Advances in Lactobacillus Restoration for β-Lactam Antibiotic-Induced Dysbiosis: A System Review in Intestinal Microbiota and Immune Homeostasis
Abstract
1. Introduction
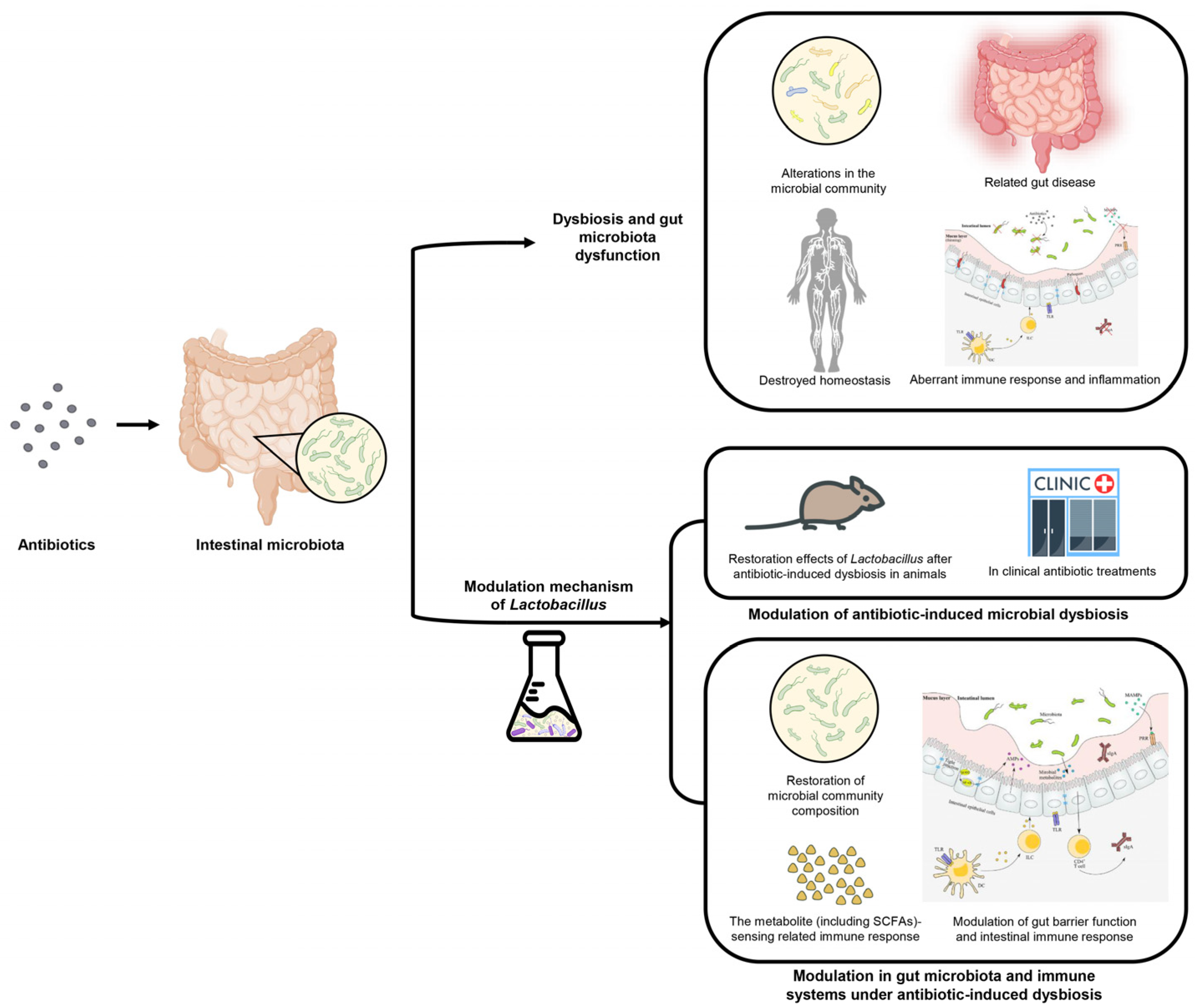
2. The Pathogenesis of Antibiotic-Induced Dysbiosis and Gut Microbiota Dysfunction
2.1. The Antibiotics Cause Diverse Gut Bacteria Community and Destroy Homeostasis
2.2. Antibiotic Treatment Induces Alterations in the Microbial Community and Metabolic System
2.2.1. Loss of Commensal Bacteria
2.2.2. Alteration in Abundance of Specific Taxa
2.2.3. Reduction of Diversity
2.2.4. Induction of the Intestinal Microbial Dysbiosis
2.2.5. Alterations in Microbial Metabolites
2.3. Antibiotic-Induced Ecological Dysfunction and Related Gut Disease
2.3.1. Dysbiosis and Inflammatory Bowel Disease (IBD)
2.3.2. Dysbiosis and Other Gut Disorders
2.3.3. Dysbiosis and Metabolic Disorders
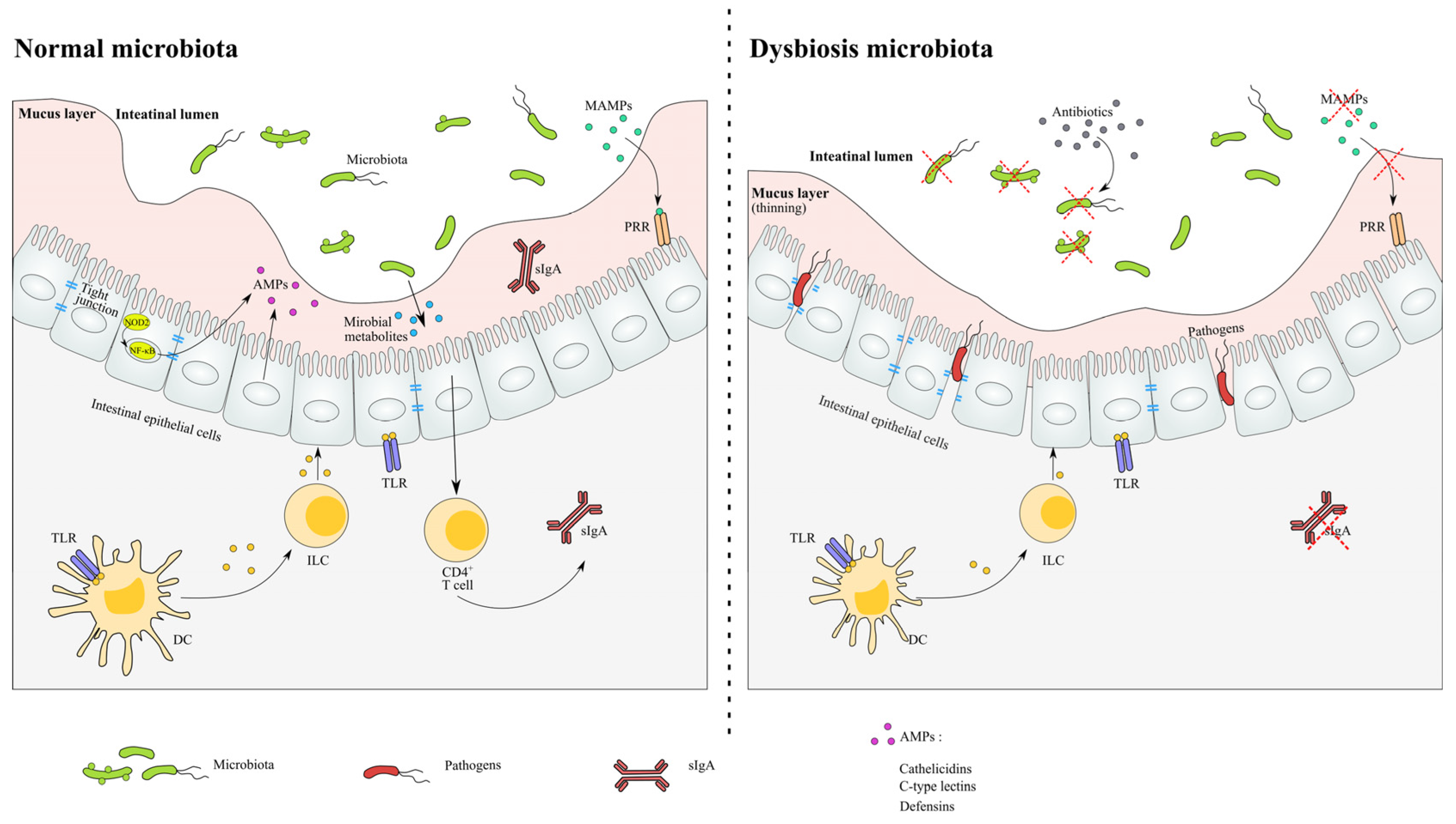
2.4. Antibiotic-Induced Aberrant Immune Response and Inflammation
- In a normal microbiota environment, the homeostasis of the microbial community and host are sustained in relatively stability. The recognition of the microbiota signals through the NF-κB pathway and induces the production of AMPs. The expression of tight junction proteins and the levels of metabolite are relatively constant.
