Abstract
The pathogenesis of periodontitis depends on a sustained feedback loop where bacterial virulence factors and immune responses both contribute to inflammation and tissue degradation. Periodontitis is a multifactorial disease that is associated with a pathogenic shift in the oral microbiome. Within this shift, low-abundance Gram-negative anaerobic pathobionts transition from harmless colonisers of the subgingival environment to a virulent state that drives evasion and subversion of innate and adaptive immune responses. This, in turn, drives the progression of inflammatory disease and the destruction of tooth-supporting structures. From an evolutionary perspective, bacteria have developed this phenotypic plasticity in order to respond and adapt to environmental stimuli or external stressors. This review summarises the available knowledge of genetic, transcriptional, and post-translational mechanisms which mediate the commensal-pathogen transition of periodontal bacteria. The review will focus primarily on Porphyromonas gingivalis.
1. Introduction
Periodontitis is a highly prevalent inflammatory condition of the tooth-supporting structures associated with bacteria of the “red complex”, which comprise Porphyromonas gingivalis, Treponema denticola and Tannerella forsythia. These bacterial species interact amongst themselves and with other species as plaque forms in the subgingival sulcus [1]. Under specific environmental conditions, periodontal bacteria produce proteases, most notoriously gingipains and dentilisin, that drive protein degradation and ultimately contribute to tissue destruction of the periodontium [2]. However, bacterial protease production is only one of a plethora of virulence mechanisms associated with periodontal bacteria, many of which are very well studied for their ability to subvert the immune system and cause local and systemic inflammation. P. gingivalis is considered a keystone bacterium in the pathogenesis of periodontitis by driving an imbalance between symbionts and pathobionts of the oral microbiota (dysbiosis) [3,4] and interfering with the complement cascade amongst other immune responses [5]. This review presents published information regarding the mechanisms by which periodontal pathobionts, in particular P. gingivalis, are able to transition from harmless colonisers of the subgingival environment to virulent pathogens.
From an evolutionary perspective, bacteria have developed a phenotypic plasticity that allows survival responses and adaptation to environmental stimuli or external stressors. This pathogenic shift may be viewed in the context of dysbiosis, a term used to broadly describe imbalances of the microbiota which cause disease [6], and therefore is likely relevant to the pathogenesis of periodontitis [7]. Dysbiosis may be induced by environmental changes such as physical disruption of the epithelial barrier, antimicrobial treatments, and immune deficiencies. At the most basic level, oral microbiota and dysbiosis studies have focused on the loss of microbial diversity, that is the increase in abundance of ‘pathobionts’ and the depletion of ’symbionts’ [8,9]. The term pathobiont is used to describe species which are present as commensal microbes but under certain environmental conditions can induce or advance disease [10]. Against a backdrop of metagenomic studies that supports periodontitis as a dysbiotic disease, the specific mechanisms by which periodontal bacteria shift from a commensal to a pathogenic state are not fully understood.
External stressors can induce bacterial phenotypic changes for their adaptation and survival. Phenotypic change may also result from the accumulation of non-synonymous single-nucleotide mutations. Such mutations are spontaneous and, at least in part, driven by aberrant DNA repair mechanisms [11]. However, variants of P. gingivalis clones generated in vitro by serial sub-culturing are characterised by loss of virulence (loss of pigmentation in blood agar growth, decreased proteolytic activity, depleted haemagglutination and higher susceptibility to complement killing) suggesting that spontaneous mutations are less likely contributors to pathogenicity [12].
Environmental factors are the major drivers of phenotypic change in most bacteria including periodontal species. For example, temperature is a well-known regulator of the activity of transcriptional regulators, kinases, and chaperones [13,14]. In the case of P. gingivalis, temperature elevation has been shown to alter the structure of Lipid A, a major component of lipopolysaccharide (LPS). P. gingivalis grown at temperatures representative of the inflammatory environment shows reduced diversity of Lipid A structural forms, namely the predominance of monophosphorylated and diphosphorylated penta-acylated forms which are more potent Toll-like receptor 4 (TLR4) agonists [15]. Similarly, haemin concentration in in vitro growth culture can modulate TLR4/2 activation and E-selectin expression by P. gingivalis through their Lipid A structure. Under elevated haemin concentration, P. gingivalis showed ‘immunosuppressive’ properties (downregulation of TLR signalling, reduced levels of TNF-α and elevated interleukin-10 production) [16,17,18]. Under haemin-limiting conditions, P. gingivalis showed ‘immune-evasive’ properties, for example, increased resistance to host cationic antimicrobial peptides and lower LPS-mediated neutrophil-priming capacity, as well as increased gingipain activity, extracellular vesicles release and nutrient storage [19]. Extracellular pH also plays a major role in driving survival and dominance of periodontal pathobionts. While T. denticola and T. forsythia show increased survival at acidic pH, P. gingivalis predominates at alkaline pH [20,21].
There is evidence that P. gingivalis can activate manganese transport in addition to iron transport as a compensatory mechanism to enhance survival in low iron environments. This mechanism is mediated by feoB1 and feoB2 encoding iron and manganese ion transporters, respectively, which also play a role in protection from oxidative stress [22]. Superoxide dismutases (sod) of P. gingivalis can utilise iron or manganese as a cofactor to catalyse a reaction that converts the superoxide radicals into hydrogen peroxide and molecular oxygen [23]. This is thought to be part of the evolutionary adaptation aiding survival of P. gingivalis in iron-depleted habitats.
The production of Reactive Oxygen Species (ROS) increases as a result of oxidative stress produced by host defences or release by early bacterial colonisers of the oral cavity, for example Streptococcus ssp. Biomarkers of oxidative stress include protein carbonylation, 8-hydroxy-2-deoxyguanosine and lipid peroxidation, which are products of oxidation produced as a result of damage to proteins, DNA and lipids, respectively [24]. Elevated levels of ROS released by neutrophils during infection induce a cytotoxic effect on both human gingival fibroblasts and periodontal bacteria. Moreover, they are secondary messengers in the regulation of cell signalling, cellular homeostasis, and cell apoptosis [25,26]. ROS are sensed by transcriptional regulators which activate a range of survival mechanisms. Fusobacterium nucleatum is considered a scaffold bacterium linking early colonisers to late colonisers of the biofilm and is essential for the continued survival of other anaerobic bacteria in the presence of ROS [27,28]. F. nucleatum uses the alkyl hydroperoxide reductase (AhpC) system for detoxification releasing less ATP, and increasing chaperone proteins to counteract oxidative stress [29]. P. gingivalis expresses similar strategies through the AhpC antioxidation system to provide cellular resistance in the earlier stages of oxidative stress. Furthermore, it can express chaperone proteins in response to oxidative stress to refold misfolded proteins [29,30].
Periodontal bacteria are capable of utilising a vast complement of mechanisms to respond to enviromental stresses and also evade/subvert host responses. In this review, we summarise the literature regarding the genetic, transcriptional, and post-translational mechanisms which underpin the phenotypic plasticity of periodontal pathobionts, with primary focus on P. gingivalis. Table 1 and Figure 1 provide an overview of all the mechanisms discussed in this review.

Table 1.
Summary of modulatory mechanisms of pathogenicity employed by P. gingivalis and other periodontal pathobionts referenced in this review.
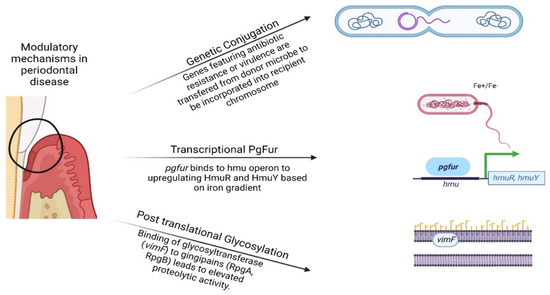
Figure 1.
Visual representation of the genetic, transcriptional, and post-translational modifications of periodontal pathobionts presented in this review. (Created by BioRender.com) accessed on 20 October 2022.
2. Genetic Mechanisms of Pathogenicity in Periodontal Bacteria
Bacterial genetic diversity enhances bacterial fitness and survival in adverse environmental conditions. This often results in enhanced bacterial pathogenicity, primarily through the ability to overcome and subvert human innate and adaptive immune responses. Genetic diversity is mediated partly by mutations and gene polymorphisms, and partly by horizontal gene transfer through conjugation or transduction. In addition, extracellular DNA may also be the source of antimicrobial resistance gene acquisition [101]. DNA transfer through outer membrane vesicles (OMVs) has also been described in periodontal bacteria [61].
2.1. Virulence Gene Polymorphisms and Genetic Diversity
The polymorphisms of several virulence genes are associated with phenotypic variation and increased pathogenicity of P. gingivalis. However, the evidence in this area is at best conflicting with many studies disputing a consistent association between gene polymorphisms and clinical disease, or even with in vitro pathogenicity. Here we briefly summarise key literature relating to the genetic diversity of genes encoding the major virulence factors of P. gingivalis.
