025. Fortune Bus. Insights 2019, 145. [Google Scholar]
- Anonymous. Biofertilizers Market by Product, Microorganism Type and Application, Crop Type: Global Opportunity Analysis and Industry Forecast, 2019–2026. Fortune Bus. Insights 2019, 199. [Google Scholar]
- Malinowski, H. The use of Bacillus thuringiensis in plant protection: Prospects and limitations. Biotechnologia 2000, 3, 81–92. (In Polish) [Google Scholar]
- Glare, T.; Caradus, J.; Gelernter, W.; Jackson, T.; Keyhani, N.; Köhl, J.; Marrone, P.; Morin, L.; Stewart, A. Have biopesticides come of age? Trends Biotechnol. 2012, 30, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Batista, B.D.; Brajesh, K.S. Realities and hopes in the application of microbial tools in agriculture. Microb. Biotechnol. 2021, 14, 1258–1268. [Google Scholar] [CrossRef]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R.; Hernandez, J.P. Advances in plant growth-promoting bacterial inoculant technology: Formulations and practical perspectives (1998–2013). Plant Soil 2014, 378, 1–33. [Google Scholar] [CrossRef]
- Timmusk, S.; Behers, L.; Muthoni, J.; Muraya, A.; Aronsson, A.C. Perspectives and Challenges of Microbial Application for Crop Improvement. Front. Plant Sci. 2017, 9, 49. [Google Scholar] [CrossRef]
- Behera, S.S.; Ray, R.C.; Das, U.; Panda, S.K.; Saranraj, P. Microorganisms in Fermentation. In Essentials in Fermentation Technology; Berenjian, A., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; pp. 1–39. [Google Scholar] [CrossRef]
- Lo, K.J.; Lee, S.K.; Liu, C.T. Development of a low-cost culture medium for the rapid production of plant growth-promoting Rhodopseudomonas palustris strain PS3. PLoS ONE 2020, 15, e0236739. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, N.; Mendes, G. Solid-state fermentation and plantbeneficial microorganisms. In Current Developments in Biotechnology and Bioengineering, Current Advances in Solid-State Fermentation; Pandey, A., Larroche, C.H., Soccol, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 402–416. [Google Scholar]
- Pertot, I.; Alabouvette, C.; Esteve, E.H.; França, S. Mini-Paper—The Use of Microbial Biocontrol Agents against Soil-Borne Diseases. EIP-Agri Focus Group Soil-Borne Diseases. 2015, pp. 1–11. Available online: https://ec.europa.eu/eip/agriculture/sites/default/files/8_eip_sbd_mp_biocontrol_final.pdf (accessed on 20 December 2022).
- Koskey, G.; Mburu, S.; Awino, R.; Maingi, J. Potential Use of Beneficial Microorganisms for Soil Amelioration, Phytopathogen Biocontrol, and Sustainable Crop Production in Smallholder Agroecosystems. Front. Sustain. Food Syst. 2021, 5, 606308. [Google Scholar] [CrossRef]
- Menge, J.A. Inoculum Production. In Va Mycorrhiza; Powel, C., Ed.; CRC Press: Boca Raton, FL, USA, 1984. [Google Scholar]
- Declerck, S.; Strullu, D.G.; Plenchette, C. In vitro mass-production of the arbuscular mycorrhizal fungus, Glomus versiforme, associated with Ri T-DNA transformed carrot roots. Mycol. Res. 1996, 100, 1237–1242. [Google Scholar] [CrossRef]
- Kar, S.; Ray, R.C. Optimization of thermostable α-amylase production by Streptomyces erumpens MTCC 7317 in solid state fermentation using cassava fibrous residue. Braz. Arch. Biol. Technol. 2010, 53, 301–309. [Google Scholar]
- Doriya, K.; Jose, N.; Gowda, M.; Kumar, D. Solid-State Fermentation vs Submerged Fermentation for the Production of l-Asparaginase. Adv. Food Nutr. Res. 2016, 78, 115–135. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, N.; Malusa, E.; Requena, A.R.; Martos, V.; López, A.; Maksimovic, I.; Vassileva, M. Potential application of glycerol in the production of plant beneficial microorganisms. J. Ind. Microbiol. Biotechnol. 2017, 44, 735–743. [Google Scholar] [CrossRef]
- Huang, Y.; Sun, L.; Zhao, J.; Huang, R.; Li, R.; Shen, Q. Utilization of different waste proteins to create a novel PGPR-containing bio-organic fertilizer. Sci. Rep. 2015, 5, 7766. [Google Scholar] [CrossRef]
- Pastor-Bueis, R.; Mulas, R.; Gómez, X.; González-Andrés, F. Innovative liquid formulation of digestates for producing a biofertilizer based on Bacillus siamensis: Field testing on sweet pepper. J. Plant Nutr. Soil Sci. 2017, 180, 748–758. [Google Scholar] [CrossRef]