- In antibiotic-induced dysbiosis, the numbers of MAMPs are decreased as a result of the loss of microbiota, and the thinned mucus layer and damaged tight junction proteins lead to the infection of opportunistic pathogens. The levels of SCFA and related immune functions are altered after antibiotic exposure.
3. Modulation of Antibiotic-Induced Microbial Dysbiosis by Lactobacillus
3.1. Restoration Effects of Lactobacillus after Antibiotic-Induced Dysbiosis in Animals
3.2. Roles of Lactobacillus Strains in Clinical Antibiotic Treatments
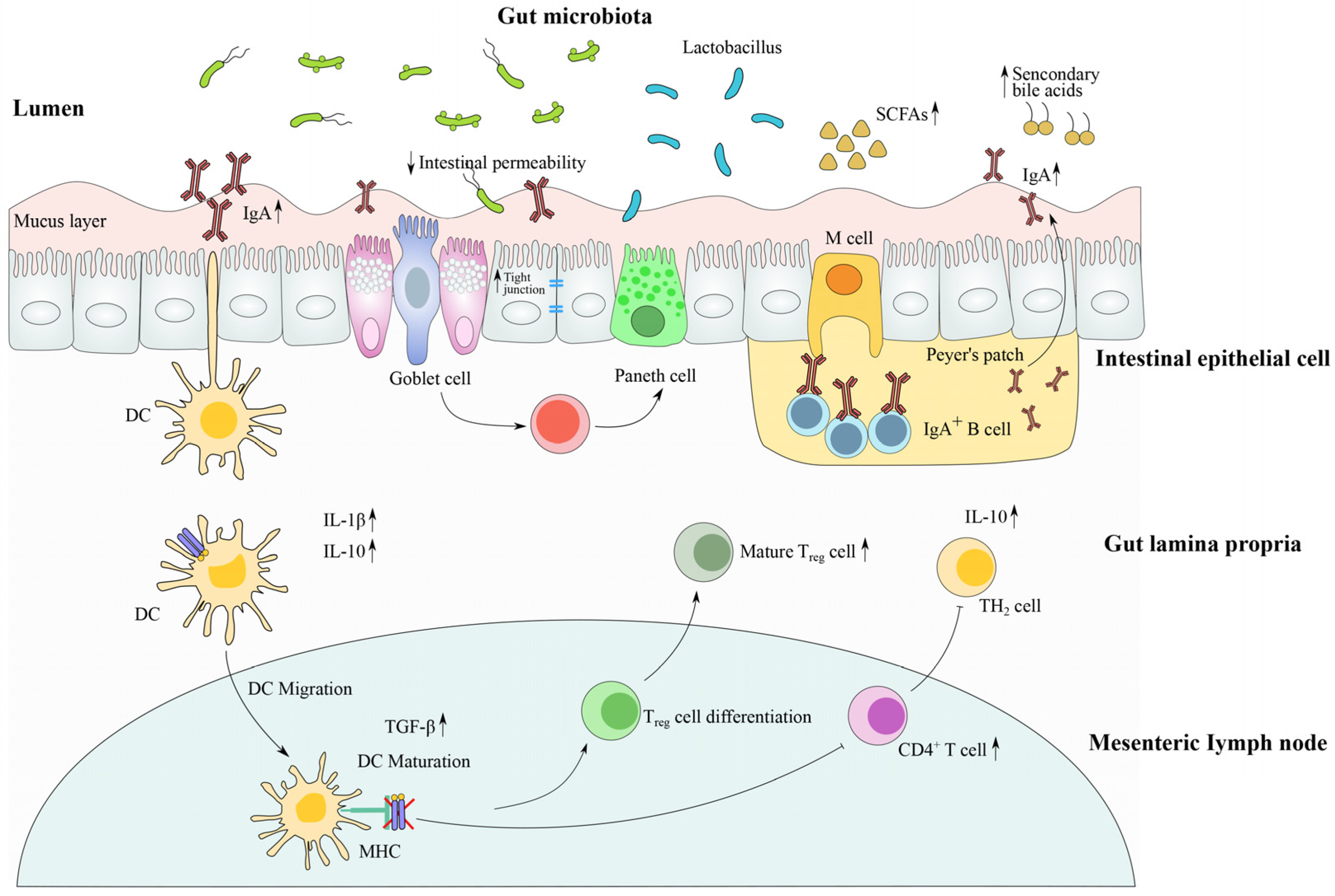
4. The Modulation Mechanism of Lactobacillus in Gut Microbiota and Immune Systems under Antibiotic-Induced Dysbiosis
4.1. Restoration of Microbial Community Composition by Lactobacillus Strains
4.2. Modulation of Gut Barrier Function and Intestinal Immune Response by Lactobacillus Strains
4.3. The Metabolite (Including SCFAs)-Sensing Related Immune Response after Antibiotic Treatment
5. Conclusions and Future Perspectives
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huttenhower, C.; Gevers, D.; Knight, R.; Abubucker, S.; Badger, J.H.; Chinwalla, A.T.; Creasy, H.H.; Earl, A.M.; FitzGerald, M.G.; Fulton, R.S.; et al. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar]
- Nobel, Y.R.; Cox, L.M.; Kirigin, F.F.; Bokulich, N.A.; Yamanishi, S.; Teitler, I.; Chung, J.; Sohn, J.; Barber, C.M.; Goldfarb, D.S.; et al. Metabolic and metagenomic outcomes from early-life pulsed antibiotic treatment. Nat. Commun. 2015, 6, 7486. [Google Scholar] [CrossRef] [PubMed]
- Cho, I.; Yamanishi, S.; Cox, L.; Methe, B.A.; Zavadil, J.; Li, K.; Gao, Z.; Mahana, D.; Raju, K.; Teitler, I.; et al. Antibiotics in early life alter the murine colonic microbiome and adiposity. Nature 2012, 488, 621–634. [Google Scholar] [CrossRef]
- Calder, P.C.; Ortega, E.F.; Meydani, S.N.; Adkins, Y.; Stephensen, C.B.; Thompson, B.; Zwickey, H. Nutrition, Immunosenescence, and Infectious Disease: An Overview of the Scientific Evidence on Micronutrients and on Modulation of the Gut Microbiota. Adv. Nutr. 2022, 13, S1–S26. [Google Scholar] [CrossRef]
- Bear, T.L.K.; Dalziel, J.E.; Coad, J.; Roy, N.C.; Butts, C.A.; Gopal, P.K. The Role of the Gut Microbiota in Dietary Interventions for Depression and Anxiety. Adv. Nutr. 2020, 11, 890–907. [Google Scholar] [CrossRef]
- Sommer, F.; Backhed, F. The gut microbiota—Masters of host development and physiology. Nat. Rev. Microbiol. 2013, 11, 227–238. [Google Scholar] [CrossRef]
- Raghunath, P. Role of Gut Microbiota and Infectious Burden in the Development of Autoimmune and Allergic Diseases. Iran. J. Allergy Asthma Immunol. 2017, 16, 77–78. [Google Scholar]
- Fujimura, K.E.; Slusher, N.A.; Cabana, M.D.; Lynch, S.V. Role of the gut microbiota in defining human health. Expert Rev. Anti Infect. Ther. 2010, 8, 435–454. [Google Scholar] [CrossRef]
- Tsou, A.; Goettel, J.A.; Biswas, A.; Kang, Y.H.; Saltzman, J.; Kelly, R.; Gringauz, J.; Shen, Z.L.; Fox, J.G.; Horwitz, B.; et al. Development of a Reductionist Model to Study Interactions between the Immune System and the Gut Microbiota in a Murine Model of Inflammatory Bowel Disease. Gastroenterology 2017, 152, S111. [Google Scholar] [CrossRef]
- Prosberg, M.; Bendtsen, F.; Vind, I.; Petersen, A.M.; Gluud, L.L. The association between the gut microbiota and the inflammatory bowel disease activity: A systematic review and meta-analysis. Scand. J. Gastroenterol. 2016, 51, 1407–1415. [Google Scholar] [CrossRef]
- Abrahamsson, T.R.; Jakobsson, H.E.; Andersson, A.F.; Bjorksten, B.; Engstrand, L.; Jenmalm, M.C. Low gut microbiota diversity in early infancy precedes asthma at school age. Clin. Exp. Allergy 2014, 44, 842–850. [Google Scholar] [CrossRef]
- Francavilla, R.; Calasso, M.; Calace, L.; Siragusa, S.; Ndagijimana, M.; Vernocchi, P.; Brunetti, L.; Mancino, G.; Tedeschi, G.; Guerzoni, E.; et al. Effect of lactose on gut microbiota and metabolome of infants with cow’s milk allergy. Pediatr. Allergy Immunol. 2012, 23, 420–427. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.B.; Johansen, L.J.; Powell, L.D.; Quig, D.; Rubin, R.A. Gastrointestinal flora and gastrointestinal status in children with autism-comparisons to typical children and correlation with autism severity. BMC Gastroenterol. 2011, 11, 22. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef]
- Korem, T.; Zeevi, D.; Suez, J.; Weinberger, A.; Avnit-Sagi, T.; Pompan-Lotan, M.; Matot, E.; Jona, G.; Harmelin, A.; Cohen, N.; et al. Growth dynamics of gut microbiota in health and disease inferred from single metagenomic samples. Science 2015, 349, 1101–1106. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Smits, S.A.; Tikhonov, M.; Higginbottom, S.K.; Wingreen, N.S.; Sonnenburg, J.L. Diet-induced extinctions in the gut microbiota compound over generations. Nature 2016, 529, 212–228. [Google Scholar] [CrossRef]
- Shi, Y.; Zhao, X.; Zhao, J.; Zhang, H.; Zhai, Q.; Narbad, A.; Chen, W. A mixture of Lactobacillus species isolated from traditional fermented foods promote recovery from antibiotic-induced intestinal disruption in mice. J. Appl. Microbiol. 2018, 124, 842–854. [Google Scholar] [CrossRef]
