Herpesvirus Screening in Childhood Hematopoietic Transplant Reveals High Systemic Inflammation in Episodes of Multiple Viral Detection and an EBV Association with Elevated IL-1β, IL-8 and Graft-Versus-Host Disease
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Statement
2.2. Patients and Clinical Samples
2.3. DNA Isolation
2.4. Viral Detection
2.5. Cytokine Detection
2.6. Sanger Sequencing
2.7. Statistical Analysis
3. Results
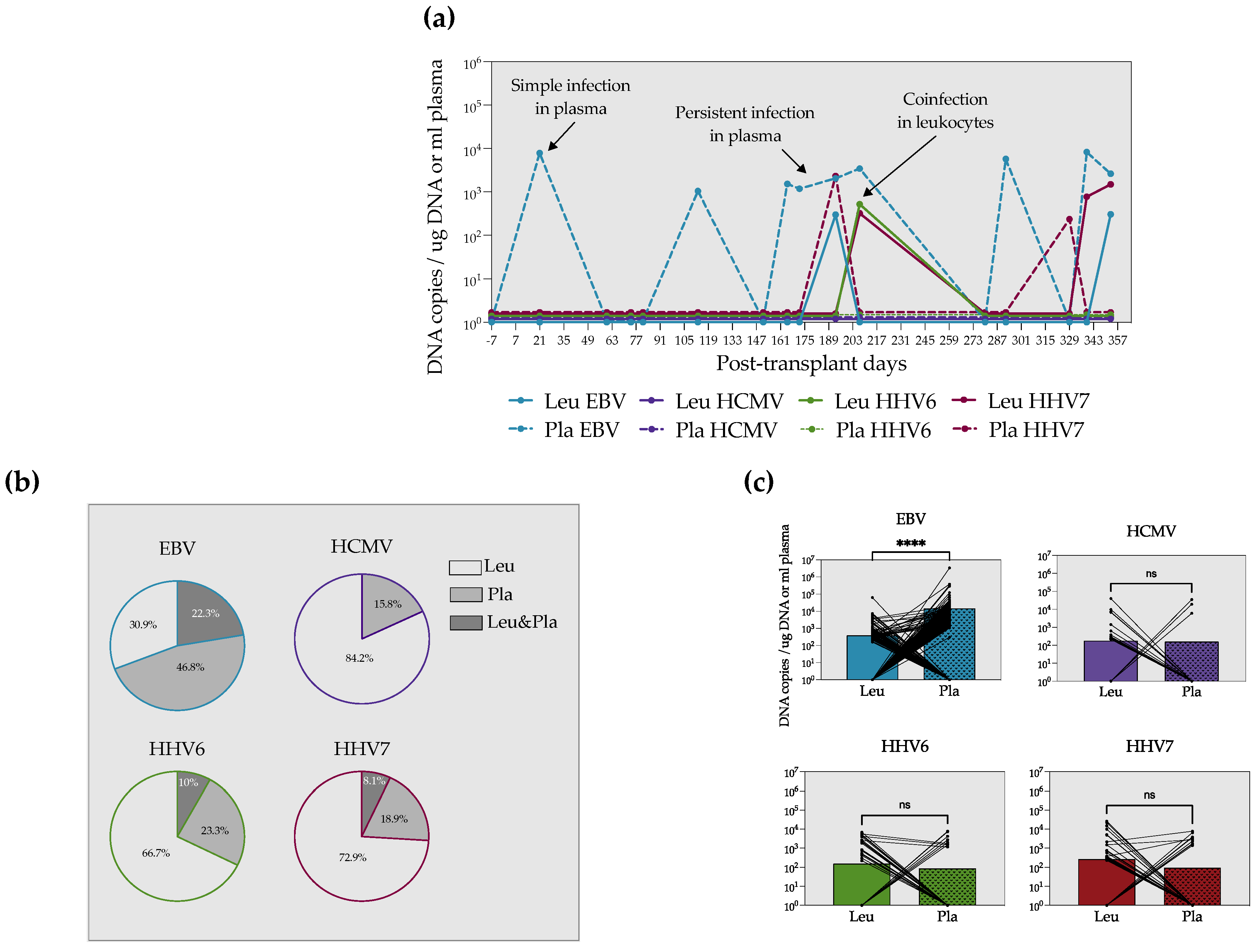
3.1. EBV, HCMV, HHV6 and HHV7 DNA Are Detected in Allogeneic and Autologous HSCT at Different Frequencies but with a Similar Burden
3.2. EBV Is Mostly Found in the Acelluar Fraction, Whereas HCMV, HHV6 and HHV7 Predominantly Expand in the Cellular Compartment
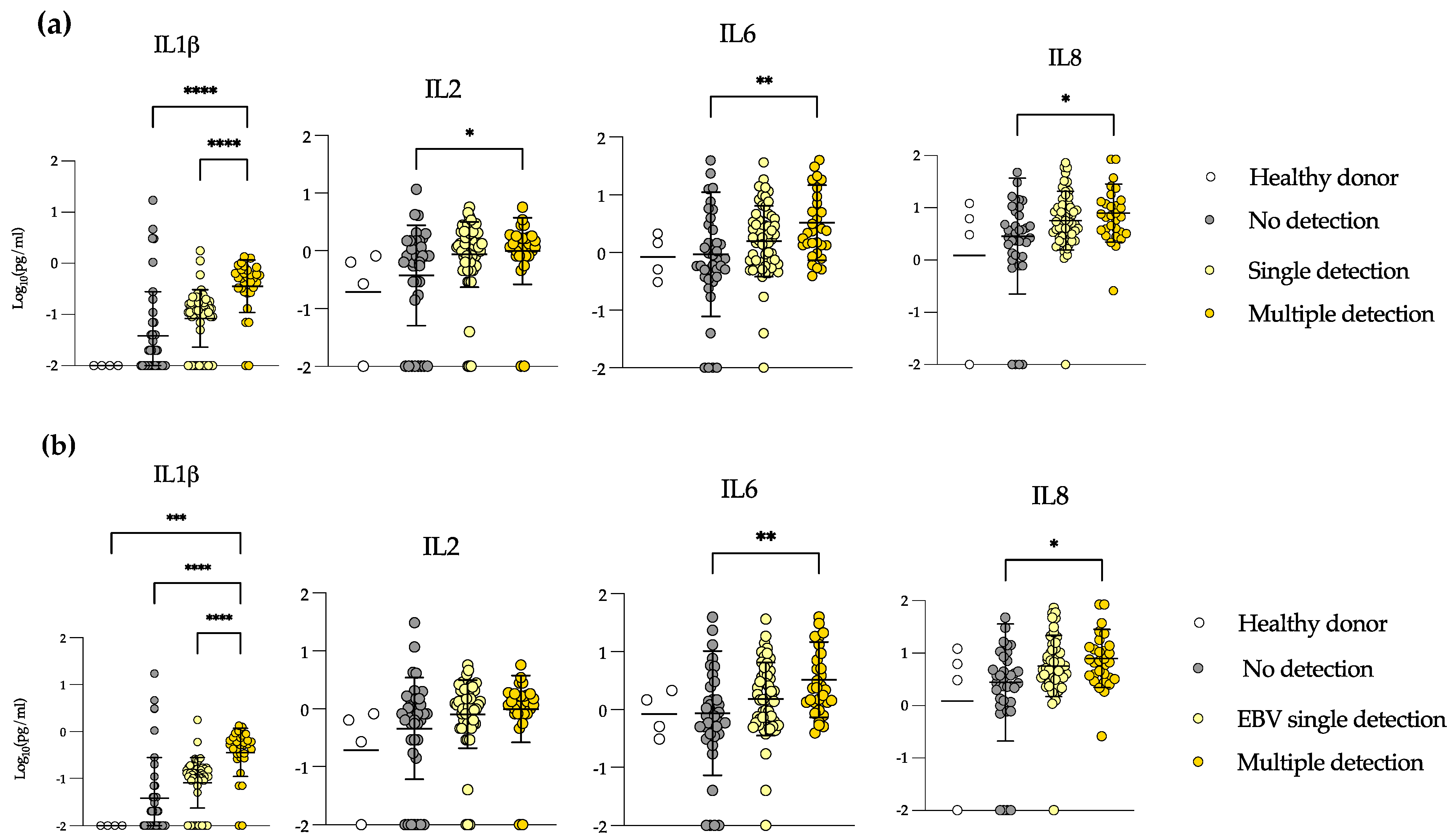
3.3. Allo-HSCT Recipients Show Increased Levels of IL-1β, IL-2, IL-6 and IL-8 during Multiple Detection Episodes
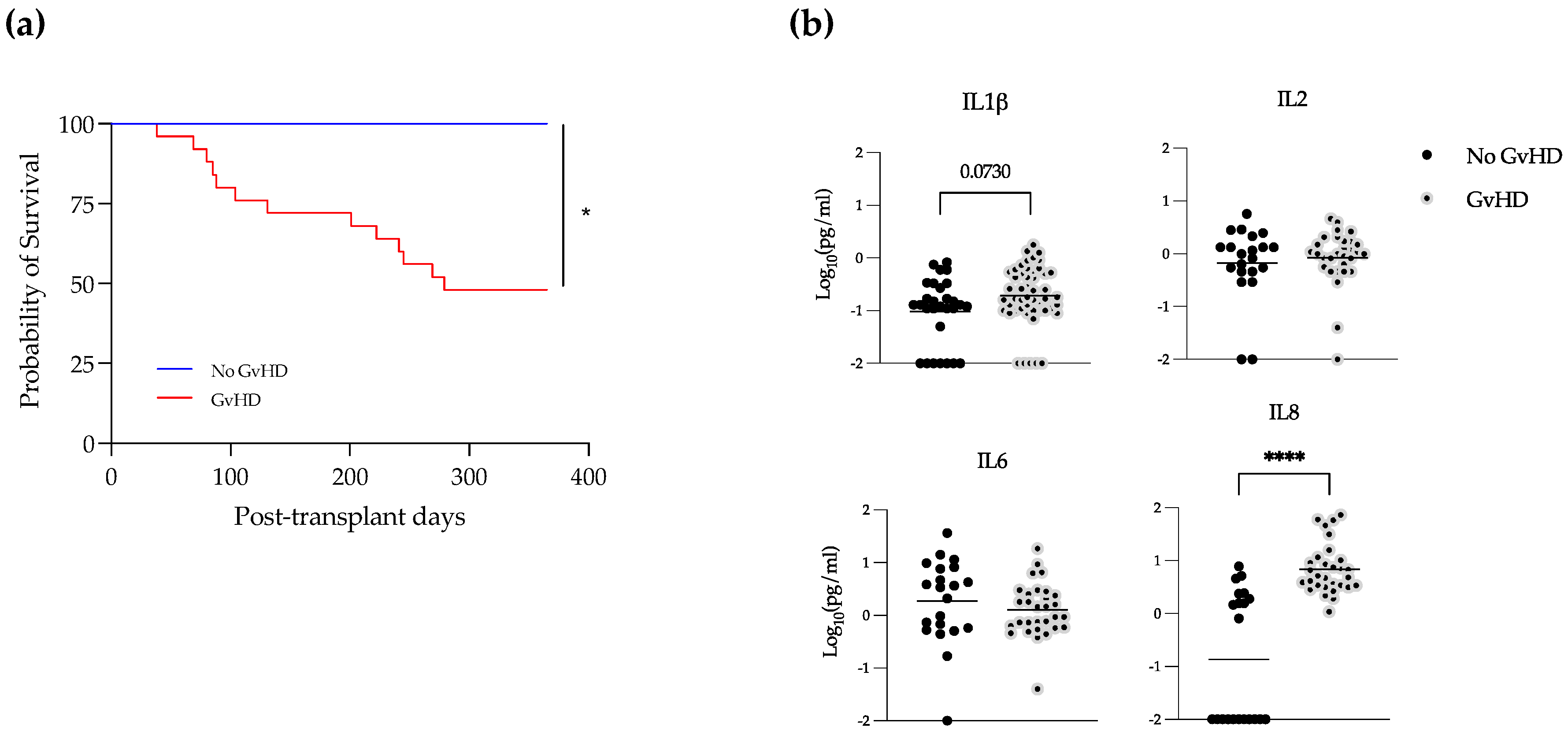
3.4. Elevated Levels of IL-1β and IL-8 in EBV-Positive Samples during GvHD
4. Discussion
5. Study Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Tomblyn, M.; Chiller, T.; Einsele, H.; Gress, R.; Sepkowitz, K.; Storek, J.; Wingard, J.R.; Young, J.A.; Boeckh, M.J. Guidelines for preventing infectious complications among hematopoietic cell transplantation recipients: A global perspective. Biol. Blood Marrow Transplant. 2009, 15, 1143–1238. [Google Scholar] [CrossRef] [PubMed]
- Jenq, R.R.; van den Brink, M.R. Allogeneic haematopoietic stem cell transplantation: Individualized stem cell and immune therapy of cancer. Nat. Rev. Cancer 2010, 10, 213–221. [Google Scholar] [CrossRef] [PubMed]
- Porrata, L.F. Autologous Graft-versus-Tumor Effect: Reality or Fiction? Adv. Hematol. 2016, 2016, 5385972. [Google Scholar] [CrossRef]
- Inazawa, N.; Hori, T.; Nojima, M.; Saito, M.; Igarashi, K.; Yamamoto, M.; Shimizu, N.; Yoto, Y.; Tsutsumi, H. Virus reactivations after autologous hematopoietic stem cell transplantation detected by multiplex PCR assay. J. Med. Virol. 2017, 89, 358–362. [Google Scholar] [CrossRef]
- Cruz-Munoz, M.E.; Fuentes-Panana, E.M. Beta and Gamma Human Herpesviruses: Agonistic and Antagonistic Interactions with the Host Immune System. Front. Microbiol. 2017, 8, 2521. [Google Scholar] [CrossRef] [PubMed]
- Hill, J.A.; Koo, S.; Guzman Suarez, B.B.; Ho, V.T.; Cutler, C.; Koreth, J.; Armand, P.; Alyea, E.P., 3rd; Baden, L.R.; Antin, J.H.; et al. Cord-blood hematopoietic stem cell transplant confers an increased risk for human herpesvirus-6-associated acute limbic encephalitis: A cohort analysis. Biol. Blood Marrow Transplant. 2012, 18, 1638–1648. [Google Scholar] [CrossRef] [PubMed]
- Agut, H. Deciphering the clinical impact of acute human herpesvirus 6 (HHV-6) infections. J. Clin. Virol. 2011, 52, 164–171. [Google Scholar] [CrossRef]
- de Oliveira, P.G.G.; Ueda, M.Y.; Real, J.M.; de Sa Moreira, E.; de Oliveira, J.S.R.; Goncalves, M.V.; Ginani, V.C.; de Oliveira, F.O.M.; Seber, A.; Novis, Y.; et al. Simultaneous Quantification of the 8 Human Herpesviruses in Allogeneic Hematopoietic Stem Cell Transplantation. Transplantation 2016, 100, 1363–1370. [Google Scholar] [CrossRef]