- Ferreira, R.D.G.; Azzoni, A.R.; Freitas, S. Techno-economic analysis of the industrial production of a low-cost enzyme using E. coli: The case of recombinant β-glucosidase. Biotechnol. Biofuels 2018, 29, 81. [Google Scholar] [CrossRef]
- Lee, S.Y.; Wayne, K.; Chew, K.W.; Show, P.L. Cell separation and disruption, product recovery, and purification. In Essentials in Fermentation Technology; Berenjian, A., Ed.; Springer Nature Switzerland AG: Cham, Switzerland, 2019; pp. 237–271. [Google Scholar] [CrossRef]
- Vassilev, N.; Vassileva, M.; Martos, V.; Garcia Del Moral, L.F.; Kowalska, J.; Tylkowski, B.; Malusà, E. Formulation of microbial inoculants by encapsulation in natural polysaccharides: Focus on beneficial properties of carrier additives and derivates. Front. Plant Sci. 2020, 11, 270. [Google Scholar] [CrossRef]
- Tylkowski, B.; Olkiewicz, M.; Montane, X.; Nogalska, A.; Haponska, M.; Kowalska, J.; Malusá, E. Encapsulation technologies in agriculture. In Microencapsulation; Tylkowski, B., Giamberini, M., Fernandez Prieto, S., Eds.; DeGruyter Publisher: Berlin, Germany; Boston, MA, USA, 2020; pp. 287–302. [Google Scholar] [CrossRef]
- Bashan, Y.; de-Bashan, L.E.; Prabhu, S.R. Superior polymeric formulations and emerging innovative products of bacterial inoculants for sustainable agriculture and the environment. In Agriculturally Important Microorganisms; Singh, H., Sarma, B., Keswani, C., Eds.; Springer: Singapore, 2016; pp. 15–46. [Google Scholar] [CrossRef]
- Martínez-Cano, B.; Mendoza-Meneses, C.J.; García-Trejo, J.F.; Macías-Bobadilla, G.; Aguirre-Becerra, H.; Soto-Zarazúa, G.M.; Feregrino-Pérez, A.A. Review and perspectives of the use of alginate as a polymer matrix for microorganisms applied in agro-industry. Molecules 2022, 27, 4248. [Google Scholar] [CrossRef] [PubMed]
- Gasperini, L.; Mano, J.F.; Reis, R.L. Natural polymers for the microencapsulation of cells. J. R. Soc. Interface 2014, 11, 20140817. [Google Scholar] [CrossRef] [PubMed]
- Pitigraisorn, P.; Srichaisupakit, K.; Wongpadungkiat, N.; Wongsasulak, S. Encapsulation of Lactobacillus acidophilus in moist-heat-resistant multilayered microcapsules. J. Food Eng. 2017, 192, 11–18. [Google Scholar] [CrossRef]
- Jeong, Y.; Irudayaraj, J. Multi-layered alginate hydrogel structures and bacteria encapsulation. Chem. Commun. 2022, 58, 8584–8587. [Google Scholar] [CrossRef]
- Bashan, Y. Alginate beads as synthetic inoculant carriers for slow release of bacteria that affect plant growth. Appl. Environ. Microbiol. 1986, 51, 1089–1098. [Google Scholar] [CrossRef]
- Wang, X.; Gao, S.; Yun, S.; Zhang, M.; Peng, L.; Li, Y.; Zhou, Y. Microencapsulating Alginate-Based Polymers for Probiotics Delivery Systems and Their Application. Pharmaceuticals 2022, 15, 644. [Google Scholar] [CrossRef] [PubMed]
- Ravensberg, W. A Roadmap to the Successful Development and Commercialization of Microbial Pest Control Products for Control of Arthropods; Springer Science+Business Media B.V.: Berlin/Heidelberg, Germany, 2011. [Google Scholar] [CrossRef]
- Świechowski, W.; Doruchowski, G.; Trzciński, P. Effect of spray application parameters on viability of bacterium Pseudomonas fluorescens used as bio-pesticide in organic fruit production. In II International Organic Fruit Research Symposium; Granatstein, D., Andrews, P., Eds.; ISHS: Leavenworth, WA, USA, 2012; pp. 18–21. [Google Scholar]
- O’Callaghan, M. Microbial inoculation of seed for improved crop performance: Issues and opportunities. Appl. Microbiol. Biotechnol. 2016, 100, 5729–5746. [Google Scholar] [CrossRef]
- Singh, S.; Patel, V.B.; Singh, A.; Verma, M.K. Mycorrhizal Fungi in Sustainable Horticultural Production under Changing Climate Situations. In Climate Dynamics in Horticultural Science Vol 2 Impact, Adaptation, and Mitigation; Apple Academic Press: Palm Bay, FL, USA, 2013; pp. 240–259. [Google Scholar] [CrossRef]
- Doruchowski, G. (The National Institute of Horticultural Research, Skierniewice, Poland). Personal communication, 2022.
