A Winter-to-Summer Transition of Bacterial and Archaeal Communities in Arctic Sea Ice
Abstract
1. Introduction
2. Materials and Methods
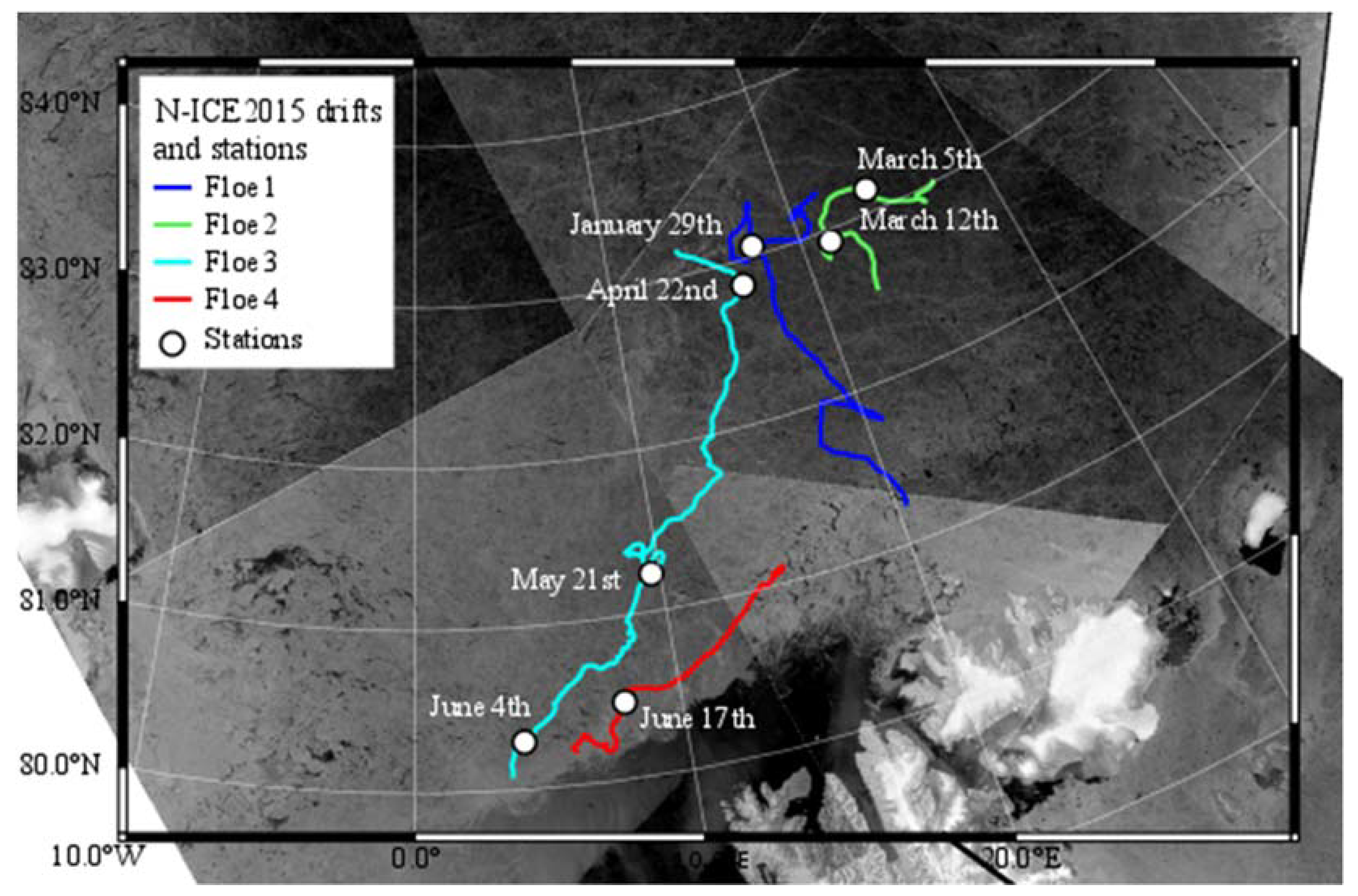
2.1. Ice Core Sampling
2.2. Bacterial Cell Numbers
2.3. DNA and RNA Extraction and Sequencing