- Sanchez-Ponce, Y.; Varela-Fascinetto, G.; Romo-Vazquez, J.C.; Lopez-Martinez, B.; Sanchez-Huerta, J.L.; Parra-Ortega, I.; Fuentes-Panana, E.M.; Morales-Sanchez, A. Simultaneous Detection of Beta and Gamma Human Herpesviruses by Multiplex qPCR Reveals Simple Infection and Coinfection Episodes Increasing Risk for Graft Rejection in Solid Organ Transplantation. Viruses 2018, 10, 730. [Google Scholar] [CrossRef]
- Cardoni, R.L.; Prigoshin, N.; Tambutti, M.L.; Ferraris, J.R. Citoquinas reguladoras de la respuesta al transplante renal alogénico. Medicina 2005, 65, 54–62. [Google Scholar]
- Samara, Y.; Mei, M. Autologous Stem Cell Transplantation in Hodgkin Lymphoma-Latest Advances in the Era of Novel Therapies. Cancers 2022, 14, 1738. [Google Scholar] [CrossRef] [PubMed]
- Puerta-Alcalde, P.; Chumbita, M.; Charry, P.; Castano-Diez, S.; Cardozo, C.; Moreno-Garcia, E.; Marco, F.; Suarez-Lledo, M.; Garcia-Pouton, N.; Morata, L.; et al. Risk Factors for Mortality in Hematopoietic Stem Cell Transplantation Recipients with Bloodstream Infection: Points To Be Addressed by Future Guidelines. Transplant. Cell Ther. 2021, 27, 501.e1–501.e6. [Google Scholar] [CrossRef] [PubMed]
- Jantunen, E.; Salonen, J.; Juvonen, E.; Koivunen, E.; Siitonen, T.; Lehtinen, T.; Kuittinen, O.; Leppa, S.; Anttila, V.J.; Itala, M.; et al. Invasive fungal infections in autologous stem cell transplant recipients: A nation-wide study of 1188 transplanted patients. Eur. J. Haematol. 2004, 73, 174–178. [Google Scholar] [CrossRef] [PubMed]
- Grow, W.B.; Moreb, J.S.; Roque, D.; Manion, K.; Leather, H.; Reddy, V.; Khan, S.A.; Finiewicz, K.J.; Nguyen, H.; Clancy, C.J.; et al. Late onset of invasive aspergillus infection in bone marrow transplant patients at a university hospital. Bone Marrow Transplant. 2002, 29, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Offidani, M.; Corvatta, L.; Olivieri, A.; Rupoli, S.; Frayfer, J.; Mele, A.; Manso, E.; Montanari, M.; Centurioni, R.; Leoni, P. Infectious complications after autologous peripheral blood progenitor cell transplantation followed by G-CSF. Bone Marrow Transplant. 1999, 24, 1079–1087. [Google Scholar] [CrossRef]
- Shi, J.M.; Pei, X.Y.; Luo, Y.; Tan, Y.M.; Tie, R.X.; He, J.S.; Zheng, W.Y.; Zhang, J.; Cai, Z.; Lin, M.F.; et al. Invasive fungal infection in allogeneic hematopoietic stem cell transplant recipients: Single center experiences of 12 years. J. Zhejiang Univ. Sci. B 2015, 16, 796–804. [Google Scholar] [CrossRef]
- Gomez, S.M.; Caniza, M.; Fynn, A.; Vescina, C.; Ruiz, C.D.; Iglesias, D.; Sosa, F.; Sung, L. Fungal infections in hematopoietic stem cell transplantation in children at a pediatric children’s hospital in Argentina. Transpl. Infect. Dis. 2018, 20, e12913. [Google Scholar] [CrossRef]
- Omer, A.K.; Ziakas, P.D.; Anagnostou, T.; Coughlin, E.; Kourkoumpetis, T.; McAfee, S.L.; Dey, B.R.; Attar, E.; Chen, Y.B.; Spitzer, T.R.; et al. Risk factors for invasive fungal disease after allogeneic hematopoietic stem cell transplantation: A single center experience. Biol. Blood Marrow Transplant. 2013, 19, 1190–1196. [Google Scholar] [CrossRef]