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant–microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Ram, R.M.; Debnath, A.; Negi, S.; Singh, H.B. Use of microbial consortia for broad spectrum protection of plant pathogens: Regulatory hurdles, present status and future prospects. In Biopesticides Volume 2: Advances in Bio-Inoculants; Woodhead Publishing: Cambridge, UK, 2022; pp. 319–335. [Google Scholar] [CrossRef]
- Bhatia, S.K.; Bhatia, R.K.; Choi, Y.K.; Kan, E.; Kim, Y.G.; Yang, Y.H. Biotechnological potential of microbial consortia and future perspectives. Crit. Rev. Biotechnol. 2018, 38, 1209–1229. [Google Scholar] [CrossRef]
- Sarma, B.K.; Yadav, S.K.; Singh, S. Microbial consortium-mediated plant defence against phytopathogens: Readdressing for enhancing efficacy. Soil Biol. Biochem. 2015, 87, 25–33. [Google Scholar] [CrossRef]
- Minchev, Z.; Kostenko, O.; Soler, R.; Pozo, M.J. Microbial consortia for effective biocontrol of root and foliar diseases in tomato. Front. Plant Sci. 2021, 12, 756368. [Google Scholar] [CrossRef] [PubMed]
- O’Callaghan, M.; Ballard, R.A.; Wright, D. Soil microbial inoculants for sustainable agriculture: Limitations and opportunities. Soil Use Manag. 2022, 38, 1340–1369. [Google Scholar] [CrossRef]
- Deepa, J.; Mathew, S.K. Compatibility studies on different endophytic microbes of tomato antagonistic to bacterial wilt pathogen. IJABR 2017, 7, 190–194. [Google Scholar]
- Faust, K. Microbial consortium design benefits from metabolic modeling. Trends Biotechnol. 2019, 37, 123–125. [Google Scholar] [CrossRef]
- Malusa, E.; Sas-Paszt, L.; Ciesielska, J. Technologies for beneficial microorganisms inocula used as biofertilizers. Sci. World J. 2012, 2012, 491206. [Google Scholar] [CrossRef]
- Mitter, B.; Brader, G.; Pfaffenbichler, N.; Sessitsch, A. Next generation microbiome applications for crop production—Limitations and the need of knowledge-based solutions. Curr. Opin. Microbiol. 2019, 49, 59–65. [Google Scholar] [CrossRef]
- Kowalska, J.; Tyburski, J.; Matysiak, K.; Tylkowski, B.; Malusá, E. Field Exploitation of Multiple Functions of Beneficial Microorganisms for Plant Nutrition and Protection: Real Possibility or Just a Hope? Front. Microbiol. 2020, 11, 1904. [Google Scholar] [CrossRef]
- Regulation (EU) 2019/1009 of the European Parliament and of the Council of 5 June 2019 Laying down Rules on the Making Available on the Market of EU Fertilising Products. Official Journal of the European Union, L170/1, 25/6/2019. Available online: http://data.europa.eu/eli/reg/2019/1009/oj (accessed on 20 December 2022).
- Kapoore, R.J.; Padmaperuma, G.; Maneein, S.; Vaidyanathan, S. Co-culturing microbial consortia: Approaches for applications in biomanufacturing and bioprocessing. Crit. Rev. Biotechnol. 2022, 42, 46–72. [Google Scholar] [CrossRef]
- Agrawal, A.A.; Ackerly, D.D.; Adler, F.; Arnold, A.E.; Cáceres, C.; Doak, D.F.; Post, E.; Hudson, P.J.; Maron, J.; Mooney, K.A.; et al. Filling key gaps in population and community ecology. Front. Ecol. Environ. 2007, 5, 145–152. [Google Scholar] [CrossRef]
- Ricklefs, R.E. Disintegration of the Ecological Community: American Society of Naturalists Sewall Wright Award Winner Address. Am. Nat. 2008, 172, 741–750. [Google Scholar] [CrossRef]
- Brooker, R.W.; Callaway, R.M. Facilitation in the conceptual melting pot. J. Ecol. 2009, 97, 1117–1120. [Google Scholar] [CrossRef]
- Haruta, S.; Yamamoto, K. Model microbial consorta as tools for understanding complex microbial communities. Curr. Genom. 2018, 19, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Mittermeier, F.; Bäumler, M.; Arulrajah, P.; de Jesus Garcia Lima, J.; Hauke, S.; Stock, A.; Weuster-Botz, D. Artificial microbial consortia for bioproduction processes. Eng. Life Sci. 2023, 23, e2100152. [Google Scholar] [CrossRef] [PubMed]
- Manfredini, A.; Malusà, E.; Costa, C.; Pallottino, F.; Mocali, S.; Pinzari, F.; Canfora, L. Current methods, common practices, and perspectives in tracking and monitoring bioinoculants in soil. Front. Microbiol. 2021, 12, 698491. [Google Scholar] [CrossRef]
- Gimeno, A.; Stanley, C.E.; Ngamenie, Z.; Hsung, M.-H.; Walder, F.; Schmieder, S.S.; Bindschedler, S.; Junier, P.; Keller, B.; Vogelgsang, S. A versatile microfluidic platform measures hyphal interactions between Fusarium graminearum and Clonostachys rosea in real-time. Commun. Biol. 2021, 4, 262. [Google Scholar] [CrossRef]
- Acin-Albiac, M.; Filannino, P.; Gobbetti, M.; Di Cagno, R. Microbial high throughput phenomics: The potential of an irreplaceable omics. Comput. Struct. Biotechnol. J. 2020, 18, 2290–2299. [Google Scholar] [CrossRef]
- Siles, J.A.; García-Sánchez, M.; Gómez-Brandón, M. Studying microbial communities through co-occurrence network analyses during processes of waste treatment and in organically amended soils: A Review. Microorganisms 2021, 9, 1165. [Google Scholar] [CrossRef]
- Wood, D.E.; Salzberg, S.L. Kraken: Ultrafast metagenomic sequence classification using exact alignments. Genome. Biol. 2014, 15, R46. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Frąc, M.; Kaczmarek, J.; Jędryczka, M. Metabolic capacity differentiates Plenodomus lingam from P. biglobosus Subclade ‘brassicae’, the causal agents of phoma leaf spotting and stem canker of oilseed rape (Brassica napus) in agricultural ecosystems. Pathogens 2022, 11, 50. [Google Scholar] [CrossRef]
- Frąc, M.; Jezierska-Tys, S.; Yaguchi, T. Occurrence, detection, and molecular and metabolic characterisation of heat-resistant fungi in soils and plants and their risk to human health. Adv. Agron. 2015, 132, 161–204. [Google Scholar]
