Virulence Factors of the Periodontal Pathogens: Tools to Evade the Host Immune Response and Promote Carcinogenesis
Abstract
1. Introduction
2. Virulence Factors of Periodontal Pathogens and Their Association with Immune Escape and Carcinogenesis
2.1. A. actinomycetemcomitans
2.2. P. gingivalis
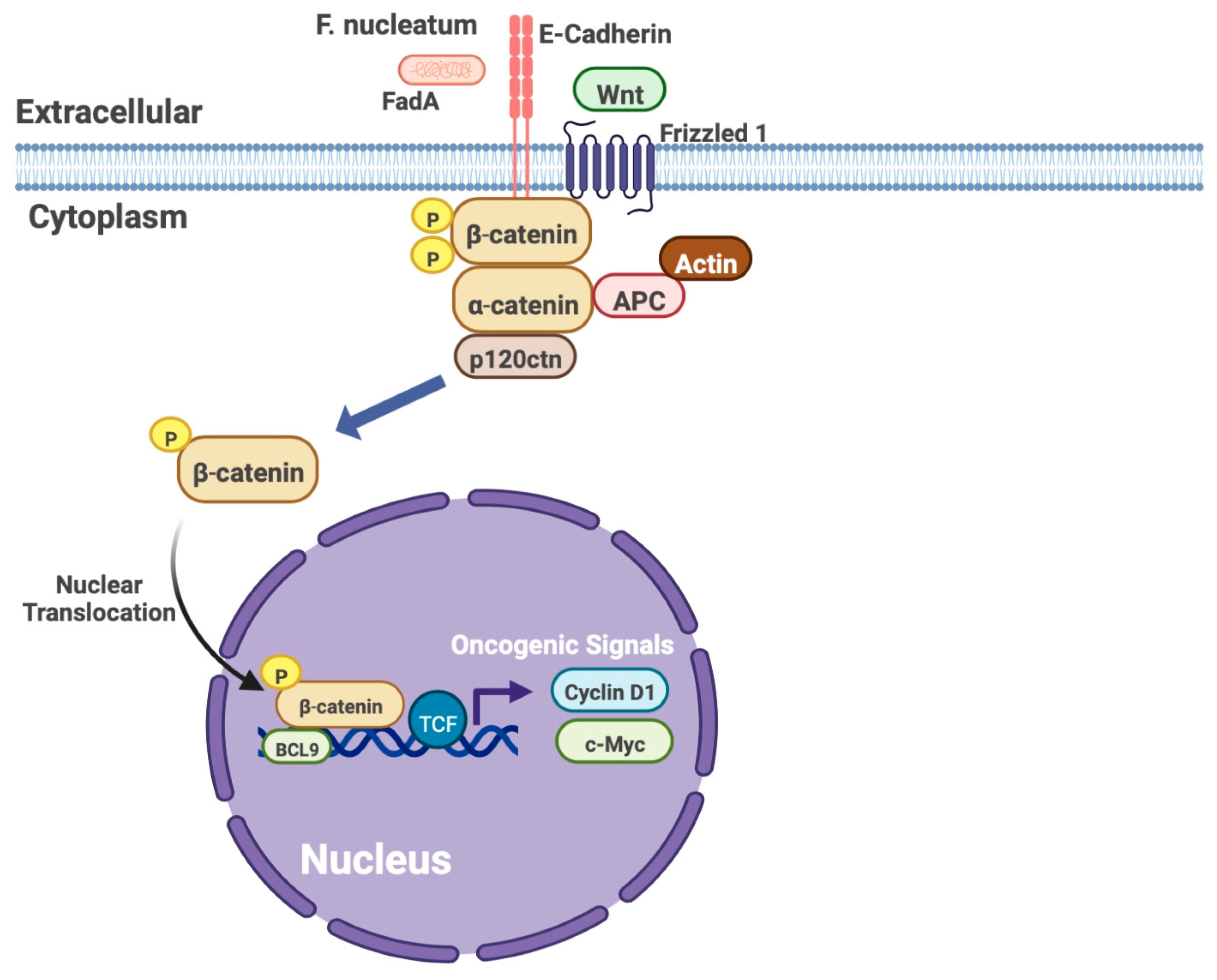
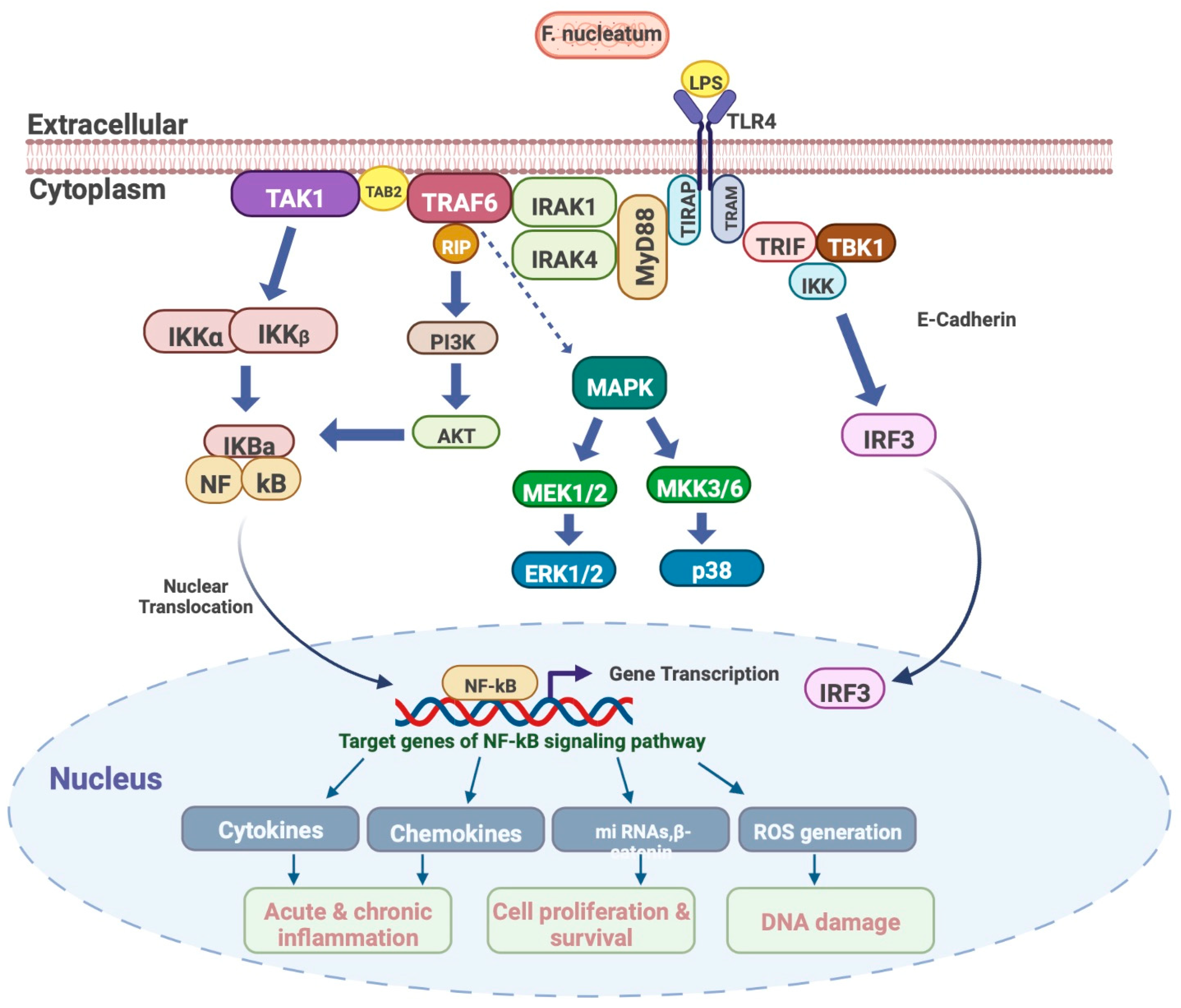
2.3. F. nucleatum
3. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Axelsson, P.A. Commentary: Periodontitis is preventable. J. Periodontol. 2014, 85, 1303–1307. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Jepsen, S.; Jin, L.; Otomo-Corgel, J. Impact of the Global Burden of Periodontal Diseases on Health, Nutrition and Wellbeing of Mankind: A Call for Global Action. J. Clin. Periodontol. 2017, 44, 456–462. [Google Scholar] [CrossRef]
- GBD. Global, Regional, and National Levels and Trends in Burden of Oral Conditions from 1990 to 2017: A Systematic Analysis for the Global Burden of Disease 2017 Study. J. Dent. Res. 2020, 99, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Nazir, M.; Al-Ansari, A.; Al-Khalifa, K.; Alhareky, M.; Gaffar, B.; Almas, K. Global Prevalence of Periodontal Disease and Lack of Its Surveillance. Sci. World J. 2020, 2020, 2146160. [Google Scholar] [CrossRef] [PubMed]
- Marcenes, W.; Kassebaum, N.J.; Bernabe, E.; Flaxman, A.; Naghavi, M.; Lopez, A.; Murray, C.J. Global burden of oral conditions in 1990–2010: A systematic analysis. J. Dent. Res. 2013, 92, 592–597. [Google Scholar] [CrossRef] [PubMed]
- Kassebaum, N.J.; Smith, A.G.C.; Bernabe, E.; Fleming, T.D.; Reynolds, A.E.; Vos, T.; Murray, C.J.L.; Marcenes, W.; Collaborators, G.B.D.O.H. Global, Regional, and National Prevalence, Incidence, and Disability-Adjusted Life Years for Oral Conditions for 195 Countries, 1990-2015: A Systematic Analysis for the Global Burden of Diseases, Injuries, and Risk Factors. J. Dent. Res. 2017, 96, 380–387. [Google Scholar] [CrossRef]
- Chen, M.X.; Zhong, Y.J.; Dong, Q.Q.; Wong, H.M.; Wen, Y.F. Global, regional, and national burden of severe periodontitis, 1990-2019: An analysis of the Global Burden of Disease Study 2019. J. Clin. Periodontol. 2021, 48, 1165–1188. [Google Scholar] [CrossRef]
- Botelho, J.; Machado, V.; Leira, Y.; Proença, L.; Chambrone, L.; Mendes, J.J. Economic burden of periodontitis in the United States and Europe—An updated estimation. J. Periodontol. 2022, 93, 373–379. [Google Scholar] [CrossRef]
- Bartold, P.M.; Van Dyke, T.E. Periodontitis: A host-mediated disruption of microbial homeostasis. Unlearning learned concepts. Periodontology 2000 2013, 62, 203–217. [Google Scholar] [CrossRef]
- Genco, R.J.; Borgnakke, W.S. Risk factors for periodontal disease. Periodontology 2000 2013, 62, 59–94. [Google Scholar] [CrossRef]
- Jepsen, S.; Caton, J.G.; Albandar, J.M.; Bissada, N.F.; Bouchard, P.; Cortellini, P.; Demirel, K.; de Sanctis, M.; Ercoli, C.; Fan, J.; et al. Periodontal manifestations of systemic diseases and developmental and acquired conditions: Consensus report of workgroup 3 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S219–S229. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, C.S.; Kinane, D.F. Nutrition, inflammation, and periodontal disease. Nutrition 2003, 19, 475. [Google Scholar] [CrossRef] [PubMed]
- Dommisch, H.; Kuzmanova, D.; Jönsson, D.; Grant, M.; Chapple, I. Effect of micronutrient malnutrition on periodontal disease and periodontal therapy. Periodontology 2000 2018, 78, 129–153. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. The Bacterial Etiology of Destructive Periodontal Disease: Current Concepts. J. Periodontol. 1992, 63 (Suppl. S4), 322–331. [Google Scholar] [CrossRef]
- Holt, S.C.; Ebersole, J.; Felton, J.; Brunsvold, M.; Kornman, K.S. Implantation of Bacteroides gingivalis in nonhuman primates initiates progression of periodontitis. Science 1988, 239, 55–57. [Google Scholar] [CrossRef]