- Swidsinski, A.; Dorffel, Y.; Loening-Baucke, V.; Theissig, F.; Ruckert, J.C.; Ismail, M.; Rau, W.A.; Gaschler, D.; Weizenegger, M.; Kuhn, S.; et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum. Gut 2011, 60, 34–40. [Google Scholar] [CrossRef]
- Ayres, J.S.; Trinidad, N.J.; Vance, R.E. Lethal inflammasome activation by a multidrug-resistant pathobiont upon antibiotic disruption of the microbiota. Nat. Med. 2012, 18, 799–811. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking Long-Term Dietary Patterns with Gut Microbial Enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Yassour, M.; Vatanen, T.; Siljander, H.; Hämäläinen, A.-M.; Härkönen, T.; Ryhänen, S.J.; Franzosa, E.A.; Vlamakis, H.; Huttenhower, C.; Gevers, D.; et al. Natural history of the infant gut microbiome and impact of antibiotic treatments on strain-level diversity and stability. Sci. Transl. Med. 2016, 8, 343–381. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Gevers, D.; Siljander, H.; Vatanen, T.; Hyotylainen, T.; Hamalainen, A.M.; Peet, A.; Tillmann, V.; Poho, P.; Mattila, I.; et al. The Dynamics of the Human Infant Gut Microbiome in Development and in Progression toward Type 1 Diabetes. Cell Host Microbe 2015, 17, 260–273. [Google Scholar] [CrossRef] [PubMed]
- Monaco, C.L.; Gootenberg, D.B.; Zhao, G.Y.; Handley, S.A.; Ghebremichael, M.S.; Lim, E.S.; Lankowski, A.; Baldridge, M.T.; Wilen, C.B.; Flagg, M.; et al. Altered Virome and Bacterial Microbiome in Human Immunodeficiency Virus-Associated Acquired Immunodeficiency Syndrome. Cell Host Microbe 2016, 19, 311–322. [Google Scholar] [CrossRef]
- Modi, S.R.; Collins, J.J.; Relman, D.A. Antibiotics and the gut microbiota. J. Clin. Investig. 2014, 124, 4212–4218. [Google Scholar] [CrossRef]
- Leffler, D.A.; Lamont, J.T. Clostridium difficile Infection. N. Engl. J. Med. 2015, 372, 1539–1548. [Google Scholar] [CrossRef]
- Cox, L.M.; Yamanishi, S.; Sohn, J.; Alekseyenko, A.V.; Leung, J.M.; Cho, I.; Kim, S.G.; Li, H.; Gao, Z.; Mahana, D.; et al. Altering the Intestinal Microbiota during a Critical Developmental Window Has Lasting Metabolic Consequences. Cell 2014, 158, 705–721. [Google Scholar] [CrossRef]
- Ivanov, I.I.; Frutos, R.D.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific Microbiota Direct the Differentiation of IL-17-Producing T-Helper Cells in the Mucosa of the Small Intestine. Cell Host Microbe 2008, 4, 337–349. [Google Scholar] [CrossRef]
- Xu, B.F.; Liang, S.N.; Zhao, J.Y.; Li, X.T.; Guo, J.Y.; Xin, B.W.; Li, B.L.; Huo, G.C.; Ma, W.W. Bifidobacterium animalis subsp. lactis XLTG11 improves antibiotic-related diarrhea by alleviating inflammation, enhancing intestinal barrier function and regulating intestinal flora. Food Funct. 2022, 13, 6404–6418. [Google Scholar] [CrossRef]
- Zhang, Y.C.; Limaye, P.B.; Renaud, H.J.; Klaassen, C.D. Effect of various antibiotics on modulation of intestinal microbiota and bile acid profile in mice. Toxicol. Appl. Pharm. 2014, 277, 138–145. [Google Scholar] [CrossRef]
- Shi, Y.; Zhai, Q.; Li, D.; Mao, B.; Liu, X.; Zhao, J.; Zhang, H.; Chen, W. Restoration of cefixime-induced gut microbiota changes by Lactobacillus cocktails and fructooligosaccharides in a mouse model. Microbiol. Res. 2017, 200, 14–24. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.J.; Wang, H.J.; Ma, X.J.; Li, Y.; Yang, H.J.; Li, H.; Su, J.R.; Zhang, C.E.; Huang, L.Q. Modulation of gut microbiota and intestinal barrier function during alleviation of antibiotic-associated diarrhea with Rhizoma Zingiber officinale (Ginger) extract. Food Funct. 2020, 11, 10839–10851. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Koenigsknecht, M.J.; Carlson, P.E., Jr.; Hatton, G.E.; Nelson, A.M.; Li, B.; Huffnagle, G.B.; Li, J.Z.; Young, V.B. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun. 2014, 5, 3114. [Google Scholar] [CrossRef]
- Den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Layden, B.T.; Angueira, A.R.; Brodsky, M.; Durai, V.; Lowe, W.L., Jr. Short chain fatty acids and their receptors: New metabolic targets. Transl. Res. 2013, 161, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Yu, D.; Schilter, H.C.; Rolph, M.S.; Mackay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Scher, J.U.; Ubeda, C.; Artacho, A.; Attur, M.; Isaac, S.; Reddy, S.M.; Marmon, S.; Neimann, A.; Brusca, S.; Patel, T. Decreased bacterial diversity characterizes the altered gut microbiota in patients with psoriatic arthritis, resembling dysbiosis in inflammatory bowel disease. Arthritis Rheumatol. 2015, 67, 128–139. [Google Scholar] [CrossRef]
- Duboc, H.; Rajca, S.; Rainteau, D.; Benarous, D.; Maubert, M.-A.; Quervain, E.; Thomas, G.; Barbu, V.; Humbert, L.; Despras, G. Connecting dysbiosis, bile-acid dysmetabolism and gut inflammation in inflammatory bowel diseases. Gut 2012, 62, 531–539. [Google Scholar] [CrossRef]
- Walker, A.W.; Sanderson, J.D.; Churcher, C.; Parkes, G.C.; Hudspith, B.N.; Rayment, N.; Brostoff, J.; Parkhill, J.; Dougan, G.; Petrovska, L. High-throughput clone library analysis of the mucosa-associated microbiota reveals dysbiosis and differences between inflamed and non-inflamed regions of the intestine in inflammatory bowel disease. BMC Microbiol. 2011, 11, 7. [Google Scholar] [CrossRef]