- Shea, A.; Wolcott, M.; Daefler, S.; Rozak, D.A. Biology Phenotype Microarrays. In Microbial Systems Biology: Methods and Protocols, Methods in Molecular Biology; Navid, A., Ed.; Springer Nature: Cham, Switzerland, 2012; p. 881. [Google Scholar] [CrossRef]
- Greetham, D. Phenotype microarray technology and its application in industrial biotechnology. Biotechn. Lett. 2014, 36, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Bochner, B.R.; Gadzinski, P.; Panomitros, E. Phenotype MicroArrays for High-Throughput Phenotypic Testing and Assay of Gene Function. Genome Res. 2001, 11, 1246–1255. [Google Scholar] [CrossRef]
- Borglin, S.; Joyner, D.; DeAngelis, K.; Khudyakov, J.; D’haeseeleer, P.; Joachimiak, M.P.; Hazen, T. Application of phenotypic microarrays to environmental microbiology. Curr. Opin. Biotechnol. 2012, 23, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Kent, A.D.; Triplett, E.W. Microbial communities and their interactions in soil and rhizosphere ecosystems. Annu. Rev. Microbiol. 2002, 56, 211–236. [Google Scholar] [CrossRef]
- Pinzari, F.; Ceci, A.; Abu-Samra, N.; Canfora, L.; Maggi, O.; Persiani, A. Phenotype MicroArrayTM system in the study of fungal functional diversity and catabolic versatility. Res. Microbiol. 2016, 167, 710–722. [Google Scholar] [CrossRef]
- Blumenstein, K.; Albrectsen, B.R.; Martin, J.A.; Hultberg, M.; Sieber, T.N.; Helander, M.; Witzell, J. Nutritional niche overlap potentiates the use of endophytes in biocontrol of a tree disease. BioControl 2015, 60, 655–667. [Google Scholar] [CrossRef]
- Canfora, L.; Abu-Samra, N.; Tartanus, M.; Łabanowska, B.H.; Benedetti, A.; Pinzari, F.; Malusà, E. Co-Inoculum of Beauveria brongniartii and B. bassiana shows in vitro different metabolic behaviour in comparison to single inoculums. Sci. Rep. 2017, 7, 13102. [Google Scholar] [CrossRef]
- Mettel, C.; Kim, Y.; Shrestha, P.M.; Liesack, W. Extraction of mRNA from soil. Appl. Environ. Microbiol. 2010, 76, 5995–6000. [Google Scholar] [CrossRef]
- Nocker, A.; Cheung, C.Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Nocker, A.; Sossa-Fernandez, P.; Burr, M.D.; Camper, A.K. Use of propidium monoazide for live/dead distinction in microbial ecology. Appl. Environ. Microbiol. 2007, 73, 5111–5117. [Google Scholar] [CrossRef] [PubMed]
- Heise, J.; Nega, M.; Alawi, M.; Wagner, D. Propidium monoazide treatment to distinguish between live and dead methanogens in pure cultures and environmental samples. J. Microbiol. Methods 2016, 121, 11–23. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Ye, Z.; Jia, Y.; Fan, J.; Hashmi, M.Z.; Shen, C. An optimised method to assess viable Escherichia coli O157:H7 in agricultural soil using combined propidium monoazide staining and quantitative PCR. Front. Microbiol. 2020, 11, 1809. [Google Scholar] [CrossRef] [PubMed]
- Matarese, F.; Sarrocco, S.; Gruber, S.; Seidl-Seiboth, V.; Vannacci, G. Biocontrol of Fusarium head blight: Interactions between Trichoderma and mycotoxigenic Fusarium. Microbiology 2012, 158, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Arellano-Caicedo, C.; Ohlsson, P.; Bengtsson, M.; Beech, J.P.; Hammer, E.C. Habitat geometry in artificial microstructure affects bacterial and fungal growth, interactions, and substrate degradation. Commun. Biol. 2021, 4, 1226. [Google Scholar] [CrossRef]
- Stanley, C.E.; van der Heijden, M.G.A. Microbiome-on-a-Chip: New frontiers in plant-microbiota research. Trends Microbiol. 2017, 25, 610–613. [Google Scholar] [CrossRef] [PubMed]
- Stanley, C.E.; Grossmann, G.; Solvas, X.C.; DeMello, A.J. Soil-on-a-Chip: Microfluidic platforms for environmental organismal studies. Lab Chip. 2016, 16, 228–241. [Google Scholar] [CrossRef] [PubMed]
- Mafla-Endara, P.M.; Arellano-Caicedo, C.; Aleklett, K.; Pucetaite, M.; Ohlsson, P.; Hammer, E.C. Microfluidic chips provide visual access to in situ soil ecology. Commun. Biol. 2021, 4, 889. [Google Scholar] [CrossRef]
- Masters-Clark, E.; Clark, A.J.; Stanley, C.E. Microfluidic tools for probing fungal-microbial interactions at the cellular level. J. Vis. Exp. 2022, 23, 184. [Google Scholar] [CrossRef]
- Baranger, C.; Fayeulle, A.; Le Goff, A. Microfluidic monitoring of the growth of individual hyphae in confined environments. R. Soc. Open Sci. 2020, 7, 191535. [Google Scholar] [CrossRef]
- Richter, F.; Bindschedler, S.; Calonne-Salmon, M.; Declerck, S.; Junier, P.; Stanley, C.E. Fungi-on-a-Chip: Microfluidic platforms for single-cell studies on fungi. FEMS Microbiol. Rev. 2022, 46, fuac039. [Google Scholar] [CrossRef] [PubMed]
- Cornell, C.; Kokkoris, V.; Richards, A.; Horst, C.; Rosa, D.; Bennett, J.A.; Hart, M.M. Do Bioinoculants Affect Resident Microbial Communities? A Meta-Analysis. Front. Agron. 2021, 3, 753474. [Google Scholar] [CrossRef]
- Trabelsi, D.; Mhamdi, R. Microbial inoculants and their impact on soil microbial communities: A review. Biomed. Res. Int. 2013, 2013, 863240. [Google Scholar] [CrossRef]
- Gougoulias, C.; Clark, J.M.; Shaw, L.J. The role of soil microbes in the global carbon cycle: Tracking the below-ground microbial processing of plant-derived carbon for manipulating carbon dynamics in agricultural systems. J. Sci. Food Agric. 2014, 94, 2362–2371. [Google Scholar] [CrossRef]
- Bohan, D.A.; Vacher, C.; Tamaddoni-Nezhad, A.; Raybould, A.; Dumbrell, A.J.; Woodward, G. Next-generation global biomonitoring: Large-scale, automated reconstruction of ecological networks. Trends Ecol. Evol. 2017, 32, 477–487. [Google Scholar] [CrossRef] [PubMed]