- Zhou, J.R.; Shi, D.Y.; Wei, R.; Wang, Y.; Yan, C.H.; Zhang, X.H.; Xu, L.P.; Liu, K.Y.; Huang, X.J.; Sun, Y.Q. Co-Reactivation of Cytomegalovirus and Epstein-Barr Virus Was Associated With Poor Prognosis After Allogeneic Stem Cell Transplantation. Front. Immunol. 2020, 11, 620891. [Google Scholar] [CrossRef]
- Ke, P.; Zhang, X.; Liu, S.; Zhu, Q.; Ma, X.; Chen, F.; Tang, X.; Han, Y.; Fu, Z.; Chen, S.; et al. The time-dependent effects of early-onset Epstein-Barr viremia on adult acute leukemia patients following allo-HSCT with ATG-containing MAC regimen. Ann. Hematol. 2021, 100, 1879–1889. [Google Scholar] [CrossRef]
- Salmona, M.; Stefic, K.; Mahjoub, N.; de Fontbrune, F.S.; Maylin, S.; Simon, F.; Scieux, C.; Socie, G.; Mazeron, M.C.; LeGoff, J. Automated quantification of Epstein-Barr virus in whole blood for post-transplant lymphoproliferative disorders monitoring. Virol. J. 2020, 17, 20. [Google Scholar] [CrossRef]
- Li, N.; Chen, Y.; He, W.; Yi, T.; Zhao, D.; Zhang, C.; Lin, C.L.; Todorov, I.; Kandeel, F.; Forman, S.; et al. Anti-CD3 preconditioning separates GVL from GVHD via modulating host dendritic cell and donor T-cell migration in recipients conditioned with TBI. Blood 2009, 113, 953–962. [Google Scholar] [CrossRef]
- Faye, A.; Vilmer, E. Post-transplant lymphoproliferative disorder in children: Incidence, prognosis, and treatment options. Paediatr. Drugs 2005, 7, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Enok Bonong, P.R.; Zahreddine, M.; Buteau, C.; Duval, M.; Laporte, L.; Lacroix, J.; Alfieri, C.; Trottier, H. Factors Associated with Post-Transplant Active Epstein-Barr Virus Infection and Lymphoproliferative Disease in Hematopoietic Stem Cell Transplant Recipients: A Systematic Review and Meta-Analysis. Vaccines 2021, 9, 288. [Google Scholar] [CrossRef] [PubMed]
- Meyers, J.D.; Flournoy, N.; Thomas, E.D. Risk factors for cytomegalovirus infection after human marrow transplantation. J. Infect. Dis. 1986, 153, 478–488. [Google Scholar] [CrossRef] [PubMed]
- Gimenez, E.; Torres, I.; Albert, E.; Pinana, J.L.; Hernandez-Boluda, J.C.; Solano, C.; Navarro, D. Cytomegalovirus (CMV) infection and risk of mortality in allogeneic hematopoietic stem cell transplantation (Allo-HSCT): A systematic review, meta-analysis, and meta-regression analysis. Am. J. Transplant. 2019, 19, 2479–2494. [Google Scholar] [CrossRef]
- Phan, T.L.; Carlin, K.; Ljungman, P.; Politikos, I.; Boussiotis, V.; Boeckh, M.; Shaffer, M.L.; Zerr, D.M. Human Herpesvirus-6B Reactivation Is a Risk Factor for Grades II to IV Acute Graft-versus-Host Disease after Hematopoietic Stem Cell Transplantation: A Systematic Review and Meta-Analysis. Biol. Blood Marrow Transplant. 2018, 24, 2324–2336. [Google Scholar] [CrossRef]
- Dadwal, S.S. Herpes Virus Infections Other than Cytomegalovirus in the Recipients of Hematopoietic Stem Cell Transplantation. Infect. Dis. Clin. N. Am. 2019, 33, 467–484. [Google Scholar] [CrossRef]
- Inazawa, N.; Hori, T.; Hatakeyama, N.; Yamamoto, M.; Yoto, Y.; Nojima, M.; Suzuki, N.; Shimizu, N.; Tsutsumi, H. Large-scale multiplex polymerase chain reaction assay for diagnosis of viral reactivations after allogeneic hematopoietic stem cell transplantation. J. Med. Virol. 2015, 87, 1427–1435. [Google Scholar] [CrossRef]