Allelic diversity of key virulence genes of P. gingivalis is thought to be partly mediated by the acquisition of extracellular DNA through a transformation-like mechanism. In this context, clonal diversity is enriched, notwithstanding that the fittest strains survive and dominate within the community [62].
P. gingivalis possesses major and minor fimbriae which play an essential role in adherence, invasion of host cells, biofilm formation and immune evasion [102]. Fimbriae enhance the intra-cellular survival of P. gingivalis in macrophages through the hijacking of the TLR2 signalling pathway and internalisation by CD11b/CD18 [31]. The fimA gene, encoding the major fimbrial protein FimA, is the best studied in terms of allelic variations. The fimA gene exists as a single copy on the P. gingivalis chromosome and is considered the basis for the definition of six genotypes of P. gingivalis. However, there are conflicting reports on the phenotypic significance and impact on the virulence of fimA variants. For example, one study that recruited 115 patients suffering from chronic periodontitis and 136 healthy controls reported the predominance of fimA type II and IV genotypes in subgingival plaque samples from deep periodontal pockets and types I and III in plaque from healthy periodontium. Additionally, the periodontal sites of fimA type II predominance displayed higher retrieval frequency of A. actinomycetemcomitans and T. forsythia [32]. Another study showed that type II fimA is associated with enhanced adherence and invasion of epithelial cells compared to type I fimA [33]. Another study followed to demonstrate that fimA type III and IV exhibit improved invasion of host gingival cells [34]. The results of these studies indicate that fimA type II is frequently associated with a diseased periodontium. On the other hand, there are conflicting reports regarding the effect of each fimA genotype on the pathogenicity of P. gingivalis [33,34,35].
The rag locus found in some strains of P. gingivalis encodes two proteins: RagA, a TonB-dependent receptor which controls substrate-specific transport through the outer membrane, and RagB, a lipoprotein that constitutes an immunodominant outer membrane antigen. There are four allele variants of the rag locus: rag-1 was first discovered in strain W50, but rag-2, rag-3 and rag-4 were subsequently characterised [36]. These alleles showed no evidence for a specific geographical predilection. In a collection of 168 isolates of P. gingivalis mainly derived from patients with periodontitis across 15 countries, rag-2 was the predominant allelic form, while rag-4 was the least [37]. In mouse models, inoculation with rag-1 positive isolates showed an increased level of virulence and soft tissue destruction compared to other rag variants [36]. On the other hand, a study that investigated rag locus variants in subgingival plaque samples from patients with chronic periodontitis showed that rag-1 and rag-3 were the most prevalent allelic forms [38]. Another study that sought to investigate the genotypes present in cases of orthodontic gingivitis and mild/moderate periodontitis reported the predominance of rag-3 and rag-4 in these conditions [39].
Six unique capsular serotypes of P. gingivalis are described based on the composition of the K-antigen capsule, each displaying different pathogenic potential, immune-evasive and pro-inflammatory properties [44]. P. gingivalis strain W83 is a virulent serotype K1 encapsulated strain, while P. gingivalis ATCC 33277 lacks a capsule and is considered avirulent [45,46]. Encapsulated P. gingivalis W50 was more resistant to phagocytosis by murine dendritic cells and macrophages compared to its isogenic nonencapsulated mutant (PgC). Additionally, reduced expression of CLCN2, CRP, TGF-α, CXCR4, IL-17, and AGT was observed 1 hr post infection with the encapsulated strain compared to nonencapsulated PgC [47].
2.2. Horizontal Gene Transfer and Genetic Rearrangements
Acquisition of new phenotypes by periodontal bacteria can be mediated by horizontal gene transfer [103,104]. Horizontal gene transfer plays a significant role in genetic diversity and evolution of P. gingivalis, and by extension in the enhancement of its survival and virulence. At the simplest level, key virulence factors and antimicrobial resistance genes of P. gingivalis are thought to be the result of intra-species and inter-species DNA acquisition.
Genetic rearrangements in P. gingivalis through mobile genetic elements are well documented. Conjunctive transposons and insertion sequences are likely incorporated in response to environmental stressors to enhance adaptability. The typical presence of antimicrobial resistance genes on conjugative transposons has also been implicated as a driver of the spread and increase of the antimicrobial resistance burden. CTnPg1 is a conjunctive transposon, the transfer of which is reported between P. gingivalis strains [48] and also between P. gingivalis and other periodontal bacteria, specifically Bacteroides thetaiotaomicron and Prevotella oralis [49]. The excision of CTnPg1 from the donor chromosome is mediated by the PGN_0094 integrase encoded within the transposon itself. CTnPg1 requires a 13-base pair sequence (TTTTCNNNNAAAA) to identify its insertion site and be incorporated within the recipient genome. Given the high variation of the target sequence across strains, the transposon may integrate across multiple sites within the P. gingivalis chromosome [49]. In addition, the recipient strain requires active RecA which is a recombinase responsible for DNA repair and maintaining genome integrity [50] to successfully receive CTnPg1. Importantly, CTnPg1 is likely a vehicle for the transfer of antimicrobial resistance as it includes genes encoding a sodium-mediated multidrug efflux pump, ABC transporters and HAE3 family efflux transporters which can be integrated into multiple sites on the recipient chromosome [49]. P. gingivalis also contains multiple insertion sequences (IS) which undergo transposition through shuffling from one genomic site to another, or retention of the original IS while producing a copy at a different site within the genome. Insertion sequences in P. gingivalis acquired several designations across studies, but they are mostly classified into ISPg1-ISPg7 [51,52]. Despite the frequency of IS shuffling, the representation of different ISPg varies greatly across strains of P. gingivalis [52]. Of note, is the identification of ISPg4 in the genome of more virulent strains of P. gingivalis, implicating ISPg4 (alias IS1598) as a unique marker of virulence [53]. However, a study later identified several duplicates of the virulent strain FDC 381 that lack the presence of ISPg4/IS1598, proving that P. gingivalis virulence is not significantly diminished by the absence of IS1598 [54]. Furthermore, miniature inverted-repeat transposable elements (MITEs) were discovered in high frequency in several P. gingivalis strains [105]. MITEs are short non-coding genetic elements that possess terminal inverted repeats and generate target site duplicates, they are usually AT-rich and preferentially insert into AT-rich regions. The role of MITEs in the modulation of gene expression is well documented [106]. A virulence-associated MITE termed E622 was identified in Pseudomonas syringae and implicated in the mobilisation of antimicrobial resistance genes through an antibiotic coupling mobility assay [107]. Another study identified MITEAba12 in Acinetobacter baumannii which confers heavy-metal resistance through mobilisation of resistance genes such as mer (mercury resistance gene) [108]. It is reasonable to expect MITEs within the P. gingivalis genome to influence gene expression in a similar mechanism. However, additional research is required to understand and confirm the full effect of these elements.
Gingipains are trypsin-like cystine proteases that contribute to most of the overall proteolytic activity of P. gingivalis during periodontal inflammation. Broadly, gingipains may be either arginine-specific or lysine-specific depending on their site of cleavage. Arginine-specific gingipains are encoded by rgpA and rgpB while lysine-specific gingipains are encoded by a single kpg gene. In addition to the proteolytic activity, arginine-specific gingipains are important mediators of pro-inflammatory responses where they directly induce NF-κB dependent production of hepatocyte growth factor (HGF) [109]. The enrichment of gingipain-encoding genes in OMVs suggests the involvement of OMVs as delivery vehicles for horizontal transfer of key virulence factors between different strains of P. gingivalis [110].
As mentioned above, the rag locus is a pathogenicity island associated with P. gingivalis virulence [40]. It is suggested that the locus is acquired from other bacteria within the microbial community such as Bacteroides species through horizontal gene transfer [111]. ISPg1 (IS1126) is present upstream of the rag locus while being flanked by 12-base pair inverted repeats; this insertion sequence consists of a single open reading frame and can generate a 5-base pair target site duplicate [111]. Multiple studies corroborated the rag locus as a horizontally transferred pathogenicity island that plays an important role in the virulence of P. gingivalis [40,111]. Subcutaneous inoculation of mice with rag-knock-out mutants resulted in reduced mortality compared to the P. gingivalis W50 wild-type inoculum [36]. Another study showed reduced spleen invasion by ΔragB P. gingivalis W83 after subcutaneous inoculation compared to P. gingivalis W83 wild-type [41]. Decreased endothelial cell invasion was also shown for P. gingivalis Δrag mutants compared to the parent wild-type as measured by antibiotic protection assays [42]. However, in a more recent study, despite the prevalence of rag-positive isolates in a periodontitis cohort over healthy controls, there was no difference in the ability to invade primary human fibroblasts by rag-positive strains over rag-negative isolates [43].