- Amann, R.I.; Wolfgang, L.; Schleifer, K.H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef] [PubMed]
- Jenkins, C.; Ling, C.L.; Ciesielczuk, H.L.; Lockwood, J.; Hopkins, S.; McHugh, T.D.; Gillespie, S.H.; Kibbler, C.C. Detection and identification of bacteria in clinical samples by 16S rRNA gene sequencing: Comparison of two different approaches in clinical practice. J. Med. Microbiol. 2012, 61, 483–488. [Google Scholar] [CrossRef]
- Schoch, C.L.; Seifert, K.A.; Huhndorf, S.; Robert, V.; Spouge, J.L.; Levesque, C.A.; Chen, W. Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 2012, 109, 6241–6246. [Google Scholar] [CrossRef]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 2007, 74, 5463–5467. [Google Scholar] [CrossRef]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Laverock, B.; Temperton, B.; Thomas, S.; Muhling, M.; Hughes, M. Metagenomics. Methods Mol. Biol. 2011, 733, 173–183. [Google Scholar] [CrossRef] [PubMed]
- Sharpton, T.J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 2014, 5, 209. [Google Scholar] [CrossRef]
- Escobar-Zepeda, A.; De León, A.V.P.; Sanchez-Flores, A. The road to metagenomics: From microbiology to DNA sequencing technologies and bioinformatics. Front. Genet. 2016, 6, 348. [Google Scholar] [CrossRef] [PubMed]
- Rondon, M.R.; August, P.R.; Bettermann, A.D.; Brady, S.F.; Grossman, T.H.; Liles, M.R.; Loiacono, K.A.; Lynch, B.A.; MacNeil, I.A.; Minor, C.; et al. Cloning the soil metagenome: A strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl. Environ. Microbiol. 2000, 66, 2541–2547. [Google Scholar] [CrossRef]
- Semenov, M.V. Metabarcoding and metagenomics in soil ecology research: Achievements, challenges, and prospects. Biol. Bull. Rev. 2011, 11, 40–53. [Google Scholar] [CrossRef]
- Jansson, J.K.; Hofmockel, K.S. The soil microbiome—From metagenomics to metaphenomics. Curr. Opin. Microbiol. 2018, 43, 162–168. [Google Scholar] [CrossRef]
- Chavan, S.; Sarangdhar, V.; Vigneshwaran, N. Nanopore-based metagenomic analysis of the impact of nanoparticles on soil microbial communities. Heliyon 2022, 9, e09693. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Balvociute, M.; Huson, D.H. SILVA, RDP, Greengenes, NCBI and OTT—How do these taxonomies compare? BMC Genome 2017, 18, 114. [Google Scholar] [CrossRef]
- Nayfach, S.; Roux, S.; Seshadri, R.; Udwary, D.; Varghese, N.; Schulz, F.; Wu, D.; Paez-Espino, D.; Chen, I.M.; Huntemann, M.; et al. A genomic catalog of Earth’s microbiomes. Nat. Biotechnol. 2021, 39, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Tsai, S.M.; Navarrete, A.A.; de Hollander, M.; van Veen, J.A.; Kuramae, E.E. Soil-borne microbiome: Linking diversity to function. Microb. Ecol. 2015, 70, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, R.C.; van Es, H.M.; Buckley, D.H. Predicting measures of soil health using the microbiome and supervised machine learning. Soil Biol. Biochem. 2021, 164, 108472. [Google Scholar] [CrossRef]
- Barberan, A.; Bates, S.T.; Casamayor, E.O.; Fierer, N. Using network analysis to explore co-occurrence patterns in soil microbial communities. ISME J. 2012, 6, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.; Mills, A.L. The effect of sample size in studies of soil microbial community structure. J. Microbiol. Methods 2006, 66, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Ji, Y.; Ashton, L.; Pedley, S.M.; Edwards, D.P.; Tang, Y.; Nakamura, A.; Kitching, R.; Dolman, P.M.; Woodcock, P.; Edwards, F.A.; et al. Reliable, verifiable and efficient monitoring of biodiversity via meta-barcoding. Ecol. Lett. 2013, 16, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.-I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018, 16, 410–422. [Google Scholar] [CrossRef]
- Lauber, C.L.; Ramirez, K.S.; Aanderud, Z.; Lennon, J.; Fierer, N. Temporal variability in soil microbial communities across land-use types. ISME J. 2013, 7, 1641–1650. [Google Scholar] [CrossRef]
- Bao, T.; Deng, S.; Yu, K.; Li, W.; Dong, A. Metagenomic insights into seasonal variations in the soil microbial community and function in a Larix gmelinii forest of Mohe, China. J. For. Res. 2021, 32, 371–383. [Google Scholar] [CrossRef]
- Nannipieri, P.; Penton, C.R.; Purahong, W.; Schloter, M.; van Elsas, J.D. Recommendations for soil microbiome analyses. Biol. Fertil. Soils 2019, 55, 765–766. [Google Scholar] [CrossRef]
- Lombard, N.; Prestat, E.; van Elsas, J.D.; Simonet, P. Soil-specific limitations for access and analysis of soil microbial communities by metagenomics. FEMS Microbiol. Ecol. 2011, 78, 31–49. [Google Scholar] [CrossRef] [PubMed]
- Morita, H.; Akao, S. The effect of soil sample size, for practical DNA extraction, on soil microbial diversity in different taxonomic ranks. PLoS ONE 2021, 16, e0260121. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson-Palme Johan. Strategies for Taxonomic and Functional Annotation of Metagenomes. In Metagenomics; Nagarajan, M., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 55–79. [Google Scholar] [CrossRef]
- Bray, R.J.; Curtis, J.T. An ordination of the upland forest communities of southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220. [Google Scholar] [PubMed]
- Clarke, K.R. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 1993, 18, 117–143. [Google Scholar] [CrossRef]
- Anderson, M.J. A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 2001, 26, 32–46. [Google Scholar]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. Vegan: Community Ecology Package. 2017. Available online: https://cran.r-project.org/web/packages/vegan/index.html (accessed on 20 December 2022).