- Miura, H.; Kawamura, Y.; Hattori, F.; Tanaka, M.; Kudo, K.; Ihira, M.; Yatsuya, H.; Takahashi, Y.; Kojima, S.; Sakaguchi, H.; et al. Human herpesvirus-6B infection in pediatric allogenic hematopoietic stem cell transplant patients: Risk factors and encephalitis. Transpl. Infect. Dis. 2020, 22, e13203. [Google Scholar] [CrossRef]
- Zallio, F.; Primon, V.; Tamiazzo, S.; Pini, M.; Baraldi, A.; Corsetti, M.T.; Gotta, F.; Bertassello, C.; Salvi, F.; Rocchetti, A.; et al. Epstein-Barr virus reactivation in allogeneic stem cell transplantation is highly related to cytomegalovirus reactivation. Clin. Transplant. 2013, 27, E491–E497. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Ponce, Y.; Fuentes-Panana, E.M. Molecular and immune interactions between beta- and gamma-herpesviruses in the immunocompromised host. J. Leukoc. Biol. 2022, 112, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Levine, J.E.; Logan, B.R.; Wu, J.; Alousi, A.M.; Bolanos-Meade, J.; Ferrara, J.L.; Ho, V.T.; Weisdorf, D.J.; Paczesny, S. Acute graft-versus-host disease biomarkers measured during therapy can predict treatment outcomes: A Blood and Marrow Transplant Clinical Trials Network study. Blood 2012, 119, 3854–3860. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, J.; Imamura, M.; Kasai, M.; Masauzi, N.; Matsuura, A.; Ohizumi, H.; Morii, K.; Kiyama, Y.; Naohara, T.; Saitho, M.; et al. Cytokine gene expression in peripheral blood mononuclear cells during graft-versus-host disease after allogeneic bone marrow transplantation. Br. J. Haematol. 1993, 85, 558–565. [Google Scholar] [CrossRef]
- Barak, V.; Levi-Schaffer, F.; Nisman, B.; Nagler, A. Cytokine dysregulation in chronic graft versus host disease. Leuk. Lymphoma 1995, 17, 169–173. [Google Scholar] [CrossRef]
- Nagasawa, M. Biomarkers of graft-vs-host disease: Understanding and applications for the future. World J. Transplant. 2021, 11, 335–343. [Google Scholar] [CrossRef]
- Zeiser, R.; Burchert, A.; Lengerke, C.; Verbeek, M.; Maas-Bauer, K.; Metzelder, S.K.; Spoerl, S.; Ditschkowski, M.; Ecsedi, M.; Sockel, K.; et al. Ruxolitinib in corticosteroid-refractory graft-versus-host disease after allogeneic stem cell transplantation: A multicenter survey. Leukemia 2015, 29, 2062–2068. [Google Scholar] [CrossRef]
- Matsuoka, K.I. Low-dose interleukin-2 as a modulator of Treg homeostasis after HSCT: Current understanding and future perspectives. Int. J. Hematol. 2018, 107, 130–137. [Google Scholar] [CrossRef]
- Chinen, T.; Kannan, A.K.; Levine, A.G.; Fan, X.; Klein, U.; Zheng, Y.; Gasteiger, G.; Feng, Y.; Fontenot, J.D.; Rudensky, A.Y. An essential role for the IL-2 receptor in Treg cell function. Nat. Immunol. 2016, 17, 1322–1333. [Google Scholar] [CrossRef]
- Koreth, J.; Matsuoka, K.; Kim, H.T.; McDonough, S.M.; Bindra, B.; Alyea, E.P., 3rd; Armand, P.; Cutler, C.; Ho, V.T.; Treister, N.S.; et al. Interleukin-2 and regulatory T cells in graft-versus-host disease. N. Engl. J. Med. 2011, 365, 2055–2066. [Google Scholar] [CrossRef]
- Cullup, H.; Stark, G. Interleukin-1 polymorphisms and graft-vs-host disease. Leuk. Lymphoma 2005, 46, 517–523. [Google Scholar] [CrossRef] [PubMed]