- Zubery, Y.; Dunstan, C.R.; Story, B.M.; Kesavalu, L.; Ebersole, J.L.; Holt, S.C.; Boyce, B.F. Bone resorption caused by three periodontal pathogens in vivo in mice is mediated in part by prostaglandin. Infect. Immun. 1998, 66, 4158–4162. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S.; Greenwell, H.; Kornman, K.S. Staging and grading of periodontitis: Framework and proposal of a new classification and case definition. J. Clin. Periodontol. 2018, 45 (Suppl. 20), S149–S161. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.S. Molecular factors associated with compartmentalization of gingival immune responses and transepithelial neutrophil migration. J. Periodontal. Res. 1997, 32, 104–109. [Google Scholar] [CrossRef]
- Jiang, Y.; Graves, D.T. Periodontal pathogens stimulate CC-chemokine production by mononuclear and bone-derived cells. J. Periodontol. 1999, 70, 1472–1478. [Google Scholar] [CrossRef]
- Genco, R.J.; Sanz, M. Clinical and public health implications of periodontal and systemic diseases: An overview. Periodontology 2000 2020, 83, 7–13. [Google Scholar] [CrossRef]
- Söder, B.; Jin, L.; Klinge, B.; Söder, P.Ö. Periodontitis and premature death: A 16-year longitudinal study in a Swedish urban population. J. Periodontal. Res. 2007, 42, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Williams, R.C.; Offenbacher, S. Periodontal medicine: The emergence of a new branch of periodontology. Periodontology 2000 2000, 23, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Kapila, Y.L. Oral health’s inextricable connection to systemic health: Special populations bring to bear multimodal relationships and factors connecting periodontal disease to systemic diseases and conditions. Periodontology 2000 2021, 87, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Tonetti, M.; Van Dyke, T.E.; Working Group 1 of the Joint EFP/AAP Workshop. Periodontitis and atherosclerotic cardiovascular disease: Consensus report of the Joint EFP. AAP Workshop on Periodontitis and Systemic Diseases. J. Clin. Periodontol. 2013, 40, S24–S29. [Google Scholar] [CrossRef]
- Chapple, I.; Genco, R. Workshop WGotJEA. Diabetes and periodontal diseases: Consensus report of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J. Clin. Periodontol. 2013, 40, S106–S112. [Google Scholar] [CrossRef]
- Sanz, M.; Kornman, K.; Working Group 3 of the Joint EFP/AAP Workshop. Periodontitis and adverse pregnancy outcomes: Consensus report of the Joint EFP/AAP Workshop on Periodontitis and Systemic Diseases. J. Periodontol. 2013, 84, S164–S169. [Google Scholar] [CrossRef]
- Hajishengallis, G. Periodontitis: From microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 2015, 15, 30–44. [Google Scholar] [CrossRef]
- Fitzpatrick, S.G.; Katz, J. The association between periodontal disease and cancer: A review of the literature. J. Dent. 2010, 38, 83–95. [Google Scholar] [CrossRef]
- Hormia, M.; Willberg, J.; Ruokonen, H.; Syrjanen, S. Marginal periodontium as a potential reservoir of human papillomavirus in oral mucosa. J. Periodontol. 2005, 76, 358–363. [Google Scholar] [CrossRef]
- Saygun, I.; Kubar, A.; Ozdemir, A.; Slots, J. Periodontitis lesions are a source of salivary cytomegalovirus and Epstein-Barr virus. J. Periodontal. Res. 2005, 40, 187–191. [Google Scholar] [CrossRef]
- Katz, J.; Onate, M.D.; Pauley, K.M.; Bhattacharyya, I.; Cha, S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int. J. Oral. Sci. 2011, 3, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.; Park, Y.; Hasegawa, Y.; Tribble, G.D.; James, C.E.; Handfield, M.; Stavropoulos, M.F.; Yilmaz, O.; Lamont, R.J. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007, 9, 1997–2007. [Google Scholar] [CrossRef]
- Sayehmiri, F.; Sayehmiri, K.; Asadollahi, K.; Soroush, S.; Bogdanovic, L.; Jalilian, F.A.; Emaneini, M.; Taherikalani, M. The prevalence rate of Porphyromonas gingivalis and its association with cancer: A systematic review and meta-analysis. Int. J. Immunopathol. Pharm. 2015, 28, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Pierotti, M.A. Cancer and inflammation: A complex relationship. Cancer Lett. 2008, 267, 180–181. [Google Scholar] [CrossRef] [PubMed]