- Baumgart, D.C.; Carding, S.R. Inflammatory bowel disease: Cause and immunobiology. Lancet 2007, 369, 1627–1640. [Google Scholar] [CrossRef] [PubMed]
- Joossens, M.; Huys, G.; Cnockaert, M.; De Preter, V.; Verbeke, K.; Rutgeerts, P.; Vandamme, P.; Vermeire, S. Dysbiosis of the faecal microbiota in patients with Crohn’s disease and their unaffected relatives. Gut 2011, 60, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef]
- Carroll, I.M.; Chang, Y.H.; Park, J.; Sartor, R.B.; Ringel, Y. Luminal and mucosal-associated intestinal microbiota in patients with diarrhea-predominant irritable bowel syndrome. Gut Pathog. 2010, 2, 19. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Nadal, I.; Medina, M.; Donat, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010, 10, 63. [Google Scholar] [CrossRef]
- Shen, X.J.; Rawls, J.F.; Randall, T.; Burcal, L.; Mpande, C.N.; Jenkins, N.; Jovov, B.; Abdo, Z.; Sandler, R.S.; Keku, T.O. Molecular characterization of mucosal adherent bacteria and associations with colorectal adenomas. Gut Microbes 2010, 1, 138–147. [Google Scholar] [CrossRef]
- Olivares, M.; Neef, A.; Castillejo, G.; De Palma, G.; Varea, V.; Capilla, A.; Palau, F.; Nova, E.; Marcos, A.; Polanco, I.; et al. The HLA-DQ2 genotype selects for early intestinal microbiota composition in infants at high risk of developing coeliac disease. Gut 2015, 64, 406–417. [Google Scholar] [CrossRef]
- Kaliannan, K.; Wang, B.; Li, X.-Y.; Bhan, A.K.; Kang, J.X. Omega-3 fatty acids prevent early-life antibiotic exposure-induced gut microbiota dysbiosis and later-life obesity. Int. J. Obes. 2016, 40, 1039–1042. [Google Scholar] [CrossRef]
- Den Besten, G.; Bleeker, A.; Gerding, A.; van Eunen, K.; Havinga, R.; van Dijk, T.H.; Oosterveer, M.H.; Jonker, J.W.; Groen, A.K.; Reijngoud, D.J.; et al. Short-Chain Fatty Acids Protect Against High-Fat Diet-Induced Obesity via a PPAR-Dependent Switch From Lipogenesis to Fat Oxidation. Diabetes 2015, 64, 2398–2408. [Google Scholar] [CrossRef]
- Teixeira, T.F.S.; Collado, M.C.; Ferreira, C.L.L.F.; Bressan, J.; Peluzio, M.D.G. Potential mechanisms for the emerging link between obesity and increased intestinal permeability. Nutr. Res. 2012, 32, 637–647. [Google Scholar] [CrossRef]
- Qin, J.J.; Li, Y.R.; Cai, Z.M.; Li, S.H.; Zhu, J.F.; Zhang, F.; Liang, S.S.; Zhang, W.W.; Guan, Y.L.; Shen, D.Q.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.W.; Ince, J.; Duncan, S.H.; Webster, L.M.; Holtrop, G.; Ze, X.L.; Brown, D.; Stares, M.D.; Scott, P.; Bergerat, A.; et al. Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J. 2011, 5, 220–230. [Google Scholar] [CrossRef] [PubMed]
- Vrieze, A.; Van Nood, E.; Holleman, F. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 2013, 144, 250. [Google Scholar] [CrossRef] [PubMed]
- Larsson, E.; Tremaroli, V.; Lee, Y.S.; Koren, O.; Nookaew, I.; Fricker, A.; Nielsen, J.; Ley, R.E.; Backhed, F. Analysis of gut microbial regulation of host gene expression along the length of the gut and regulation of gut microbial ecology through MyD88. Gut 2012, 61, 1124–1131. [Google Scholar] [CrossRef]
- Dessein, R.; Gironella, M.; Vignal, C.; Peyrinbiroulet, L.; Sokol, H.; Secher, T.; Lacasgervais, S.; Gratadoux, J.J.; Lafont, F.; Dagorn, J.C. Toll-like receptor 2 is critical for induction of Reg3 beta expression and intestinal clearance of Yersinia pseudotuberculosis. Gut 2009, 58, 771–776. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. The gut microbiota shapes intestinal immune responses during health and disease. Nat. Rev. Immunol. 2009, 9, 600–610. [Google Scholar] [CrossRef]
- Brandl, K.; Plitas, G.; Mihu, C.N.; Ubeda, C.; Jia, T.; Fleisher, M.; Schnabl, B.; DeMatteo, R.P.; Pamer, E.G. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 2008, 455, 804–808. [Google Scholar] [CrossRef]
- Deshmukh, H.S.; Liu, Y.H.; Menkiti, O.R.; Mei, J.J.; Dai, N.; O’Leary, C.E.; Oliver, P.M.; Kolls, J.K.; Weiser, J.N.; Worthen, G.S. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 2014, 20, 528–534. [Google Scholar] [CrossRef]
- Hill, D.A.; Siracusa, M.C.; Abt, M.C.; Kim, B.S.; Kobuley, D.; Kubo, M.; Kambayashi, T.; LaRosa, D.F.; Renner, E.D.; Orange, J.S.; et al. Commensal bacteria-derived signals regulate basophil hematopoiesis and allergic inflammation. Nat. Med. 2012, 18, 538–546. [Google Scholar] [CrossRef]
- Ichinohe, T.; Pang, I.K.; Kumamoto, Y.; Peaper, D.R.; Ho, J.H.; Murray, T.S.; Iwasaki, A. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc. Natl. Acad. Sci. USA 2011, 108, 5354–5359. [Google Scholar] [CrossRef]
- Abt, M.C.; Osborne, L.C.; Monticelli, L.A.; Doering, T.A.; Alenghat, T.; Sonnenberg, G.F.; Paley, M.A.; Antenus, M.; Williams, K.L.; Erikson, J.; et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity 2012, 37, 158–170. [Google Scholar] [CrossRef]
- Hill, D.A.; Hoffmann, C.; Abt, M.C.; Du, Y.; Kobuley, D.; Kirn, T.J.; Bushman, F.D.; Artis, D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010, 3, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.M.; Gurry, T.; Lampe, J.W.; Chakrabarti, A.; Dam, V.; Everard, A.; Goas, A.; Gabriele, G.; Kleerebez, M.; Lane, J.; et al. Perspective: Leveraging the Gut Microbiota to Predict Personalized Responses to Dietary, Prebiotic, and Probiotic Interventions. Adv. Nutr. 2022, 13, 1450–1461. [Google Scholar] [CrossRef]