- Abhyankar, S.; Gilliland, D.G.; Ferrara, J.L. Interleukin-1 is a critical effector molecule during cytokine dysregulation in graft versus host disease to minor histocompatibility antigens. Transplantation 1993, 56, 1518–1523. [Google Scholar] [CrossRef] [PubMed]
- Antin, J.H.; Weisdorf, D.; Neuberg, D.; Nicklow, R.; Clouthier, S.; Lee, S.J.; Alyea, E.; McGarigle, C.; Blazar, B.R.; Sonis, S.; et al. Interleukin-1 blockade does not prevent acute graft-versus-host disease: Results of a randomized, double-blind, placebo-controlled trial of interleukin-1 receptor antagonist in allogeneic bone marrow transplantation. Blood 2002, 100, 3479–3482. [Google Scholar] [CrossRef] [PubMed]
- Kroger, N.; Shahnaz Syed Abd Kadir, S.; Zabelina, T.; Badbaran, A.; Christopeit, M.; Ayuk, F.; Wolschke, C. Peritransplantation Ruxolitinib Prevents Acute Graft-versus-Host Disease in Patients with Myelofibrosis Undergoing Allogenic Stem Cell Transplantation. Biol. Blood Marrow Transplant. 2018, 24, 2152–2156. [Google Scholar] [CrossRef] [PubMed]
- Ferrara, J.L. Advances in the clinical management of GVHD. Best Pract. Res. Clin. Haematol. 2008, 21, 677–682. [Google Scholar] [CrossRef][Green Version]
- Paczesny, S.; Krijanovski, O.I.; Braun, T.M.; Choi, S.W.; Clouthier, S.G.; Kuick, R.; Misek, D.E.; Cooke, K.R.; Kitko, C.L.; Weyand, A.; et al. A biomarker panel for acute graft-versus-host disease. Blood 2009, 113, 273–278. [Google Scholar] [CrossRef]
- Jin, Y.B.; Zhang, G.Y.; Lin, K.R.; Chen, X.P.; Cui, J.H.; Wang, Y.J.; Luo, W. Changes of plasma cytokines and chemokines expression level in nasopharyngeal carcinoma patients after treatment with definitive intensity-modulated radiotherapy (IMRT). PLoS ONE 2017, 12, e0172264. [Google Scholar] [CrossRef]
- Zhu, Y.; Xu, Y.; Wei, Y.; Liang, W.; Liao, M.; Zhang, L. Association of IL-1B gene polymorphisms with nasopharyngeal carcinoma in a Chinese population. Clin. Oncol. 2008, 20, 207–211. [Google Scholar] [CrossRef]
- Broderick, G.; Katz, B.Z.; Fernandes, H.; Fletcher, M.A.; Klimas, N.; Smith, F.A.; O’Gorman, M.R.; Vernon, S.D.; Taylor, R. Cytokine expression profiles of immune imbalance in post-mononucleosis chronic fatigue. J. Transl. Med. 2012, 10, 191. [Google Scholar] [CrossRef]
- Watanabe, N.; Nodomi, K.; Koike, R.; Kato, A.; Takeichi, O.; Kotani, A.I.; Kaneko, T.; Sakagami, H.; Takei, M.; Ogata, Y.; et al. EBV LMP1 in Gingival Epithelium Potentially Contributes to Human Chronic Periodontitis via Inducible IL8 Production. Vivo 2019, 33, 1793–1800. [Google Scholar] [CrossRef]
- Zhang, J.Y.; Du, Y.; Gong, L.P.; Shao, Y.T.; Wen, J.Y.; Sun, L.P.; He, D.; Guo, J.R.; Chen, J.N.; Shao, C.K. EBV-Induced CXCL8 Upregulation Promotes Vasculogenic Mimicry in Gastric Carcinoma via NF-kappaB Signaling. Front. Cell Infect. Microbiol. 2022, 12, 780416. [Google Scholar] [CrossRef] [PubMed]
- Sisay, S.; Lopez-Lozano, L.; Mickunas, M.; Quiroga-Fernandez, A.; Palace, J.; Warnes, G.; Alvarez-Lafuente, R.; Dua, P.; Meier, U.C. Untreated relapsing remitting multiple sclerosis patients show antibody production against latent Epstein Barr Virus (EBV) antigens mainly in the periphery and innate immune IL-8 responses preferentially in the CNS. J. Neuroimmunol. 2017, 306, 40–45. [Google Scholar] [CrossRef] [PubMed]