- Coussens, L.M.; Werb, Z. Inflammation and cancer. Nature 2002, 420, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q.W.; Zhou, D.S.; Peng, H.J.; Ji, P.; Liu, D.S. Association of periodontal disease with oral cancer: A meta-analysis. Tumour Biol. 2014, 35, 7073–7077. [Google Scholar] [CrossRef] [PubMed]
- Åberg, C.H.; Kwamin, F.; Claesson, R.; Johansson, A.; Haubek, D. Presence of JP2 and Non-JP2 Genotypes of Aggregatibacter actinomycetemcomitans and attachment loss in adolescents in Ghana. J. Periodontol. 2012, 83, 1520–1528. [Google Scholar] [CrossRef]
- Bandhaya, P.; Saraithong, P.; Likittanasombat, K.; Hengprasith, B.; Torrungruang, K. Aggregatibacter actinomycetemcomitans serotypes, the JP2 clone and cytolethal distending toxin genes in a Thai population. J. Clin. Periodontol. 2012, 39, 519–525. [Google Scholar] [CrossRef]
- Chen, C.; Wang, T.; Chen, W. Occurrence of Aggregatibacter actinomycetemcomitans serotypes in subgingival plaque from United States subjects. Mol. Oral Microbiol. 2010, 25, 207–214. [Google Scholar] [CrossRef]
- Cortelli, J.R.; Aquino, D.R.; Cortelli, S.C.; Roman-Torres, C.V.; Franco, G.C.; Gomez, R.S.; Batista, L.H.; Costa, F.O. Aggregatibacter actinomycetemcomitans serotypes infections and periodontal conditions: A two-way assessment. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1311–1318. [Google Scholar] [CrossRef]
- Roman-Torres, C.V.; Aquino, D.R.; Cortelli, S.C.; Franco, G.C.; Dos Santos, J.G.; Corraini, P.; Holzhausen, M.; Diniz, M.G.; Gomez, R.S.; Cortelli, J.R. Prevalence and distribution of serotype-specific genotypes of Aggregatibacter actinomycetemcomitans in chronic periodontitis Brazilian subjects. Arch. Oral Biol. 2010, 55, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Zúñiga, J.; Melgar-Rodríguez, S.; Alvarez, C.; Monasterio, G.; Benítez, A.; Ciuchi, P.; Díaz, C.; Mardones, J.; Escobar, A.; Sanz, M.; et al. T-lymphocyte phenotype and function triggered by Aggregatibacter actinomycetemcomitans is serotype-dependent. J. Periodontal. Res. 2015, 50, 824–835. [Google Scholar] [CrossRef] [PubMed]
- Linhartová, I.; Bumba, L.; Mašín, J.; Basler, M.; Osička, R.; Kamanová, J.; Procházková, K.; Adkins, I.; Hejnová-Holubová, J.; Sadílková, L.; et al. RTX proteins: A highly diverse family secreted by a common mechanism. FEMS Microbiol. Rev. 2010, 34, 1076–1112. [Google Scholar] [CrossRef] [PubMed]
- Mazzone, A.; Ricevuti, G. Leukocyte CD11/CD18 integrins: Biological and clinical relevance. Haematologica 1995, 80, 161–175. [Google Scholar]
- Johansson, A. Aggregatibacter actinomycetemcomitans leukotoxin: A powerful tool with capacity to cause imbalance in the host inflammatory response. Toxins 2011, 3, 242–259. [Google Scholar] [CrossRef]
- Johansson, A.; Buhlin, K.; Sorsa, T.; Pussinen, P.J. Systemic Aggregatibacter actinomycetemcomitans Leukotoxin-Neutralizing Antibodies in Periodontitis. J. Periodontol. 2017, 88, 122–129. [Google Scholar] [CrossRef]
- Claesson, R.; Johansson, A.; Belibasakis, G.; Hänström, L.; Kalfas, S. Release and activation of matrix metalloproteinase 8 from human neutrophils triggered by the leukotoxin of Actinobacillus actinomycetemcomitans. J. Periodontal. Res. 2002, 37, 353–359. [Google Scholar] [CrossRef]
- Johansson, A.; Claesson, R.; Hänström, L.; Sandström, G.; Kalfas, S. Polymorphonuclear leukocyte degranulation induced by leukotoxin from Actinobacillus actinomycetemcomitans. J. Periodontal. Res. 2000, 35, 85–92. [Google Scholar] [CrossRef]
- Kelk, P.; Abd, H.; Claesson, R.; Sandström, G.; Sjöstedt, A.; Johansson, A. Cellular and molecular response of human macrophages exposed to Aggregatibacter actinomycetemcomitans leukotoxin. Cell Death Dis. 2011, 2, e126. [Google Scholar] [CrossRef]
- Ahlstrand, T.; Tuominen, H.; Beklen, A.; Torittu, A.; Oscarsson, J.; Sormunen, R.; Pöllänen, M.T.; Permi, P.; Ihalin, R. A novel intrinsically disordered outer membrane lipoprotein of Aggregatibacter actinomycetemcomitans binds various cytokines and plays a role in biofilm response to interleukin-1β and interleukin-8. Virulence 2017, 8, 115–134. [Google Scholar] [CrossRef]