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Schlenker, E. Tips and tricks for successful application of statistical methods to biological data. Methods Mol. Biol. 2016, 1366, 271–285. [Google Scholar]
- Guseva, K.; Darcy, S.; Simon, E.; Alteio, L.V.; Montesinos-Navarro, A.; Kaiser, C. From diversity to complexity: Microbial networks in soils. Soil Biol. Biochem. 2022, 169, 108604. [Google Scholar] [CrossRef]
- Leite, M.F.A.; van den Broek, S.W.E.B.; Kuramae, E.E. Current Challenges and Pitfalls in Soil Metagenomics. Microorganisms 2022, 10, 1900. [Google Scholar] [CrossRef]
- Lahlali, R.; Ezrari, S.; Radouane, N.; Kenfaoui, J.; Esmaeel, Q.; El Hamss, H.; Belabess, Z.; Barka, E.A. Biological control of plant pathogens: A global perspective. Microorganisms 2022, 10, 596. [Google Scholar] [CrossRef] [PubMed]
- Hegyi, A.; Nguyen, T.B.K.; Posta, K. Metagenomic Analysis of Bacterial Communities in Agricultural Soils from Vietnam with Special Attention to Phosphate Solubilizing Bacteria. Microorganisms 2021, 9, 1796. [Google Scholar] [CrossRef] [PubMed]
- Vassileva, M.; Flor-Peregrin, E.; Malusà, E.; Vassilev, N. Towards better understanding of the interactions and efficient application of plant beneficial prebiotics, probiotics, postbiotics and synbiotics. Front. Plant Sci. 2020, 11, 1068. [Google Scholar] [CrossRef] [PubMed]
- Mendes, R.; Kruijt, M.; de Bruijn, I.; Dekkers, E.; van der Voort, M.; Schneider, J.H.; Piceno, Y.M.; De Santis, T.Z.; Andersen, G.L.; Bakker, P.A.; et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011, 332, 1097–1100. [Google Scholar] [CrossRef] [PubMed]
- Yakhin, O.I.; Lubyanov, A.A.; Yakhin, I.A.; Brown, P.H. Biostimulants in plant science: A global perspective. Front. Plant Sci. 2017, 7, 2049. [Google Scholar] [CrossRef]
- De Corato, U.; Patruno, L.; Avella, N.; Lacolla, G.; Cucci, G. Composts from green sources show an increased suppressiveness to soilborne plant pathogenic fungi: Relationships between physicochemical properties, disease suppression, and the microbiome. Crop Prot. 2019, 124, 104870. [Google Scholar] [CrossRef]
- Bellini, A.; Gilardi, G.; Idbella, M.; Zotti, M.; Pugliese, M.; Bonanomi, G.; Gullino, M.L. Trichoderma enriched compost, BCAs and potassium phosphite control Fusarium wilt of lettuce without affecting soil microbiome at genus level. Appl. Soil Ecol. 2023, 182, 104678. [Google Scholar] [CrossRef]
- Cucu, M.A.; Gilardi, G.; Pugliese, M.; Ferrocino, I.; Gullino, M.L. Effects of biocontrol agents and compost against the Phytophthora capsici of zucchini and their impact on the rhizosphere microbiota. Appl. Soil Ecol. 2020, 154, 103659. [Google Scholar] [CrossRef]
- Cucu, M.A.; Gilardi, G.; Pugliese, M.; Matic, S.; Ulrich, G.; Gullino, M.L.; Garibaldi, A. Influence of different biological control agents and compost on total and nitrification driving microbial communities at rhizosphere and soil level in a lettuce—Fusarium oxysporum f. sp. lactucae pathosystem. J. Appl. Microbiol. 2018, 126, 905–918. [Google Scholar] [CrossRef]
- Haas, D.; Keel, C. Regulation of antibiotic production in root-colonizing Pseudomonas spp. and relevance for biological control of plant disease. Ann. Rev. Phytopathol. 2003, 41, 117–153. [Google Scholar] [CrossRef] [PubMed]
- Dukare, A.S.; Prasana, R.; Dubey, S.C.; Nain, L.; Chaudhary, V.; Singh, R.; Saxena, A.K. Evaluating novel microbe amended composts as biocontrol agents in tomato. Crop Prot. 2011, 30, 436–442. [Google Scholar] [CrossRef]
- Vassilev, N.; Martos, E.; Mendes, G.; Martos, V.; Vassileva, M. Biochar of animal origin: A sustainable solution of the high-grade rock phosphate scarcity. J. Sci. Food Agric. 2013, 93, 1799–1804. [Google Scholar] [CrossRef] [PubMed]
- Adam, E.; Groenenboom, A.E.; Kurm, V.; Rajewska, M.; Schmidt, R.; Tyc, O.; Weidner, S.; Berg, G.; De Boer, W.; Salles, J.F. Controlling the microbiome: Microhabitat adjustments for successful biocontrol strategies in soil and human gut. Front. Microbiol. 2016, 7, 1079. [Google Scholar] [CrossRef] [PubMed]
- Bellini, A.; Ferrocino, I.; Cucu, M.A.; Pugliese, M.; Garibaldi, A.; Gullino, M.L. A Compost treatment acts as a suppressive agent in Phytophthora capsici—Cucurbita pepo pathosystem by modifying the rhizosphere microbiota. Front. Plant Sci. 2020, 11, 885. [Google Scholar] [CrossRef]