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-associated genes are found in P. gingivalis. As in other bacteria, CRISPR represents a critical protective mechanism from invasion by bacteriophages [55]. The genome editing activity of CRISPR-Cas systems occurs in three stages: (i) spacer acquisition through recognition of the spacer sequence and its integration into the CRISPR array; (ii) production of CRISPR RNA through cleavage of pre-CRISPR RNA; (iii) interference through mature CRISPR RNA recognition of foreign DNA [56,57]. CRISPR spacers are direct repeats separated by short DNA stretches, which act as a memory pool of past events and have possible targets within the P. gingivalis genome. These potential targets include coding sequences of multiple functions and sequences related to exogenous elements, such as bacteriophages and conjugative transposons [58]. Four CRISPR arrays were identified in P. gingivalis W83, namely CRISPR 30, CRISPR 36.1, CRISPR 36.2 and CRISPR 37 [59]. CRISPR arrays are responsible for regulating mobile genetic element acquisition in P. gingivalis, and a recent report suggested an important role for cas3 in the modulation of potentially pathogenic phenotypes. Cas3 is considered as the signature protein of type I CRISPR-Cas systems. Cas3-deletion mutants of P. gingivalis co-cultured with the monocytic cell line THP-1 showed significantly higher expression of rgpA and several genes associated with response to oxidative stress and iron uptake in P. gingivalis. On the other hand, genes encoding Tra proteins involved in pili formation and ribosomal proteins of both the large and small subunits were downregulated [60]. Cas3-deletion mutants also displayed a modest enhancement of pro-inflammatory properties (IL-1β, IL-6, and IL-10), suggesting that cas3 plays a role in immune homeostasis [60]. Collectively, these findings suggest an overall pathogenicity downregulation by cas3, notwithstanding that the molecular mechanisms underpinning these effects remain unknown.
In addition to the well-characterised mechanisms of horizontal gene transfer (conjugation, transformation or transduction), vesicle-mediated DNA transfer has also been described in P. gingivalis strains [61,62]. OMVs are usually produced by budding from the cell surface of bacteria and enclose periplasmic content. As in other Gram-negative bacteria, P. gingivalis vesicles are 50–250 nm in diameter and have a protein profile that is similar to the outer membrane [63]. Indeed, proteins of the outer membrane of P. gingivalis that have been discovered in OMVs include PorV, LptO, RgpA and Kgp, all major contributors to the pathogenicity of P. gingivalis [64]. As a result, OMVs inherited the functions of these outer membrane proteins and exhibit features such as coaggregation with other oral bacteria, invasion of host cells and pro-inflammatory potential [65]. The molecular mechanism underpinning the formation and release of P. gingivalis vesicles remains poorly defined, but different mechanisms of OMV biogenesis were proposed [66,67]. In the biogenesis of OMV outer membrane proteins and lipoproteins are involved in peptidoglycan cross-linking and anchorage, specifically Lpp, NlpI, OmpA (Pgm6/7) [68]. Tol-Pal members are thought to stabilise the cellular envelope by linking the peptidoglycan layer with the inner membrane in Gram-negative bacteria [68]. It follows that any mutations or deletions of genes encoding Tol-Pal proteins can be reasonably expected to lead to the formation and release of OMVs. The O polysaccharide of LPS has also been implicated in the formation and protein packing of OMVs, mainly mediated by LPS-protein ionic interactions. Indeed, LPS modifications are known to upregulate OMV formation and release in other bacteria such as Salmonella typhimurium [69]. DNA packaging into outer vesicles may occur via uptake of extracellular DNA by free vesicles or DNA uptake into vesicles during cell death. However, neither of these mechanisms has been confirmed. Virulent factors and antibiotic resistance genes packaged into OMVs may play an important role in enhancement of bacterial survival and virulence mediated by horizontal gene transfer [70]. Important P. gingivalis genes such fimA, superoxide dismutase (sod), hmuY and rprY were over-represented in OMVs, implying that virulence factor DNA may be preferentially packaged by P. gingivalis into vesicles [61,62].
3. Transcriptional Mechanisms of Pathogenicity in Periodontal Bacteria
P. gingivalis induces the release of haem in the extracellular space through proteolytic activity such as that mediated by gingipains. Free haem is then sequestered by a haemophore-like protein (HmuY) through which it is delivered to the TonB-dependent outer-membrane receptor responsible for haemoglobin binding (HmuR) [71]. Ferric uptake regulator homologue (PgFur) binds to the hmu operon promoter and triggers the expression of hmuR and hmuY [72]. The expression of pgfur depends on the growth phase and iron/haem gradient in the growth environment. To better understand the role of PgFur in the virulence of P. gingivalis, a study compared the effect of pgfur inactivation in the avirulent strain ATCC 33277 versus the virulent strain A7436. Under high haem conditions the pgfur A7436 mutant strain displayed higher cell density during planktonic growth compared to the wild-type, while wild-type and the pgfur mutant strain of ATCC 33277 showed similar growth rates. Additionally, mutant strains of both virulent and avirulent P. gingivalis were highly sensitive to oxidative stress compared to the respective wild-types [73]. Moreover, the deactivation of the pgfur gene results in decreased adherence, reduced invasion of host cells and reduced intracellular survival [74]. HumY is resistant to several proteases including both gingipain and host proteases such as neutrophil elastase. This property contributes to bacterial persistence in adverse host environments and is reflected in the elevated levels of anti-HmuY antibodies observed in patients suffering from chronic periodontitis [75].
PgRsp is a member of the Crp/Fnr superfamily transcription regulators responsible for haem-binding and redox state sensing through haem-catalysed oxidation [76,77]. In support of these roles, pgrsp mutants showed a reduced uptake of haem mediated by HmuY in biofilm-forming conditions [76,78]. Further, PgRsp has been implicated in modulation of bacterial virulence. For example, upregulation of the expression of kgp and downregulation of the expression of rgpB and rgpA were observed when comparing the pgrsp knock-out mutant to the wild-type [76]. Inactivation of the PgRsp-encoding gene in P. gingivalis resulted in increased coaggregation with both Prevotella intermedia and T. forsythia whilst coaggregation with Streptococcus gordonii was not affected by mutagenesis [76]. The inactivation mutant displayed enhanced biofilm formation but diminished fimA expression and reduced survival in macrophages compared to the wild-type, suggesting an important role for pgrsp in regulation of pathogenicity [76].
Environmental stresses such as oxidative and nitrate stresses trigger transcriptional mechanisms of bacterial adaptation and survival which, in turn, may be deemed important for pathogenicity. For example, the transcriptional regulator RprY of P. gingivalis, induced as a response to oxidative stress, binds to the target promoter and represses the toxic effect of ROS by regulating the activity of the sodium-dependent ubiquinone oxidoreductase system. This system utilises the sodium gradient to produce energy for the cytochrome d oxidase (cydAB) operon which converts oxidative radicals into water. In aerobic conditions, rprY inactivation is lethal, presumably due to the decreased activity of the sodium-dependent ubiquinone oxidoreductase system regulated by RprY [79].
Extracytoplasmic function (ECF) sigma factors comprise a large group of transcriptional factors that regulate gene expression in response to environmental stresses. Six ECF sigma factors were described in P. gingivalis ATCC 33277: PGN_0274, PGN_0319, PGN_0450, PGN_0970, PGN_1108, and PGN_1740 [80], while in P. gingivalis W83 ECFs identified are PG0162, PG0214, PG0985, PG1318, PG1660, and PG1827 [81]. The P. gingivalis W83 isogenic single inactivation mutants of PG0985, PG1660 and PG1827 displayed reduced growth and survival upon exposure to hydrogen peroxide when compared to P. gingivalis W83 wild-type. Moreover, Rgp gingipain activity was reduced by 50% in the PG0162 knock-out and by 60% in the PG1660 knock-out mutant, while Kgp activity was reduced by 20% and 50% in the PG0162 and PG1660 mutants, respectively [81]. Synergy can occur between ECF sigma factors as observed for PG0162 and PG1660, jointly contributing to overall bacterial resistance to oxidative stress. However, the mechanism by which oxidative stress is sensed in this scenario and how ECF sigma factors are activated requires further study [81,82]. The PorX/PorY two-component system controls the expression of por genes encoding the type IX secretion system and ECF sigma factor SigP. The PorX/PorY system upregulates the transcription of the sigP gene, which in turn mediates the transcriptional activation of the por genes by binding to their promoters [83]. The sigP-knock-out of P. gingivalis ATCC 33277 showed reduced auto-aggregation compared to the respective wild-type, while sigP and sigH knock-outs both exhibited a drop in haemagglutination activity. When testing the effect of sigP on P. gingivalis virulence in a mouse infection model, the sigP mutant exhibited lower mortality rates compared to the W83 parent strain [80]. Another study confirmed this finding and showed a similar effect as a result of deletion of porX [84]. Additionally, the two-component system in P. gingivalis modulates the transcription of genes associated with biofilm formation, LPS modification and host cell invasion [83].