- Lally, E.T.; Boesze-Battaglia, K.; Dhingra, A.; Gomez, N.M.; Lora, J.; Mitchell, C.H.; Giannakakis, A.; Fahim, S.A.; Benz, R.; Balashova, N. Aggregatibacter actinomycetemcomitans LtxA Hijacks Endocytic Trafficking Pathways in Human Lymphocytes. Pathogens 2020, 9, 74. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.; Sandström, G.; Claesson, R.; Hänström, L.; Kalfas, S. Anaerobic neutrophil-dependent killing of Actinobacillus actinomycetemcomitans in relation to the bacterial leukotoxicity. Eur. J. Oral Sci. 2000, 108, 136–146. [Google Scholar] [CrossRef]
- Johnson, W.; Lior, H. Response of Chinese hamster ovary cells to a cytolethal distending toxin (CDT) of Escherichia coli and possible misinterpretation as heat-labile (LT) enterotoxin. FEMS Microbiol. Lett. 1987, 43, 19–23. [Google Scholar] [CrossRef]
- Johnson, W.; Lior, H. Production of Shiga toxin and a cytolethal distending toxin (CLDT) by serogroups of Shigella spp. FEMS Microbiol. Lett. 1987, 48, 235–238. [Google Scholar] [CrossRef]
- Johnson, W.M.; Lior, H. A new heat-labile cytolethal distending toxin (CLDT) produced by Campylobacter spp. Microb. Pathog. 1988, 4, 115–126. [Google Scholar] [CrossRef]
- Mayer, M.P.; Bueno, L.C.; Hansen, E.J.; DiRienzo, J.M. Identification of a cytolethal distending toxin gene locus and features of a virulence-associated region in Actinobacillus actinomycetemcomitans. Infect. Immun. 1999, 67, 1227–1237. [Google Scholar] [CrossRef]
- Sugai, M.; Kawamoto, T.; Pérès, S.Y.; Ueno, Y.; Komatsuzawa, H.; Fujiwara, T.; Kurihara, H.; Suginaka, H.; Oswald, E. The cell cycle-specific growth-inhibitory factor produced by Actinobacillus actinomycetemcomitans is a cytolethal distending toxin. Infect. Immun. 1998, 66, 5008–5019. [Google Scholar] [CrossRef]
- Scott, D.A.; Kaper, J.B. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 1994, 62, 244–251. [Google Scholar] [CrossRef]
- Thelestam, M.; Frisan, T. Cytolethal distending toxins. Rev. Physiol. Biochem. Pharm. 2004, 152, 111–133. [Google Scholar] [CrossRef]
- Cortes-Bratti, X.; Frisan, T.; Thelestam, M. The cytolethal distending toxins induce DNA damage and cell cycle arrest. Toxicon 2001, 39, 1729–1736. [Google Scholar] [CrossRef]
- DiRienzo, J.M. Uptake and processing of the cytolethal distending toxin by mammalian cells. Toxins 2014, 6, 3098–3116. [Google Scholar] [CrossRef] [PubMed]
- Ohguchi, M.; Ishisaki, A.; Okahashi, N.; Koide, M.; Koseki, T.; Yamato, K.; Noguchi, T.; Nishihara, T. Actinobacillus actinomycetemcomitans toxin induces both cell cycle arrest in the G2/M phase and apoptosis. Infect. Immun. 1998, 66, 5980–5987. [Google Scholar] [CrossRef] [PubMed]
- Shenker, B.J.; Hoffmaster, R.H.; Zekavat, A.; Yamaguchi, N.; Lally, E.T.; Demuth, D.R. Induction of apoptosis in human T cells by Actinobacillus actinomycetemcomitans cytolethal distending toxin is a consequence of G2 arrest of the cell cycle. J. Immunol. 2001, 167, 435–441. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Chahwan, C.; Bailis, J.; Hunter, T.; Russell, P. ATM activation and its recruitment to damaged DNA require binding to the C terminus of Nbs1. Mol. Cell Biol. 2005, 25, 5363–5379. [Google Scholar] [CrossRef] [PubMed]
- Coppé, J.P.; Desprez, P.Y.; Krtolica, A.; Campisi, J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu. Rev. Pathol. 2010, 5, 99–118. [Google Scholar] [CrossRef] [PubMed]
- Genco, C.A.; Schifferle, R.E.; Njoroge, T.; Forng, R.Y.; Cutler, C.W. Resistance of a Tn4351-generated polysaccharide mutant of Porphyromonas gingivalis to polymorphonuclear leukocyte killing. Infect. Immun. 1995, 63, 393–401. [Google Scholar] [CrossRef]
- Cutler, C.W.; Kalmar, J.R.; Arnold, R.R. Phagocytosis of virulent Porphyromonas gingivalis by human polymorphonuclear leukocytes requires specific immunoglobulin G. Infect. Immun. 1991, 59, 2097–2104. [Google Scholar] [CrossRef]
- Samaranayake, L.P. Essential Microbiology for Dentistry, 2nd ed.; Elsevier Health Sciences: Amsterdam, The Netherlands, 2002. [Google Scholar]
- Cutler, C.W.; Kalmar, J.R.; Genco, C.A. Pathogenic strategies of the oral anaerobe, Porphyromonas gingivalis. Trends Microbiol. 1995, 3, 45–51. [Google Scholar] [CrossRef]