2.4. Sequence Analyses
3. Results
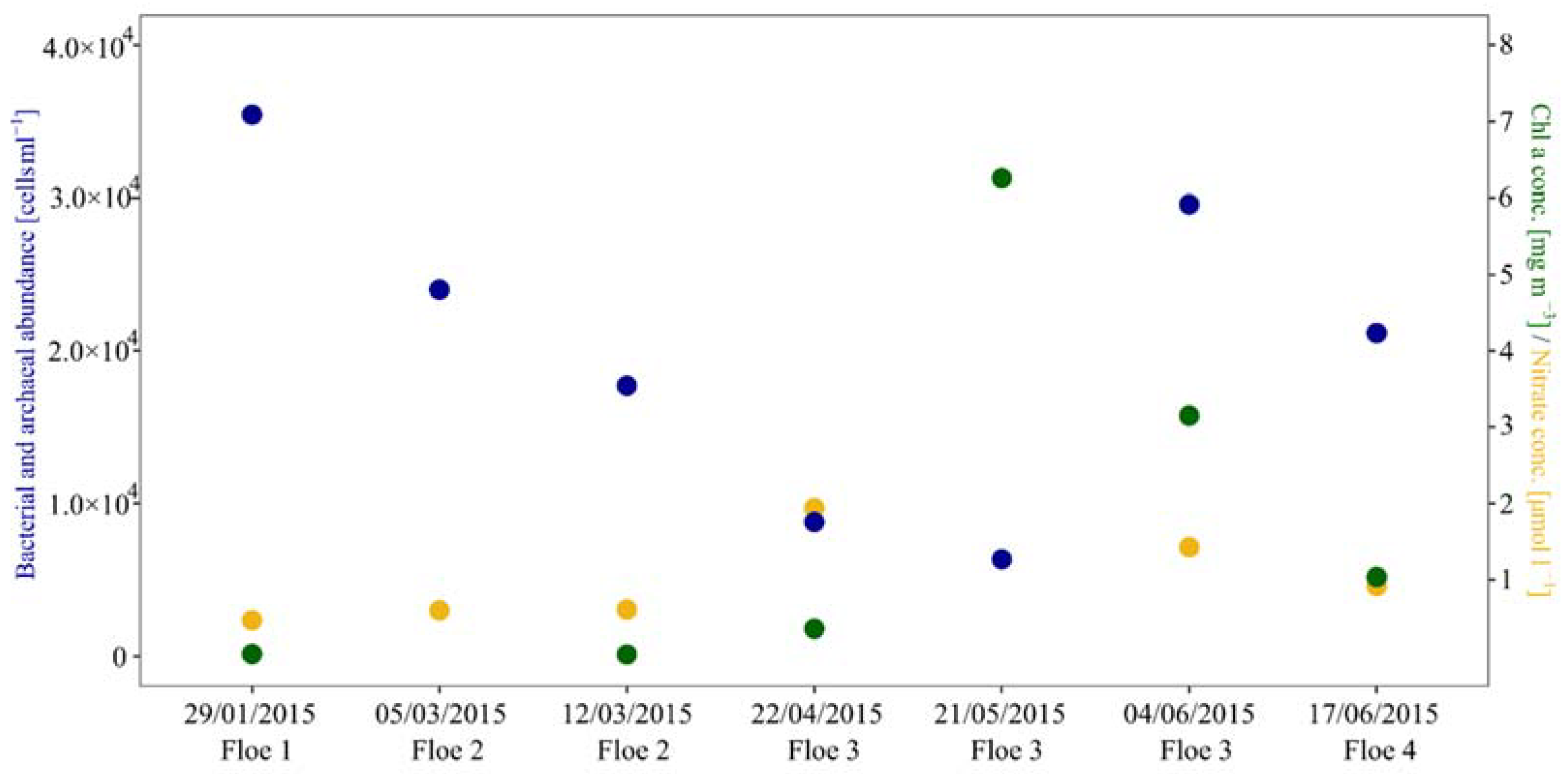
3.1. Cell Numbers and Environmental Data