4. Post-Translational Mechanisms of Pathogenicity in Periodontal Bacteria
Post-translational modification of proteins mediates important structural and functional properties. A variety of protein post-translational changes can be induced by environmental changes and affect the cellular mechanisms that enhance bacterial survival and pathogenicity. Post-translational mechanisms deemed particularly important for pathogenicity of P.gingivalis include glycosylation and acetylation [112].
4.1. Acetylation
Lysine acetylation is achieved by lysine acetyltransferases and acetyl-coenzyme A. It generally mediates changes in the secondary structure of a protein and resulting binding properties. The presence of lysine acetylated enzymes in P. gingivalis was linked to carbohydrate, amino acid and lipid metabolism but not glycan biosynthesis [85]. These metabolic pathways are essential for the continued growth and persistence of P. gingivalis in the microbial community as they produce ATP and promote the release of short fatty acids required for bacterial growth and survival [85]. Protein acetylation leads to the activation of proteases which aids pathogenesis in periodontitis through the degradation of host extracellular matrix components, degradation of antimicrobial peptides and cytokines [86]. Proteins such as Mfa1 fimbrilin, Haemagglutinin protein HagA and Methionine gamma-lyase related to P. gingivalis pigmentation were identified as targets of acetylation. Acetylation of the Ferritin and Putative universal stress protein (UspA) has also been associated with P. gingivalis oxidative stress resistance [87]. Lysine acetylation plays a major role in the enhancement of bacterial survival and virulence as all three types of gingipains are acetylated [88].
RprY mediates oxidative stress response in P. gingivalis. RprY plays an essential role in the regulation of P. gingivalis virulence through type IX secretion system, which releases gingipains and peptidyl-arginine deiminase (PPAD), both considered major mediators of periodontal tissue destruction. Moreover, the reduction of RprY decreases Mfa1 fimbriae-mediated adhesion as observed by the retarded transcription of mfa1 in ∆rprY mutants when compared to the wild-type [89]. RprY is acetylated in vivo in conjunction with its co-transcriptional partner downstream of RprY which is known as protein acetyltransferase (Pat) [90].
There is extensive overlap between lysine acetylation and lysine succinylation in P. gingivalis ribosomal proteins. It was observed that ten proteins responsible for glutamate and aspartate catabolism in P. gingivalis experience both acetylation and succinylation [87]. This overlap is consistent with findings in other bacteria, but its significance in the regulation of survival and virulence has not been well explored.
4.2. Glycosylation
Glycosyltransferases break down monosaccharides or oligosaccharides from an activated sugar donor (UDP-sugar) to different substrates, including carbohydrates, proteins and glycoproteins. P. gingivalis glycosyltransferases play a role in the synthesis of A-LPS and O-LPS lipopolysaccharide [91]. A-LPS anchors virulence proteins such as gingipain RgpB and Haemin-binding protein 35 (HBP35). The latter binds thioredoxin and haemin supporting bacterial haem acquisition especially in iron-depleted environments [92], hence affecting pathogenicity [91,92,93,94,95,96]. Gingipains themselves are also substrates of glycosyltransferases. Glycosylation of the arginine-specific proteinase and adhesin (RgpA) protects the enzyme against proteolytic degradation in adverse conditions induced by host responses at inflammed periodontal sites [97]. Virulence-modulating gene F (vimF) is a putative group 1 glycosyltransferase which regulates virulence and haem acquisition. Indeed, vimF inactivation results in reduced proteolytic activity and lower haemagglutination in P. gingivalis W83 cultured in vitro [93]. This is attributed to a glycosylation defect of haemagglutinin HagA and retarded gingipain maturation [93].
The glycosylation of surface proteins is critical to the regulation of host-microbial interactions. OMP85 is a highly conserved outer membrane protein of P. gingivalis, the glycosylation of which is mediated by the galE gene [96]. Investigations of the functional role of galE in P. gingivalis revealed a critical role in the modulation of biofilm formation. In a study where P. gingivalis ATCC 33277 and the isogenic galE knock-out mutant were incubated with an antibody targeting the outer-loop peptide of OMP85, it was observed that the antibody had an enhanced inhibitory effect on bacterial attachment and biofilm formation in the galE mutant over the wild-type. This suggested an imporant role of galE-mediated glycosylation in the maturation of adhesins and biofilm-forming factors [96].
O-glycosylation systems in T. forsythia mediate glycosylation of cell surface S-layer glycoproteins (TfsA and TfsB), which play an important role in bacterial virulence through promoting biofilm formation and bacterial aggregation [98]. A study measured the function of human monocyte-derived dendritic cells stimulated with three single gene knock-outs of glycosyltransferases gtfS, gtfI and gtfE in comparison with the respective wild-type. Here, T. forsythia gtfE knock-outs induced marked elevation of IL-1β, IL-12, IL-10 and IL-23, suggesting a role for gtfE in Th17 activation. On the other hand, these cytokines were significantly reduced in gftS and gftI single mutants compared to the wild-type suggesting Th17 suppression. These findings suggest a complex immunomodulatory role for T. forsythia [99].
Flagellar proteins (FlaA, FlaB and FlaB1) of T. denticola are heavily glycosylated. Pse (Pseudaminic Acid Bacterial Glycans) and Pse-like glycans have also been identified as an obligate precursor for glycosylation reactions involved in flagellar production. PseI (TDE0960) in T. denticola is critical for flaA and flaB expression and flagellin biosynthesis [100]. Reduction of FlaA and FlaB proteins in deletion mutants resulted in impaired assembly of flagellar filaments, ultimately leading to aberrant morphogenesis, lack of motility, reduced adherence and host cell invasion [100].
5. Conclusions and Recommendations for Future Research
An enhanced understanding of the mechanisms which drive the transition of periodontal pathobionts from commensals to pathogens is critical for the development of prevention strategies and therapies for periodontitis. This review has identified genetic, transcriptional and post-translational mechanisms that regulate the pathogenic potential of P. gingivalis. There is a distinct lack of literature regarding equivalent mechanisms in other periodontal bacteria which are considered equally critical to the pathogenesis of periodontal disease, namely Treponema denticola, Tannerella forsythia, Prevotella intermedia and Aggregatibacter Actinomycetemcomitans. Oral bacteria interact and synergise to enhance their pathogenicity under less favourable conditions. Furthermore, horizontal gene transfer occurs between different bacterial species that colonise the periodontal environment. Therefore, the scope of identifying modulatory mechanisms of bacterial virulence involved in periodontal disease must be expanded to include other periodontal pathobionts.
Moving forward, additional research is needed to establish the mechanisms by which periodontal pathobionts alter their virulence and cause disease. On the backdrop of long-standing literature investigating the genetic diversity of P. gingivalis, the evidence on the relationship between genetic diversity and altered pathogenicity of P. gingivalis is conflicting. On the other hand, the literature concerning transcriptional regulation and post-translational modifications of virulence factors is relatively limited and merits further study.
The field of epigenetics is of significant interest in the study of bacterial pathogenicity. Bacteria such as Streptococcus ssp. [113], Helicobacter pylori [114] and Neisseria gonorrhoeae [115] utilise epigenetic modifications for the modulation of virulence and immune evasion by switching to allelic forms that permits their adaptation to changing environments. In bacterial phase variation, DNA methylation driven by environmental change modulates gene expression and phenotypic diversity without mutations or genetic rearrangements of sequences encoding the virulence factors themselves. Epigenetic phase variations may be equally important in periodontal pathogens, as these mechanisms mediate virulence and immune evasion to aid the survival of a range of bacterial species under environmental stresses [116,117].