- Dzink, J.L.; Socransky, S.S.; Haffajee, A.D. The predominant cultivable microbiota of active and inactive lesions of destructive periodontal diseases. J. Clin. Periodontol. 1988, 15, 316–323. [Google Scholar] [CrossRef]
- Holt, S.C.; Kesavalu, L.; Walker, S.; Genco, C.A. Virulence factors of Porphyromonas gingivalis. Periodontology 2000 1999, 20, 168–238. [Google Scholar] [CrossRef]
- Genco, C.A.; Kapczynski, D.R.; Cutler, C.W.; Arko, R.J.; Arnold, R.R. Influence of immunization on Porphyromonas gingivalis colonization and invasion in the mouse chamber model. Infect. Immun. 1992, 60, 1447–1454. [Google Scholar] [CrossRef] [PubMed]
- Gibson, F.C., 3rd; Hong, C.; Chou, H.H.; Yumoto, H.; Chen, J.; Lien, E.; Wong, J.; Genco, C.A. Innate immune recognition of invasive bacteria accelerates atherosclerosis in apolipoprotein E-deficient mice. Circulation 2004, 109, 2801–2806. [Google Scholar] [CrossRef] [PubMed]
- Reddy, M.S. Reaching a better understanding of non-oral disease and the implication of periodontal infections. Periodontology 2000 2007, 44, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Gatz, M.; Mortimer, J.A.; Fratiglioni, L.; Johansson, B.; Berg, S.; Reynolds, C.A.; Pedersen, N.L. Potentially modifiable risk factors for dementia in identical twins. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2006, 2, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Kamer, A.R.; Pirraglia, E.; Tsui, W.; Rusinek, H.; Vallabhajosula, S.; Mosconi, L.; Yi, L.; McHugh, P.; Craig, R.G.; Svetcov, S.; et al. Periodontal disease associates with higher brain amyloid load in normal elderly. Neurobiol. Aging 2015, 36, 627–633. [Google Scholar] [CrossRef]
- Kaye, E.K.; Valencia, A.; Baba, N.; Spiro, A., 3rd; Dietrich, T.; Garcia, R.I. Tooth loss and periodontal disease predict poor cognitive function in older men. J. Am. Geriatr. Soc. 2010, 58, 713–718. [Google Scholar] [CrossRef]
- Noble, J.M.; Borrell, L.N.; Papapanou, P.N.; Elkind, M.S.; Scarmeas, N.; Wright, C.B. Periodontitis is associated with cognitive impairment among older adults: Analysis of NHANES-III. J. Neurol. Neurosurg. Psychiatry 2009, 80, 1206–1211. [Google Scholar] [CrossRef]
- Stein, P.S.; Desrosiers, M.; Donegan, S.J.; Yepes, J.F.; Kryscio, R.J. Tooth loss, dementia and neuropathology in the Nun study. J. Am. Dent. Assoc. 2007, 138, 1314–1322; quiz 1381–1312. [Google Scholar] [CrossRef]
- Dominy, S.S.; Lynch, C.; Ermini, F.; Benedyk, M.; Marczyk, A.; Konradi, A.; Nguyen, M.; Haditsch, U.; Raha, D.; Griffin, C.; et al. Porphyromonas gingivalis in Alzheimer’s disease brains: Evidence for disease causation and treatment with small-molecule inhibitors. Sci. Adv. 2019, 5, eaau3333. [Google Scholar] [CrossRef]
- Zeituni, A.E.; McCaig, W.; Scisci, E.; Thanassi, D.G.; Cutler, C.W. The native 67-kilodalton minor fimbria of Porphyromonas gingivalis is a novel glycoprotein with DC-SIGN-targeting motifs. J. Bacteriol. 2010, 192, 4103–4110. [Google Scholar] [CrossRef]
- Ezzo, P.J.; Cutler, C.W. Microorganisms as risk indicators for periodontal disease. Periodontology 2000 2003, 32, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Zeituni, A.E.; Jotwani, R.; Carrion, J.; Cutler, C.W. Targeting of DC-SIGN on human dendritic cells by minor fimbriated Porphyromonas gingivalis strains elicits a distinct effector T cell response. J. Immunol. 2009, 183, 5694–5704. [Google Scholar] [CrossRef] [PubMed]
- Hamada, N.; Sojar, H.T.; Cho, M.I.; Genco, R.J. Isolation and characterization of a minor fimbria from Porphyromonas gingivalis. Infect. Immun. 1996, 64, 4788–4794. [Google Scholar] [CrossRef]
- Carrion, J.; Scisci, E.; Miles, B.; Sabino, G.J.; Zeituni, A.E.; Gu, Y.; Bear, A.; Genco, C.A.; Brown, D.L.; Cutler, C.W. Microbial carriage state of peripheral blood dendritic cells (DCs) in chronic periodontitis influences DC differentiation, atherogenic potential. J. Immunol. 2012, 189, 3178–3187. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Lamont, R.J. Promoter architecture of the Porphyromonas gingivalis fimbrillin gene. Infect. Immun. 1999, 67, 3227–3235. [Google Scholar] [CrossRef] [PubMed]