- Cresci, G.; Nagy, L.E.; Ganapathy, V. Lactobacillus GG and Tributyrin Supplementation Reduce Antibiotic-Induced Intestinal Injury. J. Parenter. Enteral Nutr. 2013, 37, 763–774. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Kellingray, L.; Le Gall, G.; Zhao, J.; Zhang, H.; Narbad, A.; Zhai, Q.; Chen, W. The divergent restoration effects of Lactobacillus strains in antibiotic-induced dysbiosis. J. Funct. Foods 2018, 51, 142–152. [Google Scholar] [CrossRef]
- Crouzet, L.; Derrien, M.; Cherbuy, C.; Plancade, S.; Foulon, M.; Chalin, B.; Vlieg, J.E.T.V.; Grompone, G.; Rigottier-Gois, L.; Serror, P. Lactobacillus paracasei CNCM I-3689 reduces vancomycin-resistant Enterococcus persistence and promotes Bacteroidetes resilience in the gut following antibiotic challenge. Sci. Rep. 2018, 8, 5098. [Google Scholar] [CrossRef] [PubMed]
- Schepper, J.D.; Collins, F.L.; Rios-Arce, N.D.; Raehtz, S.; Schaefer, L.; Gardinier, J.D.; Britton, R.A.; Parameswaran, N.; McCabe, L.R. Probiotic Lactobacillus reuteri prevents postantibiotic bone loss by reducing intestinal dysbiosis and preventing barrier disruption. J. Bone Miner. Res. 2019, 34, 681–698. [Google Scholar] [CrossRef]
- Daisley, B.A.; Pitek, A.P.; Chmiel, J.A.; Gibbons, S.; Chernyshova, A.M.; Al, K.F.; Faragalla, K.M.; Burton, J.P.; Thompson, G.J.; Reid, G. Lactobacillus spp. attenuate antibiotic-induced immune and microbiota dysregulation in honey bees. Commun. Biol. 2020, 3, 534. [Google Scholar] [CrossRef]
- Cimperman, L.; Bayless, G.; Best, K.; Diligente, A.; Mordarski, B.; Oster, M.; Smith, M.; Vatakis, F.; Wiese, D.; Steiber, A.; et al. A randomized, double-blind, placebo-controlled pilot study of Lactobacillus reuteri ATCC 55730 for the prevention of antibiotic-associated diarrhea in hospitalized adults. J. Clin. Gastroenterol. 2011, 45, 785–789. [Google Scholar] [CrossRef]
- Do, A.D.; Su, C.H.; Hsu, Y.M. Antagonistic Activities of Lactobacillus rhamnosus JB3 Against Helicobacter pylori Infection Through Lipid Raft Formation. Front. Immunol. 2022, 12, 796177. [Google Scholar] [CrossRef]
- Lonnermark, E.; Friman, V.; Lappas, G.; Sandberg, T.; Berggren, A.; Adlerberth, I. Intake of Lactobacillus plantarum Reduces Certain Gastrointestinal Symptoms During Treatment With Antibiotics. J. Clin. Gastroenterol. 2010, 44, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Suez, J.; Zmora, N.; Zilberman-Schapira, G.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.; Federici, S.; et al. Post-antibiotic gut mucosal microbiome reconstitution is impaired by probiotics and improved by autologous FMT. Cell 2018, 174, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Barker, A.K.; Duster, M.; Valentine, S.; Hess, T.; Archbald-Pannone, L.; Guerrant, R.; Safdar, N. A randomized controlled trial of probiotics for Clostridium difficile infection in adults (PICO). J. Antimicrob. Chemother. 2017, 72, 3177–3180. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.W.; Mubasher, M.; Fang, C.Y.; Reifer, C.; Miller, L.E. Dose-Response Efficacy of a Proprietary Probiotic Formula of Lactobacillus acidophilus CL1285 and Lactobacillus casei LBC80R for Antibiotic-Associated Diarrhea and Clostridium difficile-Associated Diarrhea Prophylaxis in Adult Patients. Am. J. Gastroenterol. 2010, 105, 1636–1641. [Google Scholar] [CrossRef] [PubMed]
- Helps, A.; Bell, E.; Mactier, R. Prospective randomized doubleblind study of efficacy of probiotic milk drink in reducing the incidence of antibiotic-associated diarrhoea and Clostridium difficile diarrhoea. Int. J. Probiotics Prebiotics 2015, 10, 145–152. [Google Scholar]
- Francavilla, R.; Polimeno, L.; Demichina, A.; Maurogiovanni, G.; Principi, B.; Scaccianoce, G.; Ierardi, E.; Russo, F.; Riezzo, G.; Di Leo, A.; et al. Lactobacillus reuteri Strain Combination in Helicobacter pylori Infection A Randomized, Double-Blind, Placebo-Controlled Study. J. Clin. Gastroenterol. 2014, 48, 407–413. [Google Scholar] [CrossRef]
- Yang, B.J.; Liu, C.Y.; Huang, Y.N.; Wu, Q.W.; Xiong, Y.X.; Yang, X.F.; Hu, S.L.; Jiang, Z.Y.; Wang, L.; Yi, H.B. The Responses of Lactobacillus reuteri LR1 or Antibiotic on Intestinal Barrier Function and Microbiota in the Cecum of Pigs. Front. Microbiol. 2022, 13, 877297. [Google Scholar] [CrossRef]
- Folwarski, M.; Dobosz, M.; Malgorzewicz, S.; Skonieczna-Zydecka, K.; Kazmierczak-Siedlecka, K. Effects of Lactobacillus rhamnosus GG on early postoperative outcome after pylorus-preserving pancreatoduodenectomy: A randomized trial. Eur. Rev. Med. Pharmacol. Sci. 2021, 25, 397–405. [Google Scholar]
- Liang, H.; Miao, Z.; Guo, J.; Jiang, F.; Cheng, R.; Shen, X.; Li, M.; He, F. Alleviation and recovery effects of Lactobacillus on the antibiotic-induced intestinal dysbiosis in early life stages of mice. J. Hyg. Res. 2020, 49, 873–880. [Google Scholar]
- Wan, Q.-Y.; Zhao, R.; Wang, Y.; Wu, Y.; Wu, X.-T. Antibiotic use and risk of colorectal cancer: A meta-analysis of 412,450 participants. Gut 2020, 69, 2059–2060. [Google Scholar] [CrossRef]