Author Contributions
Conceptualisation, K.H.; writing—original draft preparation, S.S.; writing—review and editing, K.H. and S.S.; supervision, K.H. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- Hajishengallis, G.; Lamont, R.J. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 2012, 27, 409–419. [Google Scholar] [CrossRef]
- Suzuki, N.; Yoneda, M.; Hirofuji, T. Mixed red-complex bacterial infection in periodontitis. Int. J. Dent. 2013, 587279. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Lambris, J.D. Complement and dysbiosis in periodontal disease. Immunobiology 2012, 217, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Hajishengallis, G.; Liang, S.; Payne, M.A.; Hashim, A.; Jotwani, R.; Eskan, M.A.; McIntosh, M.L.; Alsam, A.; Kirkwood, K.L.; Lambris, J.D.; et al. Low-Abundance Biofilm Species Orchestrates Inflammatory Periodontal Disease through the Commensal Microbiota and Complement. Cell Host Microbe 2011, 10, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Hornef, M. Pathogens, Commensal Symbionts, and Pathobionts: Discovery and Functional Effects on the Host. ILAR J. 2015, 56, 159–162. [Google Scholar] [CrossRef] [PubMed]
- Kawamoto, D.; Borges, R.; Ribeiro, R.A.; de Souza, R.F.; Amado, P.P.P.; Saraiva, L.; Horliana, A.C.R.T.; Faveri, M.; Mayer, M.P.A. Oral Dysbiosis in Severe Forms of Periodontitis Is Associated with Gut Dysbiosis and Correlated with Salivary Inflammatory Mediators: A Preliminary Study. Front. Oral Health 2021, 2, 722495. [Google Scholar] [CrossRef]
- Payne, M.; Hashim, A.; Alsam, A.; Joseph, S.; Aduse-Opoku, J.; Wade, W.; A Curtis, M. Horizontal and Vertical Transfer of Oral Microbial Dysbiosis and Periodontal Disease. J. Dent. Res. 2019, 98, 1503–1510. [Google Scholar] [CrossRef]
- Deng, Z.-L.; Szafrański, S.P.; Jarek, M.; Bhuju, S.; Wagner-Döbler, I. Dysbiosis in chronic periodontitis: Key microbial players and interactions with the human host. Sci. Rep. 2017, 7, 3703. [Google Scholar] [CrossRef]
- Hijazi, K.; Morrison, R.W.; Mukhopadhya, I.; Martin, B.; Gemmell, M.; Shaw, S.; Santoro, F. Oral bacterial diversity is inversely correlated with mucosal inflammation. Oral Dis. 2020, 26, 1566–1575. [Google Scholar] [CrossRef]
- Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends Immunol. 2014, 35, 3–11. [Google Scholar] [CrossRef]
- Schroeder, J.W.; Yeesin, P.; Simmons, L.A.; Wang, J.D. Sources of spontaneous mutagenesis in bacteria. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 29–48. [Google Scholar] [CrossRef] [PubMed]
- Aduse-Opoku, J.; Joseph, S.; A Devine, D.; Marsh, P.D.; A Curtis, M. Molecular basis for avirulence of spontaneous variants of Porphyromonas gingivalis: Genomic analysis of strains W50, BE1 and BR1. Mol. Oral Microbiol. 2022, 37, 122–132. [Google Scholar] [CrossRef] [PubMed]
- Shapiro Rebecca, S.; Cowen Leah, E. Thermal Control of Microbial Development and Virulence: Molecular Mechanisms of Microbial Temperature Sensing. mBio 2012, 3, e00238-12. [Google Scholar] [PubMed]
- Chopra, A.; Bhat, S.G.; Sivaraman, K. Porphyromonas gingivalis adopts intricate and unique molecular mechanisms to survive and persist within the host: A critical update. J. Oral Microbiol. 2020, 12, 1801090. [Google Scholar] [CrossRef]
- Curtis, M.A.; Percival, R.S.; Devine, D.; Darveau, R.P.; Coats, S.R.; Rangarajan, M.; Tarelli, E.; Marsh, P.D. Temperature-Dependent Modulation of Porphyromonas gingivalis Lipid A Structure and Interaction with the Innate Host Defenses. Infect. Immun. 2011, 79, 1187–1193. [Google Scholar] [CrossRef]
- Al-Qutub, M.N.; Braham, P.H.; Karimi-Naser, L.M.; Liu, X.; Genco, C.A.; Darveau, R.P. Hemin-Dependent Modulation of the Lipid A Structure of Porphyromonas gingivalis Lipopolysaccharide. Infect. Immun. 2006, 74, 4474–4485. [Google Scholar] [CrossRef]
- Portnoy, D.A. Manipulation of Innate Immunity by Bacterial Pathogens. Curr. Opin. Immunol. 2005, 17, 25–28. [Google Scholar] [CrossRef]
- Darveau, R.P.; Pham, T.T.; Lemley, K.; Reife, R.A.; Bainbridge, B.W.; Coats, S.R.; Howald, W.N.; Way, S.S.; Hajjar, A.M. Porphyromonas gingivalis Lipopolysaccharide Contains Multiple Lipid A Species That Functionally Interact with Both Toll-Like Receptors 2 and 4. Infect. Immun. 2004, 72, 5041–5051. [Google Scholar] [CrossRef]
- Kesavalu, L.; Holt, S.C.; Ebersole, J.L. In vitro environmental regulation of Porphyromonas gingivalis growth and virulence. Oral Microbiol. Immunol. 2003, 18, 226–233. [Google Scholar] [CrossRef]
- Schultze, L.B.; Maldonado, A.; Lussi, A.; Sculean, A.; Eick, S. The Impact of the pH Value on Biofilm Formation. Monogr. Oral Sci. 2020, 19–29. [Google Scholar]
- Boisen, G.; Davies, J.R.; Neilands, J. Acid tolerance in early colonizers of oral biofilms. BMC Microbiol. 2021, 21, 45. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Miyazaki, H.; Anaya, C.; Yu, F.; Yeudall, W.A.; Lewis, J.P. Role of Porphyromonas gingivalis FeoB2 in Metal Uptake and Oxidative Stress Protection. Infect. Immun. 2006, 74, 4214–4223. [Google Scholar] [CrossRef] [PubMed]
- Dashper, S.; Butler, C.; Lissel, J.P.; Paolini, R.A.; Hoffmann, B.; Veith, P.; O'Brien-Simpson, N.; Snelgrove, S.L.; Tsiros, J.T.; Reynolds, E. A Novel Porphyromonas gingivalis FeoB Plays a Role in Manganese Accumulation. J. Biol. Chem. 2005, 280, 28095–28102. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Mo, L.; Niu, Y.; Li, X.; Zhou, X.; Xu, X. The Role of Reactive Oxygen Species and Autophagy in Periodontitis and Their Potential Linkage. Front. Physiol. 2017, 8, 439. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.-Q.; Lu, W.; Chen, Y.; Cheng, X.-F.; Qiu, J.-Y.; Xu, Y.; Sun, Y. Effects of Porphyromonas gingivalis Lipopolysaccharide Tolerized Monocytes on Inflammatory Responses in Neutrophils. PLoS ONE 2016, 11, e0161482. [Google Scholar]
- Tomofuji, T.; Irie, K.; Sanbe, T.; Azuma, T.; Ekuni, D.; Tamaki, N.; Yamamoto, T.; Morita, M. Periodontitis and increase in circulating oxidative stress. Jpn. Dent. Sci. Rev. 2009, 45, 46–51. [Google Scholar] [CrossRef]
- Thurnheer, T.; Karygianni, L.; Flury, M.; Belibasakis, G.N. Fusobacterium Species and Subspecies Differentially Affect the Composition and Architecture of Supra- and Subgingival Biofilms Models. Front. Microbiol. 2019, 10, 1716. [Google Scholar] [CrossRef] [PubMed]
- Zilm, P.S.; Rogers, A.H. Co-adhesion and biofilm formation by Fusobacterium nucleatum in response to growth pH. Anaerobe 2007, 13, 146–152. [Google Scholar] [CrossRef]
- Steeves, C.H.; Potrykus, J.; Barnett, D.A.; Bearne, S.L. Oxidative stress response in the opportunistic oral pathogen Fusobacterium nucleatum. Proteomics 2011, 11, 2027–2037. [Google Scholar] [CrossRef]