- Meghil, M.M.; Tawfik, O.K.; Elashiry, M.; Rajendran, M.; Arce, R.M.; Fulton, D.J.; Schoenlein, P.V.; Cutler, C.W. Disruption of Immune Homeostasis in Human Dendritic Cells via Regulation of Autophagy and Apoptosis by Porphyromonas gingivalis. Front. Immunol. 2019, 10, 2286. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of cells and tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef]
- Lamb, C.A.; Yoshimori, T.; Tooze, S.A. The autophagosome: Origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 2013, 14, 759–774. [Google Scholar] [CrossRef]
- El-Awady, A.R.; Miles, B.; Scisci, E.; Kurago, Z.B.; Palani, C.D.; Arce, R.M.; Waller, J.L.; Genco, C.A.; Slocum, C.; Manning, M.; et al. Porphyromonas gingivalis evasion of autophagy and intracellular killing by human myeloid dendritic cells involves DC-SIGN-TLR2 crosstalk. PLoS Pathog. 2015, 10, e1004647. [Google Scholar] [CrossRef]
- Thurston, T.L.; Wandel, M.P.; von Muhlinen, N.; Foeglein, A.; Randow, F. Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 2012, 482, 414–418. [Google Scholar] [CrossRef]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the autophagy receptor optineurin restricts Salmonella growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, T.; Fujita, N.; Jang, M.H.; Uematsu, S.; Yang, B.G.; Satoh, T.; Omori, H.; Noda, T.; Yamamoto, N.; Komatsu, M.; et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature 2008, 456, 264–268. [Google Scholar] [CrossRef] [PubMed]
- Paludan, C.; Schmid, D.; Landthaler, M.; Vockerodt, M.; Kube, D.; Tuschl, T.; Munz, C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005, 307, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Loi, M.; Muller, A.; Steinbach, K.; Niven, J.; Barreira da Silva, R.; Paul, P.; Ligeon, L.A.; Caruso, A.; Albrecht, R.A.; Becker, A.C.; et al. Macroautophagy Proteins Control MHC Class I Levels on Dendritic Cells and Shape Anti-viral CD8(+) T Cell Responses. Cell Rep. 2016, 15, 1076–1087. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Long, L.; Yang, K.; Guy, C.; Shrestha, S.; Chen, Z.; Wu, C.; Vogel, P.; Neale, G.; Green, D.R.; et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat. Immunol. 2016, 17, 277–285. [Google Scholar] [CrossRef] [PubMed]
- Miles, B.; Scisci, E.; Carrion, J.; Sabino, G.J.; Genco, C.A.; Cutler, C.W. Noncanonical dendritic cell differentiation and survival driven by a bacteremic pathogen. J. Leukoc. Biol. 2013, 94, 281–289. [Google Scholar] [CrossRef] [PubMed]
- Patterson, S.D.; Spahr, C.S.; Daugas, E.; Susin, S.A.; Irinopoulou, T.; Koehler, C.; Kroemer, G. Mass spectrometric identification of proteins released from mitochondria undergoing permeability transition. Cell Death Differ. 2000, 7, 137–144. [Google Scholar] [CrossRef]
- Liu, X.; Kim, C.N.; Yang, J.; Jemmerson, R.; Wang, X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 1996, 86, 147–157. [Google Scholar] [CrossRef]
- Chinnaiyan, A.M.; O’Rourke, K.; Tewari, M.; Dixit, V.M. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell 1995, 81, 505–512. [Google Scholar] [CrossRef]
- Kischkel, F.C.; Hellbardt, S.; Behrmann, I.; Germer, M.; Pawlita, M.; Krammer, P.H.; Peter, M.E. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. EMBO J. 1995, 14, 5579–5588. [Google Scholar] [CrossRef]
- Muzio, M.; Chinnaiyan, A.M.; Kischkel, F.C.; O’Rourke, K.; Shevchenko, A.; Ni, J.; Scaffidi, C.; Bretz, J.D.; Zhang, M.; Gentz, R.; et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death--inducing signaling complex. Cell 1996, 85, 817–827. [Google Scholar] [CrossRef] [PubMed]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Bugueno, I.M.; Batool, F.; Korah, L.; Benkirane-Jessel, N.; Huck, O. Porphyromonas gingivalis Differentially Modulates Apoptosome Apoptotic Peptidase Activating Factor 1 in Epithelial Cells and Fibroblasts. Am. J. Pathol. 2018, 188, 404–416. [Google Scholar] [CrossRef] [PubMed]