- Gilardi, G.; Pugliese, M.; Gullino, M.L.; Garibaldi, A. Nursery treatments with resistant inducers, soil amendments and biocontrol agents for the management of the Fusarium wilt of lettuce under glasshouse and field conditions. J. Phytopathol. 2019, 167, 98–110. [Google Scholar] [CrossRef]
- Bonanomi, G.; Antignani, V.; Pane, C.; Scala, F. Suppression of soilborne fungal diseases with organic amendments. J. Plant Pathol. 2007, 8, 311–324. [Google Scholar]
- Srivastava, R.; Khalid, A.; Singh, U.S.; Sharma, A.K. Evaluation of arbuscular mycorrhizal fungus, fluorescent Pseudomonas and Trichoderma harzianum formulation against Fusarium oxysporum f. sp. Lycopersici for the management of tomato wilt. Biol. Control 2010, 53, 24–31. [Google Scholar] [CrossRef]
- Angelopoulou, D.J.; Naska, E.J.; Paplomatas, E.J.; Tjamos, S.E. Biological control agents (BCAs) of V. dahliae wilt: Infuence of application rates and delivery method on plant protection, triggering of host defense mechanisms and rhizosphere populations of BCAs. Plant Pathol. 2014, 63, 1062–1069. [Google Scholar] [CrossRef]
- Berg, G.; Kusstatscher, P.; Abdelfattah, A.; Cernava, T.; Smalla, K. Microbiome modulation—Toward a better understanding of plant microbiome response to microbial inoculants. Front. Microbiol. 2021, 12, 650610. [Google Scholar] [CrossRef]
- Snelders, N.C.; Rovenich, H.; Petti, G.C.; Rocafort, M.; van den Berg, G.; Vorholt, J.A.; Mesters, J.R.; Seidl, M.F.; Nijland, R.; Thomma, B.P. Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins. Nat. Plants 2020, 6, 1365–1374. [Google Scholar] [CrossRef] [PubMed]
- Sommermann, L.; Babin, D.; Behr, J.H.; Chowdhury, S.P.; Sandmann, M.; Windisch, S.; Neumann, G.; Nesme, J.; Sørensen, S.J.; Schellenberg, I.; et al. Long-term fertilization strategy impacts Rhizoctonia solani–microbe interactions in soil and rhizosphere and defense responses in lettuce. Microorganisms 2022, 10, 1717. [Google Scholar] [CrossRef] [PubMed]
- Raaijmakers, J.M.; Mazzola, M. Soil immune responses. Science 2016, 352, 1392–1393. [Google Scholar] [CrossRef] [PubMed]
- Schlatter, D.; Kinkel, L.L.; Thomashow, L.S.; Weller, D.M.; Paulitz, T.C. Disease suppressive soils: New insights from the soil microbiome. Phytopathology 2017, 107, 1284–1297. [Google Scholar] [CrossRef]
- Dandurand, L.M.; Knudsen, G.R. Influence of Pseudomonas fluorescens on hyphal growth and biocontrol activity of Trichoderma harzianum in the spermosphere and rhizosphere of pea. Phytopathology 1993, 83, 265–270. [Google Scholar] [CrossRef]
- Hubbard, J.P.; Harman, G.E.; Hadar, Y. Effects of soilborne Pseudomonas spp. on the biological control agent Trichoderma hamatum, on pea seeds. Phytopathology 1983, 73, 655–659. [Google Scholar] [CrossRef]
- Freeman, S.; Minz, D.; Kolesnik, I.; Barbul, O.; Zveibil, A.; Maymon, M. Trichoderma biocontrol of Colletotrichum acutatum and Botrytis cinerea and survival in strawberry. Eur. J. Plant Pathol. 2004, 110, 361–370. [Google Scholar] [CrossRef]
- Hu, J.; Wei, Z.; Friman, V.P.; Gu, S.H.; Wang, X.F.; Eisenhauer, N.; Yang, T.J.; Ma, J.; Shen, Q.R.; Xu, Y.C.; et al. Probiotic diversity enhances rhizosphere microbiome function and plant disease suppression. mBio 2016, 7, e01790-16. [Google Scholar] [CrossRef]
- Jain, A.; Singh, S.; Sarma, B.K.; Singh, H.B. Microbial consortium-mediated reprogramming of defence network in pea to enhance tolerance against Sclerotinia sclerotiorum. J. Appl. Microbiol. 2012, 112, 537–550. [Google Scholar] [CrossRef]
- Guetsky, R.; Shtienberg, D.; Elad, Y.; Dinoor, A. Combining biocontrol agents to reduce the variability of biological control. Biol. Control 2001, 91, 621–627. [Google Scholar] [CrossRef]
- Kong, H.G.; Song, G.C.; Sim, H.J.; Ryu, C.M. Achieving similar root microbiota composition in neighbouring plants through airborne signalling. ISME J. 2020, 15, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.-C.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef] [PubMed]
- Fredrickson, J.K. Ecology communities by design. Science 2015, 348, 1425–1427. [Google Scholar] [CrossRef]