- Imadome, K.; Yajima, M.; Arai, A.; Nakazawa, A.; Kawano, F.; Ichikawa, S.; Shimizu, N.; Yamamoto, N.; Morio, T.; Ohga, S.; et al. Novel mouse xenograft models reveal a critical role of CD4+ T cells in the proliferation of EBV-infected T and NK cells. PLoS Pathog. 2011, 7, e1002326. [Google Scholar] [CrossRef] [PubMed]
- Tang, S.; Zhao, C.; Chen, W. Aggressive diffuse large B-cell lymphoma with hemophagocytic lymphohistiocytosis: Report of one case. Int. J. Clin. Exp. Pathol. 2020, 13, 2392–2396. [Google Scholar] [PubMed]
- Hsiao, J.R.; Chang, K.C.; Chen, C.W.; Wu, S.Y.; Su, I.J.; Hsu, M.C.; Jin, Y.T.; Tsai, S.T.; Takada, K.; Chang, Y. Endoplasmic reticulum stress triggers XBP-1-mediated up-regulation of an EBV oncoprotein in nasopharyngeal carcinoma. Cancer Res. 2009, 69, 4461–4467. [Google Scholar] [CrossRef]
- Hsu, M.; Wu, S.Y.; Chang, S.S.; Su, I.J.; Tsai, C.H.; Lai, S.J.; Shiau, A.L.; Takada, K.; Chang, Y. Epstein-Barr virus lytic transactivator Zta enhances chemotactic activity through induction of interleukin-8 in nasopharyngeal carcinoma cells. J. Virol. 2008, 82, 3679–3688. [Google Scholar] [CrossRef]
- Li, Z.; Tsai, M.H.; Shumilov, A.; Baccianti, F.; Tsao, S.W.; Poirey, R.; Delecluse, H.J. Epstein-Barr virus ncRNA from a nasopharyngeal carcinoma induces an inflammatory response that promotes virus production. Nat. Microbiol. 2019, 4, 2475–2486. [Google Scholar] [CrossRef]
- Ren, Q.; Sato, H.; Murono, S.; Furukawa, M.; Yoshizaki, T. Epstein-Barr virus (EBV) latent membrane protein 1 induces interleukin-8 through the nuclear factor-kappa B signaling pathway in EBV-infected nasopharyngeal carcinoma cell line. Laryngoscope 2004, 114, 855–859. [Google Scholar] [CrossRef]
- Xie, L.Q.; Bian, L.J.; Li, Z.; Li, Y.; Liang, Y.J. Co-elevated expression of hepatocyte growth factor and Interleukin-8 contributes to poor prognosis of patients with primary nasopharyngeal carcinoma. Oncol. Rep. 2010, 23, 141–150. [Google Scholar]
- Yoshizaki, T. Promotion of metastasis in nasopharyngeal carcinoma by Epstein-Barr virus latent membrane protein-1. Histol. Histopathol. 2002, 17, 845–850. [Google Scholar] [CrossRef]
- Savitri, E.; Haryana, M.S. Expression of interleukin-8, interleukin-10 and Epstein-Barr viral-load as prognostic indicator in nasopharyngeal carcinoma. Glob. J. Health Sci. 2015, 7, 364–372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zong, J.; Ji, P.; Lin, C.; Zhang, R.; Chen, Y.; Lu, Q.; Peng, X.; Pan, J.; Lin, S. Plasma Epstein-Barr viral DNA load after completion of two cycles of induction chemotherapy predicts outcomes for patients with advanced-stage nasopharyngeal carcinoma. Oral Oncol. 2022, 131, 105972. [Google Scholar] [CrossRef] [PubMed]
- Hendrix, P.; Tang, Z.; Silasi, M.; Racicot, K.E.; Mor, G.; Abrahams, V.M.; Guller, S. Herpesvirus-infected Hofbauer cells activate endothelial cells through an IL-1beta-dependent mechanism. Placenta 2020, 91, 59–65. [Google Scholar] [CrossRef] [PubMed]
- Takamatsu, A.; Nakashima, Y.; Haji, S.; Tsuda, M.; Masuda, T.; Kimura, D.; Shiratsuchi, M.; Ogawa, Y. Circulating endothelial cells and endothelial progenitor cells as potential predictors of acute GVHD after allogeneic hematopoietic stem cell transplantation. Eur. J. Haematol. 2022, 109, 146–153. [Google Scholar] [CrossRef]