- Okano, S.; Shibata, Y.; Shiroza, T.; Abiko, Y. Proteomics-based analysis of a counter-oxidative stress system in Porphyromonas gingivalis. Proteomics 2006, 6, 251–258. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Wang, M.; Harokopakis, E.; Triantafilou, M.; Triantafilou, K. Porphyromonas gingivalis fimbriae proactively modulate beta2 integrin adhesive activity and promote binding to and internalization by macrophages. Infect Immun. 2006, 74, 5658–5666. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Wu, Y.-F.; Meng, S.; Yang, H.; Ouyang, Y.-L.; Zhou, X.-D. Prevalence of fimA genotypes of Porphyromonas gingivalis and periodontal health status in Chinese adults. J. Periodontal Res. 2007, 42, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kawai, S.; Nakano, K.; Inaba, H.; Kuboniwa, M.; Nakagawa, I.; Tsuda, K.; Omori, H.; Ooshima, T.; Yoshimori, T.; et al. Virulence of Porphyromonas gingivalis is altered by substitution of fimbria gene with different genotype. Cell Microbiol. 2007, 9, 753–765. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.E.; Abramian, J.R.; Dao, D.-H.V.; Rigney, T.W.; Fritz, J.; Pham, T.; Gay, I.; Parthasarathy, K.; Wang, B.-Y.; Zhang, W.; et al. Genetic Exchange of Fimbrial Alleles Exemplifies the Adaptive Virulence Strategy of Porphyromonas gingivalis. PLoS ONE 2014, 9, e91696. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tribble, G.D.; Kerr, J.E.; Wang, B.-Y. Genetic diversity in the oral pathogen Porphyromonas gingivalis: Molecular mechanisms and biological consequences. Future Microbiol. 2013, 8, 607–620. [Google Scholar] [CrossRef]
- Shi, X.; Hanley, S.A.; Faray-Kele, M.-C.; Fawell, S.C.; Aduse-Opoku, J.; Whiley, R.A.; Curtis, M.A.; Hall, L.M.C. The rag Locus of Porphyromonas gingivalis Contributes to Virulence in a Murine Model of Soft Tissue Destruction. Infect. Immun. 2007, 75, 2071–2074. [Google Scholar] [CrossRef][Green Version]
- Hall Lucinda, M.C.; Fawell, S.C.; Shi, X.; Faray-Kele, M.; Aduse-Opoku, J.; Whiley, R.A.; Curtis, M.A. Sequence Diversity and Antigenic Variation at the rag Locus of Porphyromonas gingivalis. Infect. Immun. 2005, 73, 4253–4262. [Google Scholar] [CrossRef]
- Wang, G.H.; Zhang, D.M.; Pan, Y.P. Distribution of rag genotypes of Porphyromonas gingivalis in patients with chronic periodontitis. West China J. Stomatol. 2009, 27, 168–171. [Google Scholar]
- Liu, Y.; Zhang, Y.; Wang, L.; Guo, Y.; Xiao, S. Prevalence of Porphyromonas gingivalis Four rag Locus Genotypes in Patients of Orthodontic Gingivitis and Periodontitis. PLoS ONE 2013, 8, e61028. [Google Scholar] [CrossRef]
- Curtis, M.A.; Hanley, S.A.; Aduse-Opoku, J. The rag locus of Porphyromonas gingivalis: A novel pathogenicity island. J. Periodontal Res. 1999, 34, 400–405. [Google Scholar] [CrossRef]
- Nagano, K.; Murakami, Y.; Nishikawa, K.; Sakakibara, J.; Shimozato, K.; Yoshimura, F. Characterization of RagA and RagB in Porphyromonas gingivalis: Study using gene-deletion mutants. J. Med. Microbiol. 2007, 56, 1536–1548. [Google Scholar] [CrossRef] [PubMed]
- Dolgilevich, S.; Rafferty, B.; Luchinskaya, D.; Kozarov, E. Genomic comparison of invasive and rare non-invasive strains reveals Porphyromonas gingivalis genetic polymorphisms. J. Oral Microbiol. 2011, 3, 5764. [Google Scholar] [CrossRef] [PubMed]
- Bunte, K.; Kuhn, C.; Walther, C.; Peters, U.; Aarabi, G.; Smeets, R.; Beikler, T. Clinical significance of ragA, ragB, and PG0982 genes in Porphyromonas gingivalis isolates from periodontitis patients. Eur. J. Oral Sci. 2021, 129, e12776. [Google Scholar] [CrossRef] [PubMed]
- Aduse-Opoku, J.; Slaney, J.M.; Hashim, A.; Gallagher, A.; Gallagher, R.P.; Rangarajan, M.; Boutaga, K.; Laine, M.L.; Van Winkelhoff, A.J.; Curtis, M.A. Identification and Characterization of the Capsular Polysaccharide (K-Antigen) Locus of Porphyromonas gingivalis. Infect. Immun. 2006, 74, 449–460. [Google Scholar] [CrossRef] [PubMed]
- Yoshino, T.; Laine, M.L.; jan van Winkelhoff, A.; Dahlén, G. Genotype variation and capsular serotypes of Porphyromonas gingivalis from chronic periodontitis and periodontal abscesses. FEMS Microbiol. Lett. 2007, 270, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Hosogi, Y.; Nishikawa, K.; Abbey, K.; Fleischmann, R.D.; Walling, J.; Duncan, M.J. Comparative Whole-Genome Analysis of Virulent and Avirulent Strains of Porphyromonas gingivalis. J. Bacteriol. 2004, 186, 5473–5479. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Wyant, T.; Anaya-Bergman, C.; Aduse-Opoku, J.; Brunner, J.; Laine, M.L.; A Curtis, M.; Lewis, J.P. The Capsule of Porphyromonas gingivalis Leads to a Reduction in the Host Inflammatory Response, Evasion of Phagocytosis, and Increase in Virulence. Infect. Immun. 2011, 79, 4533–4542. [Google Scholar] [CrossRef]
- Naito, M.; Hirakawa, H.; Yamashita, A.; Ohara, N.; Shoji, M.; Yukitake, H.; Nakayama, K.; Toh, H.; Yoshimura, F.; Kuhara, S.; et al. Determination of the Genome Sequence of Porphyromonas gingivalis Strain ATCC 33277 and Genomic Comparison with Strain W83 Revealed Extensive Genome Rearrangements in P. gingivalis. DNA Res. 2008, 15, 215–225. [Google Scholar] [CrossRef]
- Naito, M.; Sato, K.; Shoji, M.; Yukitake, H.; Ogura, Y.; Hayashi, T.; Nakayama, K. Characterization of the Porphyromonas gingivalis conjugative transposon CTnPg1: Determination of the integration site and the genes essential for conjugal transfer. Microbiology 2011, 157, 2022–2032. [Google Scholar] [CrossRef]
- del Val, E.; Nasser, W.; Abaibou, H.; Reverchon, S. RecA and DNA recombination: A review of molecular mechanisms. Biochem. Soc. Trans. 2019, 47, 1511–1531. [Google Scholar] [CrossRef]
- Califano, J.V.; Arimoto, T.; Kitten, T. The genetic relatedness of Porphyromonas gingivalis clinical and laboratory strains assessed by analysis of insertion sequence (IS) element distribution. J. Periodontal Res. 2003, 38, 411–416. [Google Scholar] [CrossRef] [PubMed]
- Acuña-Amador, L.; Primot, A.; Cadieu, E.; Roulet, A.; Barloy-Hubler, F. Genomic repeats, misassembly and reannotation: A case study with long-read resequencing of Porphyromonas gingivalis reference strains. BMC Genom. 2018, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Sawada, K.; Kokeguchi, S.; Hongyo, H.; Sawada, S.; Miyamoto, M.; Maeda, H.; Nishimura, F.; Takashiba, S.; Murayama, Y. Identification by Subtractive Hybridization of a Novel Insertion Sequence Specific for Virulent Strains of Porphyromonas gingivalis. Infect. Immun. 1999, 67, 5621–5625. [Google Scholar] [CrossRef] [PubMed]
- Frandsen, E.V.G.; Poulsen, K.; Curtis, M.A.; Kilian, M. Evidence of Recombination in Porphyromonas gingivalis and Random Distribution of Putative Virulence Markers. Infect. Immun. 2001, 69, 4479–4485. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Olsen, I. Porphyromonas gingivalis and its CRISPR-Cas system. J. Oral Microbiol. 2019, 11, 1638196. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, M.A.B.; Shabbir, M.Z.; Wu, Q.; Mahmood, S.; Sajid, A.; Maan, M.K.; Ahmed, S.; Naveed, U.; Hao, H.; Yuan, Z. CRISPR-cas system: Biological function in microbes and its use to treat antimicrobial resistant pathogens. Ann. Clin. Microbiol. Antimicrob. 2019, 18, 21. [Google Scholar] [CrossRef] [PubMed]
- Shabbir, M.A.B.; Hao, H.; Shabbir, M.Z.; Hussain, H.I.; Iqbal, Z.; Ahmed, S.; Sattar, A.; Iqbal, M.; Li, J.; Yuan, Z. Survival and Evolution of CRISPR–Cas System in Prokaryotes and Its Applications. Front Immunol. 2016, 7. [Google Scholar] [CrossRef]
- Watanabe, T.; Shibasaki, M.; Maruyama, F.; Sekizaki, T.; Nakagawa, I. Investigation of potential targets of Porphyromonas CRISPRs among the genomes of Porphyromonas species. PLoS ONE 2017, 12, e0183752. [Google Scholar] [CrossRef]
- Burmistrz, M.; Dudek, B.; Staniec, D.; Martinez, J.I.R.; Bochtler, M.; Potempa, J.; Pyrc, K. Functional Analysis of Porphyromonas gingivalis W83 CRISPR-Cas Systems. J. Bacteriol. 2015, 197, 2631–2641. [Google Scholar] [CrossRef]