- Patrick, D.M.; Sbihi, H.; Dai, D.L.Y.; Al Mamun, A.; Rasali, D.; Rose, C.; Marra, F.; Boutin, R.C.T.; Petersen, C.; Stiemsma, L.T.; et al. Decreasing antibiotic use, the gut microbiota, and asthma incidence in children: Evidence from population-based and prospective cohort studies. Lancet Respir. Med. 2020, 20, 2213–2600. [Google Scholar] [CrossRef] [PubMed]
- Keerthisinghe, T.P.; Wang, F.; Wang, M.; Yang, Q.; Li, J.; Yang, J.; Xi, L.; Dong, W.; Fang, M. Long-term exposure to TET increases body weight of juvenile zebrafish as indicated in host metabolism and gut microbiome. Environ. Int. 2020, 139, 105705. [Google Scholar] [CrossRef]
- Kappel, B.A.; De Angelis, L.; Heiser, M.; Ballanti, M.; Stoehr, R.; Goettsch, C.; Mavilio, M.; Artati, A.; Paoluzi, O.A.; Adamski, J.; et al. Cross-omics analysis revealed gut microbiome-related metabolic pathways underlying atherosclerosis development after antibiotics treatment. Mol. Metab. 2020, 36, 100976. [Google Scholar] [CrossRef]
- Zhang, W.D.; Zhu, B.; Xu, J.H.; Liu, Y.Y.; Qiu, E.Q.; Li, Z.J.; Li, Z.C.; He, Y.; Zhou, H.W.; Bai, Y.; et al. Bacteroides fragilis Protects Against Antibiotic-Associated Diarrhea in Rats by Modulating Intestinal Defenses. Front. Immunol. 2018, 9, 1040. [Google Scholar] [CrossRef] [PubMed]
- Grazul, H.; Kanda, L.L.; Gondek, D. Impact of probiotic supplements on microbiome diversity following antibiotic treatment of mice. Gut Microbes 2016, 7, 101–114. [Google Scholar] [CrossRef]
- Evans, M.; Salewski, R.P.; Christman, M.C.; Girard, S.A.; Tompkins, T.A. Effectiveness of Lactobacillus helveticus and Lactobacillus rhamnosus for the management of antibiotic-associated diarrhoea in healthy adults: A randomised, double-blind, placebo-controlled trial. Br. J. Nutr. 2016, 116, 94–103. [Google Scholar] [CrossRef] [PubMed]
- Mekonnen, S.A.; Merenstein, D.; Fraser, C.M.; Marco, M.L. Molecular mechanisms of probiotic prevention of antibiotic-associated diarrhea. Curr. Opin. Biotechnol. 2020, 61, 226–234. [Google Scholar] [CrossRef]
- Henry, D.E.; Venkateswara Rao, V. Antibiotic-associated diarrhea and update on probiotics recommendations. In Probiotic Research in Therapeutics: Volume 2: Modulation of Gut Flora: Management of Inflammation and Infection Related Gut Etiology; Pawar, S.V., Rishi, P., Eds.; Springer: Singapore, 2021; pp. 141–166. [Google Scholar]
- Maier, L.; Goemans, C.V.; Wirbel, J.; Kuhn, M.; Eberl, C.; Pruteanu, M.; Muller, P.; Garcia-Santamarina, S.; Cacace, E.; Zhang, B.Y.; et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature 2021, 599, 120–134. [Google Scholar] [CrossRef]
- Liao, W.; Chen, C.; Wen, T.; Zhao, Q. Probiotics for the prevention of antibiotic-associated diarrhea in adults: A meta-analysis of randomized placebo-controlled trials. J. Clin. Gastroenterol. 2021, 55, 469–480. [Google Scholar] [CrossRef]
- Liu, L.; Kirst, M.E.; Zhao, L.; Li, E.; Wang, G.P.; Woodworth, M.H. Microbiome resilience despite a profound loss of minority microbiota following clindamycin challenge in humanized gnotobiotic mice. Microbiol. Spectr. 2022, 10, e01960-21. [Google Scholar] [CrossRef]
- Tan, R.; Jin, M.; Shao, Y.F.; Yin, J.; Li, H.B.; Chen, T.J.; Shi, D.Y.; Zhou, S.Q.; Li, J.W.; Yang, D. High-sugar, high-fat, and high-protein diets promote antibiotic resistance gene spreading in the mouse intestinal microbiota. Gut Microbes 2022, 14, 2022442. [Google Scholar] [CrossRef] [PubMed]
- Roodgar, M.; Good, B.H.; Garud, N.R.; Martis, S.; Avula, M.; Zhou, W.Y.; Lancaster, S.M.; Lee, H.; Babveyh, A.; Nesamoney, S.; et al. Longitudinal linked-read sequencing reveals ecological and evolutionary responses of a human gut microbiome during antibiotic treatment. Genome Res. 2021, 31, 1433–1445. [Google Scholar] [CrossRef] [PubMed]
- Pennycook, J.H.; Scanlan, P.D. Ecological and Evolutionary responses to Antibiotic Treatment in the Human Gut Microbiota. FEMS Microbiol. Rev. 2021, 45, fuab018. [Google Scholar] [CrossRef]
- Martel, J.; Chang, S.H.; Ko, Y.F.; Hwang, T.L.; Young, J.D.; Ojcius, D.M. Gut barrier disruption and chronic disease. Trends Endocrinol. Metab. 2022, 33, 247–265. [Google Scholar] [CrossRef] [PubMed]
- Konig, J.; Wells, J.; Cani, P.D.; Garcia-Rodenas, C.L.; MacDonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.J. Human intestinal barrier function in health and disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef] [PubMed]
- Vangay, P.; Ward, T.; Gerber, J.S.; Knights, D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe 2015, 17, 553–564. [Google Scholar] [CrossRef]
- Evanovich, E.; de Souza Mendonça Mattos, P.J.; Guerreiro, J.F. Comparative genomic analysis of Lactobacillus plantarum: An overview. Int. J. Genom. 2019, 2019, 4973214. [Google Scholar]
- Pratap, K.; Taki, A.C.; Johnston, E.B.; Lopata, A.L.; Kamath, S.D. A Comprehensive Review on Natural Bioactive Compounds and Probiotics as Potential Therapeutics in Food Allergy Treatment. Front. Immunol. 2020, 11, 996. [Google Scholar] [CrossRef]
- Marinelli, L.; Tenore, G.C.; Novellino, E. Probiotic species in the modulation of the anticancer immune response. Semin. Cancer Biol. 2017, 46, 182–190. [Google Scholar] [CrossRef]