- Fadeel, B.; Orrenius, S. Apoptosis: A basic biological phenomenon with wide-ranging implications in human disease. J. Intern. Med. 2005, 258, 479–517. [Google Scholar] [CrossRef]
- Graves, D.T.; Oskoui, M.; Volejnikova, S.; Naguib, G.; Cai, S.; Desta, T.; Kakouras, A.; Jiang, Y. Tumor necrosis factor modulates fibroblast apoptosis, PMN recruitment, and osteoclast formation in response to P. gingivalis infection. J. Dent. Res. 2001, 80, 1875–1879. [Google Scholar] [CrossRef]
- Hiroi, M.; Shimojima, T.; Kashimata, M.; Miyata, T.; Takano, H.; Takahama, M.; Sakagami, H. Inhibition by Porphyromonas gingivalis LPS of apoptosis induction in human peripheral blood polymorphonuclear leukocytes. Anticancer Res. 1998, 18, 3475–3479. [Google Scholar]
- Murray, D.A.; Wilton, J.M. Lipopolysaccharide from the periodontal pathogen Porphyromonas gingivalis prevents apoptosis of HL60-derived neutrophils in vitro. Infect. Immun. 2003, 71, 7232–7235. [Google Scholar] [CrossRef]
- Ozaki, K.; Hanazawa, S. Porphyromonas gingivalis fimbriae inhibit caspase-3-mediated apoptosis of monocytic THP-1 cells under growth factor deprivation via extracellular signal-regulated kinase-dependent expression of p21 Cip/WAF1. Infect. Immun. 2001, 69, 4944–4950. [Google Scholar] [CrossRef][Green Version]
- Arjunan, P.; Meghil, M.M.; Pi, W.; Xu, J.; Lang, L.; El-Awady, A.; Sullivan, W.; Rajendran, M.; Rabelo, M.S.; Wang, T.; et al. Oral Pathobiont Activates Anti-Apoptotic Pathway, Promoting both Immune Suppression and Oncogenic Cell Proliferation. Sci. Rep. 2018, 8, 16607. [Google Scholar] [CrossRef]
- Pike, R.; McGraw, W.; Potempa, J.; Travis, J. Lysine- and arginine-specific proteinases from Porphyromonas gingivalis. Isolation, characterization, and evidence for the existence of complexes with hemagglutinins. J. Biol. Chem. 1994, 269, 406–411. [Google Scholar] [CrossRef]
- Njoroge, T.; Genco, R.J.; Sojar, H.T.; Hamada, N.; Genco, C.A. A role for fimbriae in Porphyromonas gingivalis invasion of oral epithelial cells. Infect. Immun. 1997, 65, 1980–1984. [Google Scholar] [CrossRef]
- Weinberg, A.; Belton, C.M.; Park, Y.; Lamont, R.J. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 1997, 65, 313–316. [Google Scholar] [CrossRef] [PubMed]
- NM, O.B.-S.; Veith, P.D.; Dashper, S.G.; Reynolds, E.C. Porphyromonas gingivalis gingipains: The molecular teeth of a microbial vampire. Curr. Protein Pept. Sci. 2003, 4, 409–426. [Google Scholar] [CrossRef]
- Inaba, H.; Sugita, H.; Kuboniwa, M.; Iwai, S.; Hamada, M.; Noda, T.; Morisaki, I.; Lamont, R.J.; Amano, A. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell Microbiol. 2014, 16, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Mu, W.; Jia, Y.; Chen, X.; Li, H.; Wang, Z.; Cheng, B. Intracellular Porphyromonas gingivalis Promotes the Proliferation of Colorectal Cancer Cells via the MAPK/ERK Signaling Pathway. Front. Cell Infect. Microbiol. 2020, 10, 584798. [Google Scholar] [CrossRef]
- Kapatral, V.; Anderson, I.; Ivanova, N.; Reznik, G.; Los, T.; Lykidis, A.; Bhattacharyya, A.; Bartman, A.; Gardner, W.; Grechkin, G. Genome sequence and analysis of the oral bacterium Fusobacterium nucleatum strain ATCC 25586. J. Bacteriol. 2002, 184, 2005–2018. [Google Scholar] [CrossRef]
- Socransky, S.S.; Haffajee, A.D. Periodontal microbial ecology. Periodontology 2000 2005, 38, 135–187. [Google Scholar] [CrossRef]
- Pereira, A.; Tavares, A.T.; Prates, M.; Ribeiro, N.; Fonseca, L.F.; Marques, M.D.R.; Proença, F. Brain Abscess: A Rare Clinical Case with Oral Etiology. Case Rep. Infect. Dis. 2022, 2022, 5140259. [Google Scholar] [CrossRef]
- Jayasimhan, D.; Wu, L.; Huggan, P. Fusobacterial liver abscess: A case report and review of the literature. BMC Infect. Dis. 2017, 17, 440. [Google Scholar] [CrossRef]
- Gedik, A.H.; Cakir, E.; Soysal, O.; Umutoğlu, T. Endobronchial lesion due to pulmonary Fusobacterium nucleatum infection in a child. Pediatr. Pulmonol. 2014, 49, E63–E65. [Google Scholar] [CrossRef]
- Liu, C.; Jia, Q.; Wang, L.; Yang, D. A case report of severe Fusobacterium nucleatum sepsis secondary to nephrectomy. BMC Infect. Dis. 2022, 22, 309. [Google Scholar] [CrossRef] [PubMed]