- Solbiati, J.; Duran-Pinedo, A.; Rocha, F.G.; Gibson, F.C.; Frias-Lopez, J. Virulence of the Pathogen Porphyromonas gingivalis Is Controlled by the CRISPR-Cas Protein Cas3. mSystems 2020, 5, e00852-20. [Google Scholar] [CrossRef]
- Ho, M.-H.; Chen, C.-H.; Goodwin, J.S.; Wang, B.-Y.; Xie, H. Functional Advantages of Porphyromonas gingivalis Vesicles. PLoS ONE 2015, 10, e0123448. [Google Scholar] [CrossRef] [PubMed]
- Tribble, G.D.; Rigney, T.W.; Dao, D.V.; Wong, C.T.; Kerr, J.E.; Taylor, B.E.; Pacha, S.; Kaplan, H.B. Natural Competence Is a Major Mechanism for Horizontal DNA Transfer in the Oral Pathogen Porphyromonas gingivalis. mBio 2012, 3, e00231-11. [Google Scholar] [CrossRef] [PubMed]
- Nakao, R.; Takashiba, S.; Kosono, S.; Yoshida, M.; Watanabe, H.; Ohnishi, M.; Senpuku, H. Effect of Porphyromonas gingivalis outer membrane vesicles on gingipain-mediated detachment of cultured oral epithelial cells and immune responses. Microbes Infect. 2014, 16, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Veith, P.D.; Chen, Y.-Y.; Gorasia, D.G.; Chen, D.; Glew, M.D.; O’Brien-Simpson, N.M.; Cecil, J.D.; Holden, J.A.; Reynolds, E.C. Porphyromonas gingivalis Outer Membrane Vesicles Exclusively Contain Outer Membrane and Periplasmic Proteins and Carry a Cargo Enriched with Virulence Factors. J. Proteome Res. 2014, 13, 2420–2432. [Google Scholar] [CrossRef]
- Mantri, C.K.; Chen, C.; Dong, X.; Goodwin, J.S.; Pratap, S.; Paromov, V.; Xie, H. Fimbriae-mediated outer membrane vesicle production and invasion of Porphyromonas gingivalis. MicrobiologyOpen 2015, 4, 53–65. [Google Scholar] [CrossRef]
- Gui, M.J.; Dashper, S.G.; Slakeski, N.; Chen, Y.-Y.; Reynolds, E.C. Spheres of influence: Porphyromonas gingivalis outer membrane vesicles. Mol. Oral Microbiol. 2016, 31, 365–378. [Google Scholar] [CrossRef]
- Okamura, H.; Hirota, K.; Yoshida, K.; Weng, Y.; He, Y.; Shiotsu, N.; Ikegame, M.; Uchida-Fukuhara, Y.; Tanai, A.; Guo, J. Outer membrane vesicles of Porphyromonas gingivalis: Novel communication tool and strategy. Jpn. Dent. Sci. Rev. 2021, 57, 138–146. [Google Scholar] [CrossRef]
- Iwami, J.; Murakami, Y.; Nagano, K.; Nakamura, H.; Yoshimura, F. Further evidence that major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis stabilize bacterial cells. Oral Microbiol. Immunol. 2007, 22, 356–360. [Google Scholar] [CrossRef]
- Avila-Calderón, E.D.; Ruiz-Palma, M.D.S.; Aguilera-Arreola, M.G.; Velázquez-Guadarrama, N.; Ruiz, E.A.; Gomez-Lunar, Z.; Witonsky, S.; Contreras-Rodríguez, A. Outer Membrane Vesicles of Gram-Negative Bacteria: An Outlook on Biogenesis. Front. Microbiol. 2021, 12, 557902. [Google Scholar] [CrossRef]
- Berleman, J.; Auer, M. The role of bacterial outer membrane vesicles for intra- and interspecies delivery. Environ. Microbiol. 2013, 15, 347–354. [Google Scholar] [CrossRef]
- Kosno, J.; Siemińska, K.; Olczak, T. Unique Properties of Heme Binding of the Porphyromonas gingivalis HmuY Hemophore-like Protein Result from the Evolutionary Adaptation of the Protein Structure. Molecules 2022, 27, 1703. [Google Scholar] [CrossRef] [PubMed]
- Simpson, W.; Olczak, T.; Caroline, A.G. Characterization and Expression of HmuR, a TonB-Dependent Hemoglobin Receptor of Porphyromonas gingivalis. J. Bacteriol. 2000, 182, 5737–5748. [Google Scholar] [CrossRef] [PubMed]
- Śmiga, M.; Stępień, P.; Olczak, M.; Olczak, T. PgFur participates differentially in expression of virulence factors in more virulent A7436 and less virulent ATCC 33277 Porphyromonas gingivalis strains. BMC Microbiol. 2019, 19, 127. [Google Scholar] [CrossRef]
- Ciuraszkiewicz, J.; Śmiga, M.; Mackiewicz, P.; Gmiterek, A.; Bielecki, M.; Olczak, M.; Olczak, T. Fur homolog regulates Porphyromonas gingivalis virulence under low-iron/heme conditions through a complex regulatory network. Mol. Oral Microbiol. 2014, 29, 333–353. [Google Scholar] [CrossRef] [PubMed]
- Olczak, T.; Sosicka, P.; Olczak, M. HmuY is an important virulence factor for Porphyromonas gingivalis growth in the heme-limited host environment and infection of macrophages. Biochem. Biophys. Res. Commun. 2015, 467, 748–753. [Google Scholar] [CrossRef] [PubMed]
- Śmiga, M.; Olczak, T. PgRsp Is a Novel Redox-Sensing Transcription Regulator Essential for Porphyromonas gingivalis Virulence. Microorganisms 2019, 7, 623. [Google Scholar] [CrossRef]
- Pinochet-Barros, A.; Helmann, J.D. Redox Sensing by Fe2+ in Bacterial Fur Family Metalloregulators. Antioxid. Redox Signal. 2017, 29, 1858–1871. [Google Scholar] [CrossRef]
- Romero-Lastra, P.; Sánchez, M.D.C.; Llama-Palacios, A.; Figuero, E.; Herrera, D.; Sanz, M. Gene expression of Porphyromonas gingivalis ATCC 33277 when growing in an in vitro multispecies biofilm. PLoS ONE 2019, 14, e0221234. [Google Scholar] [CrossRef]
- Krishnan, K.; Duncan, M.J. Role of Sodium in the RprY-Dependent Stress Response in Porphyromonas gingivalis. PLoS ONE 2013, 8, e63180. [Google Scholar] [CrossRef]
- Fujise, K.; Kikuchi, Y.; Kokubu, E.; Okamoto-Shibayama, K.; Ishihara, K. Effect of extracytoplasmic function sigma factors on autoaggregation, hemagglutination, and cell surface properties of Porphyromonas gingivalis. PLoS ONE 2017, 12, e0185027. [Google Scholar] [CrossRef]
- Dou, Y.; Osbourne, D.; McKenzie, R.; Fletcher, H.M. Involvement of extracytoplasmic function sigma factors in virulence regulation in Porphyromonas gingivalis W83. FEMS Microbiol. Lett. 2010, 312, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Dou, Y.; Rutanhira, H.; Chen, X.; Mishra, A.; Wang, C.; Fletcher, H. Role of extracytoplasmic function sigma factor PG1660 (RpoE) in the oxidative stress resistance regulatory network of Porphyromonas gingivalis. Mol. Oral Microbiol. 2018, 33, 89–104. [Google Scholar] [CrossRef] [PubMed]
- Kadowaki, T.; Yukitake, H.; Naito, M.; Sato, K.; Kikuchi, Y.; Kondo, Y.; Shoji, M.; Nakayama, K. A two-component system regulates gene expression of the type IX secretion component proteins via an ECF sigma factor. Sci. Rep. 2016, 6, 23288. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Yang, D.; Hua, T.; Hua, Z.; Kong, W.; Shi, Y. A PorX/PorY and σP Feedforward Regulatory Loop Controls Gene Expression Essential for Porphyromonas gingivalis Virulence. mSphere 2021, 6, e00428-21. [Google Scholar] [CrossRef] [PubMed]
- Butler, C.A.; Veith, P.D.; Nieto, M.F.; Dashper, S.G.; Reynolds, E.C. Lysine acetylation is a common post-translational modification of key metabolic pathway enzymes of the anaerobe Porphyromonas gingivalis. J. Proteom. 2015, 128, 352–364. [Google Scholar] [CrossRef] [PubMed]
- Luu, J.; Carabetta Valerie, J. Contribution of Nε-lysine Acetylation towards Regulation of Bacterial Pathogenesis. mSystems 2021, 6, e00422-21. [Google Scholar] [CrossRef]
- Zeng, J.; Wu, L.; Chen, Q.; Wang, L.; Qiu, W.; Zheng, X.; Yin, X.; Liu, J.; Ren, Y.; Li, Y. Comprehensive profiling of protein lysine acetylation and its overlap with lysine succinylation in the Porphyromonas gingivalis fimbriated strain ATCC 33277. Mol. Oral Microbiol. 2020, 35, 240–250. [Google Scholar] [CrossRef]
- Mishra, A.; Roy, F.; Dou, Y.; Zhang, K.; Tang, H.; Fletcher, H.M. Role of Acetyltransferase PG1842 in Gingipain Biogenesis in Porphyromonas gingivalis. J. Bacteriol. 2018, 200, e00385-18. [Google Scholar] [CrossRef]
- Shen, D.; Perpich, J.D.; Stocke, K.S.; Yakoumatos, L.; Fitzsimonds, Z.R.; Liu, C.; Miller, D.P.; Lamont, R.J. Role of the RprY response regulator in P. gingivalis community development and virulence. Mol. Oral Microbiol. 2020, 35, 231–239. [Google Scholar] [CrossRef]