- Yao, G.Q.; Cao, C.X.; Zhang, M.; Kwok, L.Y.; Zhang, H.P.; Zhang, W.Y. Lactobacillus casei Zhang exerts probiotic effects to antibiotic-treated rats. Comput. Struct. Biotechnol. 2021, 19, 5888–5897. [Google Scholar] [CrossRef]
- Balasubramanian, D.; López-Pérez, M.; Grant, T.-A.; Ogbunugafor, C.B.; Almagro-Moreno, S. Molecular mechanisms and drivers of pathogen emergence. Trends Microbiol. 2022, 30, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly, Y.M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Sun, J.; Furio, L.; Mecheri, R.; van der Does, A.M.; Lundeberg, E.; Saveanu, L.; Chen, Y.; van Endert, P.; Agerberth, B.; Diana, J. Pancreatic beta-Cells Limit Autoimmune Diabetes via an Immunoregulatory Antimicrobial Peptide Expressed under the Influence of the Gut Microbiota. Immunity 2015, 43, 304–317. [Google Scholar] [CrossRef] [PubMed]
- Trompette, A.; Gollwitzer, E.S.; Yadava, K.; Sichelstiel, A.K.; Sprenger, N.; Ngom-Bru, C.; Blanchard, C.; Junt, T.; Nicod, L.P.; Harris, N.L.; et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 2014, 20, 159–166. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2012, 469, 543–547. [Google Scholar] [CrossRef]
- Zhu, Z.M.; Zhu, L.Y.; Jiang, L. Dynamic regulation of gut Clostridium-derived short-chain fatty acids. Trends Biotechnol. 2022, 40, 266–270. [Google Scholar] [CrossRef]
- Jiao, W.; Zhang, Z.; Xu, Y.; Gong, L.; Zhang, W.; Tang, H.; Zeng, S.; Zhang, Q.; Sun, Z.; Liu, L.; et al. Butyric acid normalizes hyperglycemia caused by the tacrolimus-induced gut microbiota. Am. J. Transplant. 2020, 20, 2413–2424. [Google Scholar] [CrossRef]
| Antibiotics | Spectrum | Effects on Microbiota | Risk of Dysbiosis | Outcome of Diseases or Immunity | Ref |
|---|---|---|---|---|---|
| Penicillin | Narrow, Gram-positive | Altered the composition, increased Lachnospiraceae | Moderate | Increased adiposity and hormone levels in mice | [3] |
| Penicillin | Narrow, Gram-positive | Altered microbial community composition, reduced Lactobacillus, Candidatus Arthromitus and Allobaculum levels | High | Enhanced the effect of high-fat diet induced obesity and affected ileal genes expression involved in immunity | [28] |
| Ampicillin | Broad, Gram-positive and some Gram-negative | Decreased microbial community diversity, loss of Akkermansia, Eubacterium, Alistipes and increase of Staphylococcus, Acinetobacter, Enterococcus | Moderate | increased gut permeability, increased the production of inflammatory cytokines including TNF-α, IFN-γ, IL-6, MCP-1 and IL-1β in the ileum | [19] |
| Ampicillin | Broad, Gram-positive and some Gram-negative | Depleted segmented filamentous bacteria and Gram-positive bacteria | Moderate | Depletion of Th 17 cells | [29] |
| Amoxycillin | Broad, Gram-positive and some Gram-negative | Changed the microbial composition, Depletion of Lactobacillus | Moderate | Decreased expression of MHC molecules and increased mast cell proteases | [30] |
| Amoxycillin | Broad, Gram-positive and some Gram-negative | Increased Clostridium clostriforme, Eubacterium desmolans, Porphyromonas, Bacillus mycoides, Helicobacter, Rumniococcus gnavus and R. schinkii | Not studied | [31] | |
| Amoxycillin | Broad, Gram-positive and some Gram-negative | Decreased richness and Shannon evenness, no significant difference in community structure | Mild | Influenced microbial oxalate-degrading capacity | [2] |
| Cefixime | Broad, Gram-positive and Gram-negative | Reduction in the diversity of the microbial community and led to decreasing to one preponderant Firmicutes | High | Decreased the production of short-chain fatty acids and induced intestinal inflammation | [32] |
| Cefoperazone | Broad, Gram-positive and Gram-negative | Reduced the total number of bacteria, allowing overgrowth of Candida albicans | High | Allergic-airways disease develops after challenge with mould spores | [33] |
| Cefoperazone | Broad, Gram-positive and Gram-negative | Induced substantial changes in gut microbial community and susceptible to C. difficile infection | High | Modified metabolic activity: decreased the levels of glucose, secondary bile acids, free fatty acids and dipeptides | [34] |
| Experimental Design | Strains, Dosage and Duration | Antibiotic | Effects of Lactobacillus in the Gut Microbiota and Metabolites | Effects of Lactobacillus in the Immune Ecology | Ref |
|---|---|---|---|---|---|
| Mice | |||||
| C57BL/6 mice aged 6-8 weeks received a chow diet for 7 days with broad-spectrum antibiotics | L. rhamnosus GG | Metronidazole, neomycin sulfate andvancomycin | ND | Minimized the decrease expression of butyrate transporter and receptor, and tight junction proteins caused by antibiotic. Decreased GPR109a, SLC5A8, AQP4, and NHE3 transcripts | [64] |
| Mice receive oral gavage with either cefixime (50 mg/kg) or high-dose cefixime (150 mg/kg) | Cocktails of L. plantarum, L. casei, L. rhamnosus, and L. helveticus | Cefixime | Recovered composition of microbiota, enhanced abundance of Firmicutes, decreased Bacteroidetes, Proteobacteria | Decreased serum C-reactive protein, complement 3, and IgG | [32] |