- Almici, C.; Skert, C.; Verardi, R.; Di Palma, A.; Bianchetti, A.; Neva, A.; Braga, S.; Malagola, M.; Turra, A.; Marini, M.; et al. Changes in circulating endothelial cells count could become a valuable tool in the diagnostic definition of acute graft-versus-host disease. Transplantation 2014, 98, 706–712. [Google Scholar] [CrossRef]
- Kimura, H.; Morita, M.; Yabuta, Y.; Kuzushima, K.; Kato, K.; Kojima, S.; Matsuyama, T.; Morishima, T. Quantitative analysis of Epstein-Barr virus load by using a real-time PCR assay. J. Clin. Microbiol. 1999, 37, 132–136. [Google Scholar] [CrossRef]
- Hoshino, Y.; Kimura, H.; Tanaka, N.; Tsuge, I.; Kudo, K.; Horibe, K.; Kato, K.; Matsuyama, T.; Kikuta, A.; Kojima, S.; et al. Prospective monitoring of the Epstein-Barr virus DNA by a real-time quantitative polymerase chain reaction after allogenic stem cell transplantation. Br. J. Haematol. 2001, 115, 105–111. [Google Scholar] [CrossRef]
- Kimura, H.; Ito, Y.; Suzuki, R.; Nishiyama, Y. Measuring Epstein-Barr virus (EBV) load: The significance and application for each EBV-associated disease. Rev. Med. Virol. 2008, 18, 305–319. [Google Scholar] [CrossRef]
- Wadowsky, R.M.; Laus, S.; Green, M.; Webber, S.A.; Rowe, D. Measurement of Epstein-Barr virus DNA loads in whole blood and plasma by TaqMan PCR and in peripheral blood lymphocytes by competitive PCR. J. Clin. Microbiol. 2003, 41, 5245–5249. [Google Scholar] [CrossRef]
- Sanchez-Ponce, Y.; Fuentes-Panana, E.M. The Role of Coinfections in the EBV-Host Broken Equilibrium. Viruses 2021, 13, 1399. [Google Scholar] [CrossRef]
- Barani, R.; Ravi, Y.; Seshan, V.; Reju, S.B.; Soundararajan, P.; Palani, G.; Srikanth, P. Epstein-Barr Virus DNAemia and co-occurrence with cytomegalovirus DNAemia in postrenal transplant recipients from a tertiary care center. Indian J. Transplant. 2018, 12, 95–102. [Google Scholar]
- Kimura, H.; Kwong, Y.L. EBV Viral Loads in Diagnosis, Monitoring, and Response Assessment. Front. Oncol. 2019, 9, 62. [Google Scholar] [CrossRef]
- Cardenas-Mondragon, M.G.; Torres, J.; Flores-Luna, L.; Camorlinga-Ponce, M.; Carreon-Talavera, R.; Gomez-Delgado, A.; Kasamatsu, E.; Fuentes-Panana, E.M. Case-control study of Epstein-Barr virus and Helicobacter pylori serology in Latin American patients with gastric disease. Br. J. Cancer 2015, 112, 1866–1873. [Google Scholar] [CrossRef] [PubMed]
- Cardenas-Mondragon, M.G.; Carreon-Talavera, R.; Camorlinga-Ponce, M.; Gomez-Delgado, A.; Torres, J.; Fuentes-Panana, E.M. Epstein Barr virus and Helicobacter pylori co-infection are positively associated with severe gastritis in pediatric patients. PLoS ONE 2013, 8, e62850. [Google Scholar] [CrossRef] [PubMed]
- Morales-Sanchez, A.; Torres, J.; Cardenas-Mondragon, M.G.; Romo-Gonzalez, C.; Camorlinga-Ponce, M.; Flores-Luna, L.; Fuentes-Panana, E.M. Detection of Epstein-Barr Virus DNA in Gastric Biopsies of Pediatric Patients with Dyspepsia. Pathogens 2020, 9, 623. [Google Scholar] [CrossRef] [PubMed]

| Type of Transplant | Allogenic | Autologous |
|---|---|---|
| No. of patients | n = 32 | n = 8 |
| No. of blood samples | 382 | 30 |
| Samples per patient, mean ± SD | 14 ± 6 | 4.6 ± 1.4 |