- Harrier, L.A.; Watson, C.A. The potential role of arbuscular mycorrhizal (AM) fungi in the bioprotection of plants against soil-borne pathogens in organic and/or other sustainable farming systems. Pest Manag. Sci. 2004, 60, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Whipps, J.M. Prospects and limitations for mycorrhizas in biocontrol of root pathogens. Can. J. Bot. 2004, 82, 1198–1227. [Google Scholar] [CrossRef]
- Schouteden, N.; De Waele, D.; Panis, B.; Vos, C.M. Arbuscular mycorrhizal fungi for the biocontrol of plant-parasitic nematodes: A review of the mechanisms involved. Front. Microbiol. 2015, 6, 1280. [Google Scholar] [CrossRef]
- Hodge, A.; Storer, K. Arbuscular mycorrhiza and nitrogen: Implications for individual plants through to ecosystems. Plant Soil 2014, 386, 1–19. [Google Scholar] [CrossRef]
- Brimner, T.A.; Boland, G.J. A review of the non-target effects of fungi used to biologically control plant diseases. Agric. Ecosyst. Environ. 2003, 100, 3–16. [Google Scholar] [CrossRef]
- Commission Regulation (EU) No 546/2011 of 10 June 2011 Implementing Regulation (EC) No 1107/2009 of the European Parliament and of the Council as Regards Uniform Principles for Evaluation and Authorisation of Plant Protection Products. Official Journal of the European Union, L 155/127, 11.6.2011. Available online: http://data.europa.eu/eli/reg/2011/546/oj (accessed on 20 December 2022).
- Zimmermann, G. Review on safety of the entomopathogenic fungi Beauveria bassiana and Beauveria brongniartii. Biocontrol Sci. Technol. 2007, 17, 553–596. [Google Scholar] [CrossRef]
- Zimmermann, G. Review on safety of the entomopathogenic fungus Metarhizium anisopliae. Biocontrol Sci. Technol. 2007, 17, 879–920. [Google Scholar] [CrossRef]
- Bass, M.; Cherrett, J.M. Leaf-cutting ants (Formicidae, Attini) prune their fungus to increase and direct its productivity. Funct. Ecol. 1996, 10, 55–61. [Google Scholar] [CrossRef]
- Demir, S.; Şensoy, S.; Ocak, E.; Tüfenkci, Ş.; Durak, E.D.; Erdinç, Ç.; Ünsal, H. Effects of arbuscular mycorrhizal fungus, humic acid, and whey on wilt diseasecaused by Verticillium dahliae Kleb. in three solanaceous crops. Turk. J. Agric. For. 2015, 39, 15. [Google Scholar] [CrossRef]
- Devi, N.O.; Tombisana Devi, R.K.; Debbarma, M.; Hajong, M.; Thokchom, S. Effect of endophytic Bacillus and arbuscular mycorrhiza fungi (AMF) against Fusarium wilt of tomato caused by Fusarium oxysporum f. sp. lycopersici. Egypt J. Biol. Pest Control 2022, 32, 1. [Google Scholar] [CrossRef]
- Kaiser, C.; Kilburn, M.; Clode, P.; Fuchslueger, L.; Koranda, M.; Cliff, J.B.; Solaiman, Z.M.; Murphy, D. Exploring the transfer of recent plant photosynthates to soil microbes: Mycorrhizal pathway vs direct root exudation. New Phytol. 2015, 205, 1537–1551. [Google Scholar] [CrossRef] [PubMed]


| Advantages | Disadvantages |
|---|---|
|
|
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ptaszek, M.; Canfora, L.; Pugliese, M.; Pinzari, F.; Gilardi, G.; Trzciński, P.; Malusà, E. Microbial-Based Products to Control Soil-Borne Pathogens: Methods to Improve Efficacy and to Assess Impacts on Microbiome. Microorganisms 2023, 11, 224. https://doi.org/10.3390/microorganisms11010224
Ptaszek M, Canfora L, Pugliese M, Pinzari F, Gilardi G, Trzciński P, Malusà E. Microbial-Based Products to Control Soil-Borne Pathogens: Methods to Improve Efficacy and to Assess Impacts on Microbiome. Microorganisms. 2023; 11(1):224. https://doi.org/10.3390/microorganisms11010224
Chicago/Turabian StylePtaszek, Magdalena, Loredana Canfora, Massimo Pugliese, Flavia Pinzari, Giovanna Gilardi, Paweł Trzciński, and Eligio Malusà. 2023. "Microbial-Based Products to Control Soil-Borne Pathogens: Methods to Improve Efficacy and to Assess Impacts on Microbiome" Microorganisms 11, no. 1: 224. https://doi.org/10.3390/microorganisms11010224
APA StylePtaszek, M., Canfora, L., Pugliese, M., Pinzari, F., Gilardi, G., Trzciński, P., & Malusà, E. (2023). Microbial-Based Products to Control Soil-Borne Pathogens: Methods to Improve Efficacy and to Assess Impacts on Microbiome. Microorganisms, 11(1), 224. https://doi.org/10.3390/microorganisms11010224