- McKinnon, A.; Black, A.Y.; Lortie, K.; Fleming, N.A. A case of adolescent pelvic inflammatory disease caused by a rare bacterium: Fusobacterium nucleatum. J. Pediatr. Adolesc. Gynecol. 2013, 26, e113–e115. [Google Scholar] [CrossRef] [PubMed]
- Parhi, L.; Abed, J.; Shhadeh, A.; Alon-Maimon, T.; Udi, S.; Ben-Arye, S.L.; Tam, J.; Parnas, O.; Padler-Karavani, V.; Goldman-Wohl, D.; et al. Placental colonization by Fusobacterium nucleatum is mediated by binding of the Fap2 lectin to placentally displayed Gal-GalNAc. Cell Rep. 2022, 38, 110537. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Yamada, M.; Li, M.; Liu, H.; Chen, S.G.; Han, Y.W. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J. Biol. Chem. 2007, 282, 25000–25009. [Google Scholar] [CrossRef]
- Fardini, Y.; Wang, X.; Témoin, S.; Nithianantham, S.; Lee, D.; Shoham, M.; Han, Y.W. Fusobacterium nucleatum adhesin FadA binds vascular endothelial cadherin and alters endothelial integrity. Mol. Microbiol. 2011, 82, 1468–1480. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 Protein of Fusobacterium nucleatum to Human Inhibitory Receptor TIGIT Protects Tumors from Immune Cell Attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Yang, Y.; Weng, W.; Peng, J.; Hong, L.; Yang, L.; Toiyama, Y.; Gao, R.; Liu, M.; Yin, M.; Pan, C.; et al. Fusobacterium nucleatum Increases Proliferation of Colorectal Cancer Cells and Tumor Development in Mice by Activating Toll-Like Receptor 4 Signaling to Nuclear Factor-κB, and Up-regulating Expression of MicroRNA-21. Gastroenterology 2017, 152, 851–866. [Google Scholar] [CrossRef]
- Kawai, T.; Akira, S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 2007, 13, 460–469. [Google Scholar] [CrossRef]
- Udayasuryan, B.; Ahmad, R.N.; Nguyen, T.T.D.; Umaña, A.; Monét Roberts, L.; Sobol, P.; Jones, S.D.; Munson, J.M.; Slade, D.J.; Verbridge, S.S. Fusobacterium nucleatum induces proliferation and migration in pancreatic cancer cells through host autocrine and paracrine signaling. Sci. Signal 2022, 15, eabn4948. [Google Scholar] [CrossRef]
- Gnanasekaran, J.; Binder Gallimidi, A.; Saba, E.; Pandi, K.; Eli Berchoer, L.; Hermano, E.; Angabo, S.; Makkawi, H.A.; Khashan, A.; Daoud, A.; et al. Intracellular Porphyromonas gingivalis Promotes the Tumorigenic Behavior of Pancreatic Carcinoma Cells. Cancers 2020, 12, 2331. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Shen, X.; Zhou, M.; Tang, B. Periodontal Pathogens Promote Oral Squamous Cell Carcinoma by Regulating ATR and NLRP3 Inflammasome. Front. Oncol. 2021, 11, 722797. [Google Scholar] [CrossRef] [PubMed]
- Groeger, S.; Wu, F.; Wagenlehner, F.; Dansranjav, T.; Ruf, S.; Denter, F.; Meyle, J. PD-L1 Up-Regulation in Prostate Cancer Cells by Porphyromonas gingivalis. Front. Cell Infect. Microbiol. 2022, 12, 935806. [Google Scholar] [CrossRef] [PubMed]

| Cancer Type | Proposed Pathomechanism |
|---|---|
| Pancreatic cancer |
|
| Head and neck SCC | |
| Prostate cancer |
|
| Colorectal cancer |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahoumi, L.A.; Saleh, M.H.A.; Meghil, M.M. Virulence Factors of the Periodontal Pathogens: Tools to Evade the Host Immune Response and Promote Carcinogenesis. Microorganisms 2023, 11, 115. https://doi.org/10.3390/microorganisms11010115
Shahoumi LA, Saleh MHA, Meghil MM. Virulence Factors of the Periodontal Pathogens: Tools to Evade the Host Immune Response and Promote Carcinogenesis. Microorganisms. 2023; 11(1):115. https://doi.org/10.3390/microorganisms11010115
Chicago/Turabian StyleShahoumi, Linah A., Muhammad H. A. Saleh, and Mohamed M. Meghil. 2023. "Virulence Factors of the Periodontal Pathogens: Tools to Evade the Host Immune Response and Promote Carcinogenesis" Microorganisms 11, no. 1: 115. https://doi.org/10.3390/microorganisms11010115
APA StyleShahoumi, L. A., Saleh, M. H. A., & Meghil, M. M. (2023). Virulence Factors of the Periodontal Pathogens: Tools to Evade the Host Immune Response and Promote Carcinogenesis. Microorganisms, 11(1), 115. https://doi.org/10.3390/microorganisms11010115