- Li, Y.; Krishnan, K.; Duncan, M.J. Post-translational regulation of a Porphyromonas gingivalis regulator. J. Oral Microbiol. 2018, 10, 1487743. [Google Scholar] [CrossRef]
- Shoji, M.; Sato, K.; Yukitake, H.; Kamaguchi, A.; Sasaki, Y.; Naito, M.; Nakayama, K. Identification of genes encoding glycosyltransferases involved in lipopolysaccharide synthesis in Porphyromonas gingivalis. Mol. Oral Microbiol. 2018, 33, 68–80. [Google Scholar] [CrossRef] [PubMed]
- Shoji, M.; Shibata, Y.; Shiroza, T.; Yukitake, H.; Peng, B.; Chen, Y.-Y.; Sato, K.; Naito, M.; Abiko, Y.; Reynolds, E.C.; et al. Characterization of hemin-binding protein 35 (HBP35) in Porphyromonas gingivalis: Its cellular distribution, thioredoxin activity and role in heme utilization. BMC Microbiol. 2010, 10, 152. [Google Scholar] [CrossRef] [PubMed]
- Vanterpool, E.; Roy, F.; Hansel, M.F. Inactivation of vimF, a Putative Glycosyltransferase Gene Downstream of vimE, Alters Glycosylation and Activation of the Gingipains in Porphyromonas gingivalis W83. Infect. Immun. 2005, 73, 3971–3982. [Google Scholar] [CrossRef] [PubMed]
- Veith, P.D.; Shoji, M.; O’Hair, R.A.J.; Leeming, M.G.; Nie, S.; Glew, M.D.; Reid, G.E.; Nakayama, K.; Reynolds, E.C. Type IX Secretion System Cargo Proteins Are Glycosylated at the C Terminus with a Novel Linking Sugar of the Wbp/Vim Pathway. mBio 2020, 11, e01497-20. [Google Scholar] [CrossRef]
- Paul, D.V.; Shoji, M.; Scott, N.E.; Reynolds, E.C. Characterization of the O-Glycoproteome of Porphyromonas gingivalis. Microbiol. Spectr. 2022, 10, e01502-21. [Google Scholar]
- Nakao, R.; Tashiro, Y.; Nomura, N.; Kosono, S.; Ochiai, K.; Yonezawa, H.; Watanabe, H.; Senpuku, H. Glycosylation of the OMP85 homolog of Porphyromonas gingivalis and its involvement in biofilm formation. Biochem. Biophys. Res. Commun. 2008, 365, 784–789. [Google Scholar] [CrossRef]
- Rangarajan, M.; Hashim, A.; Aduse-Opoku, J.; Paramonov, N.; Hounsell, E.F.; Curtis, M.A. Expression of Arg-Gingipain RgpB Is Required for Correct Glycosylation and Stability of Monomeric Arg-Gingipain RgpA from Porphyromonas gingivalis W50. Infect. Immun. 2005, 73, 4864–4878. [Google Scholar] [CrossRef]
- Posch, G.; Pabst, M.; Brecker, L.; Altmann, F.; Messner, P.; Schäffer, C. Characterization and Scope of S-layer Protein O-Glycosylation in Tannerella forsythia. J. Biol. Chem. 2011, 286, 38714–38724. [Google Scholar] [CrossRef]
- Tomek, M.B.; Maresch, D.; Windwarder, M.; Friedrich, V.; Janesch, B.; Fuchs, K.; Neumann, L.; Nimeth, I.; Zwickl, N.F.; Dohm, J.C.; et al. A General Protein O-Glycosylation Gene Cluster Encodes the Species-Specific Glycan of the Oral Pathogen Tannerella forsythia: O-Glycan Biosynthesis and Immunological Implications. Front. Microbiol. 2018, 9, 2008. [Google Scholar] [CrossRef]
- Kurniyati, K.; Kelly, J.F.; Vinogradov, E.; Robotham, A.; Tu, Y.; Wang, J.; Liu, J.; Logan, S.M.; Li, C. A novel glycan modifies the flagellar filament proteins of the oral bacterium Treponema denticola. Mol. Microbiol. 2017, 103, 67–85. [Google Scholar] [CrossRef]
- Annan, S.; Ready, D.; Jasni, A.S.; Rogers, M.; Pratten, J.; Roberts, A. Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol. Med. Microbiol. 2010, 59, 345–349. [Google Scholar]
- Jia, L.; Han, N.; Du, J.; Guo, L.; Luo, Z.; Liu, Y. Pathogenesis of Important Virulence Factors of Porphyromonas gingivalis via Toll-Like Receptors. Front. Cell. Infect. Microbiol. 2019, 9, 262. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zeng, J.; Zhou, X.; Li, Y. Drug Resistance and Gene Transfer Mechanisms in Respiratory/Oral Bacteria. J. Dent. Res. 2018, 97, 1092–1099. [Google Scholar] [CrossRef] [PubMed]
- Roberts, A.P.; Kreth, J. The impact of horizontal gene transfer on the adaptive ability of the human oral microbiome. Front. Cell. Infect. Microbiol. 2014, 4, 124. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.A.; Chen, T.; Scott, J.C.; Koenigsberg, A.L.; Duncan, M.J.; Hu, L.T. Identification and characterization of a minisatellite contained within a novel miniature inverted-repeat transposable element (MITE) of Porphyromonas gingivalis. Mob DNA 2015, 6, 18. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, F.; Li, G.; Xu, Y. A recently active miniature inverted-repeat transposable element, Chunjie, inserted into an operon without disturbing the operon structure in Geobacter uraniireducens Rf4. Genetics 2008, 179, 2291–2297. [Google Scholar] [CrossRef] [PubMed]
- Stavrinides, J.; Kirzinger, M.W.; Beasley, F.C.; Guttman, D.S. E622, a miniature, virulence-associated mobile element. J. Bacteriol. 2012, 194, 509–517. [Google Scholar] [CrossRef][Green Version]
- Adams, F.G.; Brown, M.H. MITE Aba12, a Novel Mobile Miniature Inverted-Repeat Transposable Element Identified in Acinetobacter baumannii ATCC 17978 and Its Prevalence across the Moraxellaceae Family. mSphere 2019, 4, e00028-19. [Google Scholar] [CrossRef]
- Into, T.; Inomata, M.; Kanno, Y.; Matsuyama, T.; Machigashira, M.; Izumi, Y.; Imamura, T.; Nakashima, M.; Noguchi, T.; Matsushita, K. Arginine-specific gingipains from Porphyromonas gingivalis deprive protective functions of secretory leucocyte protease inhibitor in periodontal tissue. Clin. Exp. Immunol. 2006, 145, 545–554. [Google Scholar] [CrossRef]
- Haurat, M.F.; Aduse-Opoku, J.; Rangarajan, M.; Dorobantu, L.; Gray, M.R.; Curtis, M.A.; Feldman, M.F. Selective Sorting of Cargo Proteins into Bacterial Membrane Vesicles*. J. Biol. Chem. 2011, 286, 1269–1276. [Google Scholar] [CrossRef]
- Su, Z.; Kong, F.; Wang, S.; Chen, J.; Yin, R.; Zhou, C.; Zhang, Y.; He, Z.; Shi, Y.; Xue, Y.; et al. The rag locus of Porphyromonas gingivalis might arise from Bacteroides via horizontal gene transfer. Eur. J. Clin. Microbiol. Infect. Dis. 2010, 29, 429–437. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Macek, B.; Forchhammer, K.; Hardouin, J.; Weber-Ban, E.; Grangeasse, C.; Mijakovic, I. Protein post-translational modifications in bacteria. Nat. Rev. Microbiol. 2019, 17, 651–664. [Google Scholar] [CrossRef] [PubMed]
- DebRoy, S.; Shropshire, W.C.; Tran, C.N.; Hao, H.; Gohel, M.; Galloway-Peña, J.; Hanson, B.; Flores, A.R.; Shelburne, S.A. Characterization of the Type I Restriction Modification System Broadly Conserved among Group A Streptococci. mSphere 2021, 6, e00799-21. [Google Scholar] [CrossRef] [PubMed]
- Salaün, L.; Ayraud, S.; Saunders, N. Phase variation mediated niche adaptation during prolonged experimental murine infection with Helicobacter pylori. Microbiology 2005, 151 Pt 3, 917–923. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Seib, K.L.; Jen, F.E.-C.; Scott, A.L.; Tan, A.; Jennings, M.P. Phase variation of DNA methyltransferases and the regulation of virulence and immune evasion in the pathogenic Neisseria. Pathog. Dis. 2017, 75, ftx080. [Google Scholar] [CrossRef] [PubMed]
- Phillips, Z.N.; Tram, G.; Seib, K.; Atack, J.M. Phase-variable bacterial loci: How bacteria gamble to maximise fitness in changing environments. Biochem. Soc. Trans. 2019, 47, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.; Fridman, O.; Ronin, I.; Balaban, N.Q. Systematic identification and quantification of phase variation in commensal and pathogenic Escherichia coli. Genome Med. 2014, 6, 112. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).