| Follow-up, median days (IQR) | 237 (88–338) | 49 (23–80) |
| Sampling frequency, median days (IQR) | 14 (14–24) | 19 (13–28) |
| Sex, n (%) | ||
| Female | 16 (50) | 5 (62.5) |
| Male | 16 (50) | 3 (37.5) |
| Age at transplant, median years (IQR) | 8 (5–13) | 10 (5–14.5) |
| Stem cell source, n (%) | ||
| Peripheral blood | 27 (84.4) | 8 (100) |
| Bone marrow | 5 (15.6) | 0 (0) |
| HLA compatibility, n (%) | ||
| 50% | 10 (31.3) | |
| 75% | 4 (12.5) | |
| 80% | 9 (28.1) | |
| 100% | 9 (28.1) | |
| Primary disease, n (%) | ||
| Acute lymphoblastic leukemia | 14 (43.8) | 0 (0) |
| Acute myeloid leukemia | 4 (12.5) | 0 (0) |
| Aplastic anemia | 5 (15.6) | 0 (0) |
| Ewing sarcoma | 0 (0) | 4 (50) |
| Neuroblastoma | 0 (0) | 2 (25) |
| Other | 9 (28.1) | 2 (25) |
| Detection frequency of viral DNA (%) | ||
| EBV | 43.7 | 30 |
| HCMV | 5 | 6.7 |
| HHV6 | 7.9 | 20 |
| HHV7 | 9.7 | 23.3 |
| Graft-versus-host disease, n (%) | 25 (78) | |
| Mortality, n (%) | At 365 dayspost-transplant | At 100 dayspost-transplant |
| 13 (40.6) | 3 (37.5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rojas-Rechy, M.H.; Gaytán-Morales, F.; Sánchez-Ponce, Y.; Castorena-Villa, I.; López-Martínez, B.; Parra-Ortega, I.; Escamilla-Núñez, M.C.; Méndez-Tenorio, A.; Pompa-Mera, E.N.; Martinez-Ruiz, G.U.; et al. Herpesvirus Screening in Childhood Hematopoietic Transplant Reveals High Systemic Inflammation in Episodes of Multiple Viral Detection and an EBV Association with Elevated IL-1β, IL-8 and Graft-Versus-Host Disease. Microorganisms 2022, 10, 1685. https://doi.org/10.3390/microorganisms10081685
Rojas-Rechy MH, Gaytán-Morales F, Sánchez-Ponce Y, Castorena-Villa I, López-Martínez B, Parra-Ortega I, Escamilla-Núñez MC, Méndez-Tenorio A, Pompa-Mera EN, Martinez-Ruiz GU, et al. Herpesvirus Screening in Childhood Hematopoietic Transplant Reveals High Systemic Inflammation in Episodes of Multiple Viral Detection and an EBV Association with Elevated IL-1β, IL-8 and Graft-Versus-Host Disease. Microorganisms. 2022; 10(8):1685. https://doi.org/10.3390/microorganisms10081685
Chicago/Turabian StyleRojas-Rechy, Moisés H., Félix Gaytán-Morales, Yessica Sánchez-Ponce, Iván Castorena-Villa, Briceida López-Martínez, Israel Parra-Ortega, María C. Escamilla-Núñez, Alfonso Méndez-Tenorio, Ericka N. Pompa-Mera, Gustavo U. Martinez-Ruiz, and et al. 2022. "Herpesvirus Screening in Childhood Hematopoietic Transplant Reveals High Systemic Inflammation in Episodes of Multiple Viral Detection and an EBV Association with Elevated IL-1β, IL-8 and Graft-Versus-Host Disease" Microorganisms 10, no. 8: 1685. https://doi.org/10.3390/microorganisms10081685
APA StyleRojas-Rechy, M. H., Gaytán-Morales, F., Sánchez-Ponce, Y., Castorena-Villa, I., López-Martínez, B., Parra-Ortega, I., Escamilla-Núñez, M. C., Méndez-Tenorio, A., Pompa-Mera, E. N., Martinez-Ruiz, G. U., Fuentes-Pananá, E. M., & Morales-Sánchez, A. (2022). Herpesvirus Screening in Childhood Hematopoietic Transplant Reveals High Systemic Inflammation in Episodes of Multiple Viral Detection and an EBV Association with Elevated IL-1β, IL-8 and Graft-Versus-Host Disease. Microorganisms, 10(8), 1685. https://doi.org/10.3390/microorganisms10081685