| Four-week-old male C57BL/6J mice | Cocktails of L. casei L. plantarum, L. rhamnosus, and L. helveticus | Ampicillin | Modulated the microbiota community structure and promoted the abundance of Akkermansia | Increased the expression of tight-junction proteins, reducing the production of TNF-α, IL-6, MCP-1, IFN-γ and IL-1β in the ileum and the colon | [19] |
| Four-week-old male C57BL/6 mice | L. casei CGMCC 12,435 (LacC), L. plantarum CGMCC 12436 (LacP), or L. rhamnosus GG (LacG), respectively | 500 mg/kg/day Ampicillin | LacC strain enhanced the alpha diversity and levels of Bacteriodetes and SCFAs | LacC strain enhanced the ileum ZO-1, occluding, down-regulated the expression of NF-κB p65 and modulated the ampicillin-induced inflammatory responses | [65] |
| Antibiotic-induced microbiota dysbiosis mice with enterococci overgrowth and vancomycin-resistant enterococci persistence | L. paracasei CNCM I-3689 | 1.4 mg/day of clindamycin | Recovery of members of the phylum Bacteroidetes | Increased level of lithocholate and of ileal expression of camp (human LL-37) | [66] |
| Eleven-week-old male BALB/c mice | 3.3 × 108 CFU/mL of either L. reuteri 6475 (LR) or L. rhamnosus (LGG), | Ampicillin 1.0 g/L and neomycin 0.5 g/L | L. reuteri but not L. rhamnosus GG reduced the post-antibiotic elevation of the Firmicutes: Bacteroidetes ratio | Increased intestinal permeability, and notably reduced femoral trabecular bone volume (approximately 30%, p < 0.01) | [67] |
| Other animals | |||||
| Apis mellifera | Three immunostimulatory Lactobacillus strains | Oxytetracycline | Mitigate antibiotic-associated microbiota dysbiosis | Alleviated immune deficits | [68] |
| Humans | |||||
| 31 In-patients receiving antibiotics | L. reuteri, 2 × 108 CFU, 28 days | Not specific | Decreased AAD among hospitalized adults | ND | [69] |
| 66 subjects screening positive for H. pylori infection | L. rhamnosus, 1.2 × 1010 CFU 7 days | H. pylori eradication | Prevent or minimize the gastrointestinal side-effect burden | ND | [70] |
| Patients treated for infections at an infectious diseases clinic | L. plantarum 299v, 1 × 1010 CFU, 2 weeks | Not specific | risk of developing loose or watery stools was significantly lower | ND | [71] |
| Post-antibiotic reconstitution of the gut mucosal host-microbiome niche | Strain cocktails including Lactobacillus, Bifidobacterium, Lactococcus and Streptococcus | Ciprofloxacin and metronidazole | Decreased abundance of Clostridiales, recovered microbiome structure | Enhanced expression of ileum REG3G and colon IL1B | [72] |
| 33 participants in patients with an initial mild to moderate C. difficile infection | L. acidophilus NCFM, L. paracasei Lpc-37 | Metronidazole; Vancomycinor | Decreased levels of Verrucomicrobiaceae and Bacteroides | ND | [73] |
| Double-blind, placebo-controlled dose-ranging study, 255 adult inpatients | Mixture of L. acidophilus and L. casei, 5 × 1010 or 1 × 1011 CFU, 26 d | One of penicillin, cephalosporin, or clindamycin | Lower antibiotic-associated diarrhea and Clostridium difficile-associated diarrhea incidence | ND | [74] |
| double-blind randomized placebo-controlled trial 85 inpatients | L. casei, Shirota, 1.3 × 1010 CFU, 12 weeks | Not specific | No improvement | ND | [75] |
| 100 H. pylori-positive naive patients | A combination of 2 strains of L. Reuteri 2 × 108 CFU, 7 days | Helicobacter pylori | L. reuteri combination increased eradication rate by 9.1%, and it determines a significant reduction in antibiotic-associated side effects | ND | [76] |
| Probiotic (n = 80) or placebo (n = 80) intervention in healthy adults receiving antibiotics | A combination of L. helveticus and L. rhamnosus, 0·4 × 109 and 7.6 × 109 CFU, 14 days, | Amoxicillin clavulanic acid | Probiotic supplementation reduced the duration of diarrhea-like defecations | ND | [77] |
| 302 hospitalized patients receiving antibiotics | Lactobacillus GG 1 × 1010 CFU 14 d | Not specific | Lactobacillus GG had no obvious improvement in reducing the rate of occurrence of diarrhea in this sample of 267 adult patients | ND | [78] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shi, Y.; Luo, J.; Narbad, A.; Chen, Q. Advances in Lactobacillus Restoration for β-Lactam Antibiotic-Induced Dysbiosis: A System Review in Intestinal Microbiota and Immune Homeostasis. Microorganisms 2023, 11, 179. https://doi.org/10.3390/microorganisms11010179
Shi Y, Luo J, Narbad A, Chen Q. Advances in Lactobacillus Restoration for β-Lactam Antibiotic-Induced Dysbiosis: A System Review in Intestinal Microbiota and Immune Homeostasis. Microorganisms. 2023; 11(1):179. https://doi.org/10.3390/microorganisms11010179
Chicago/Turabian StyleShi, Ying, Jiaqi Luo, Arjan Narbad, and Qihe Chen. 2023. "Advances in Lactobacillus Restoration for β-Lactam Antibiotic-Induced Dysbiosis: A System Review in Intestinal Microbiota and Immune Homeostasis" Microorganisms 11, no. 1: 179. https://doi.org/10.3390/microorganisms11010179
APA StyleShi, Y., Luo, J., Narbad, A., & Chen, Q. (2023). Advances in Lactobacillus Restoration for β-Lactam Antibiotic-Induced Dysbiosis: A System Review in Intestinal Microbiota and Immune Homeostasis. Microorganisms, 11(1), 179. https://doi.org/10.3390/microorganisms11010179