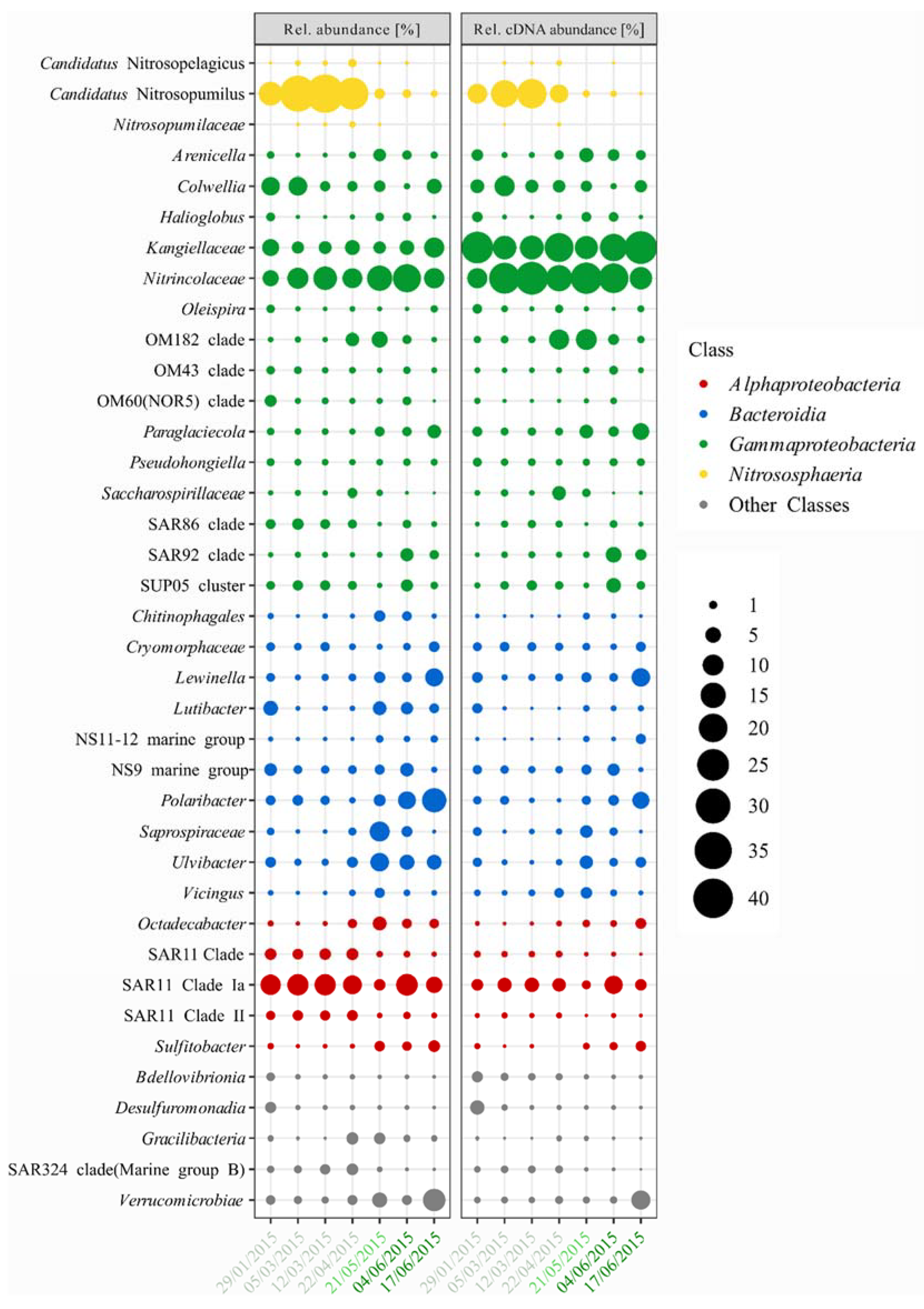
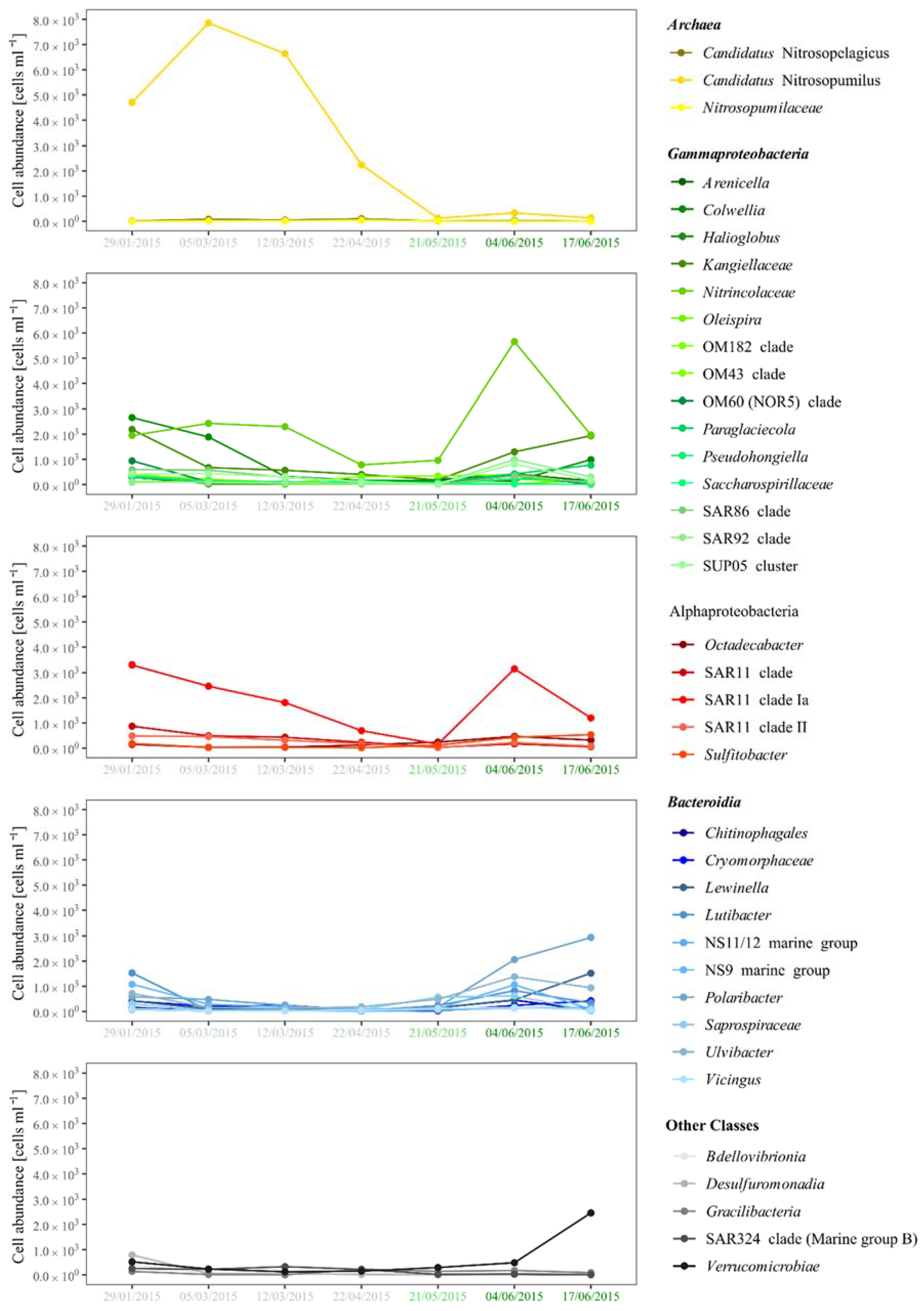
3.2. Bacterial and Archaeal Community Composition
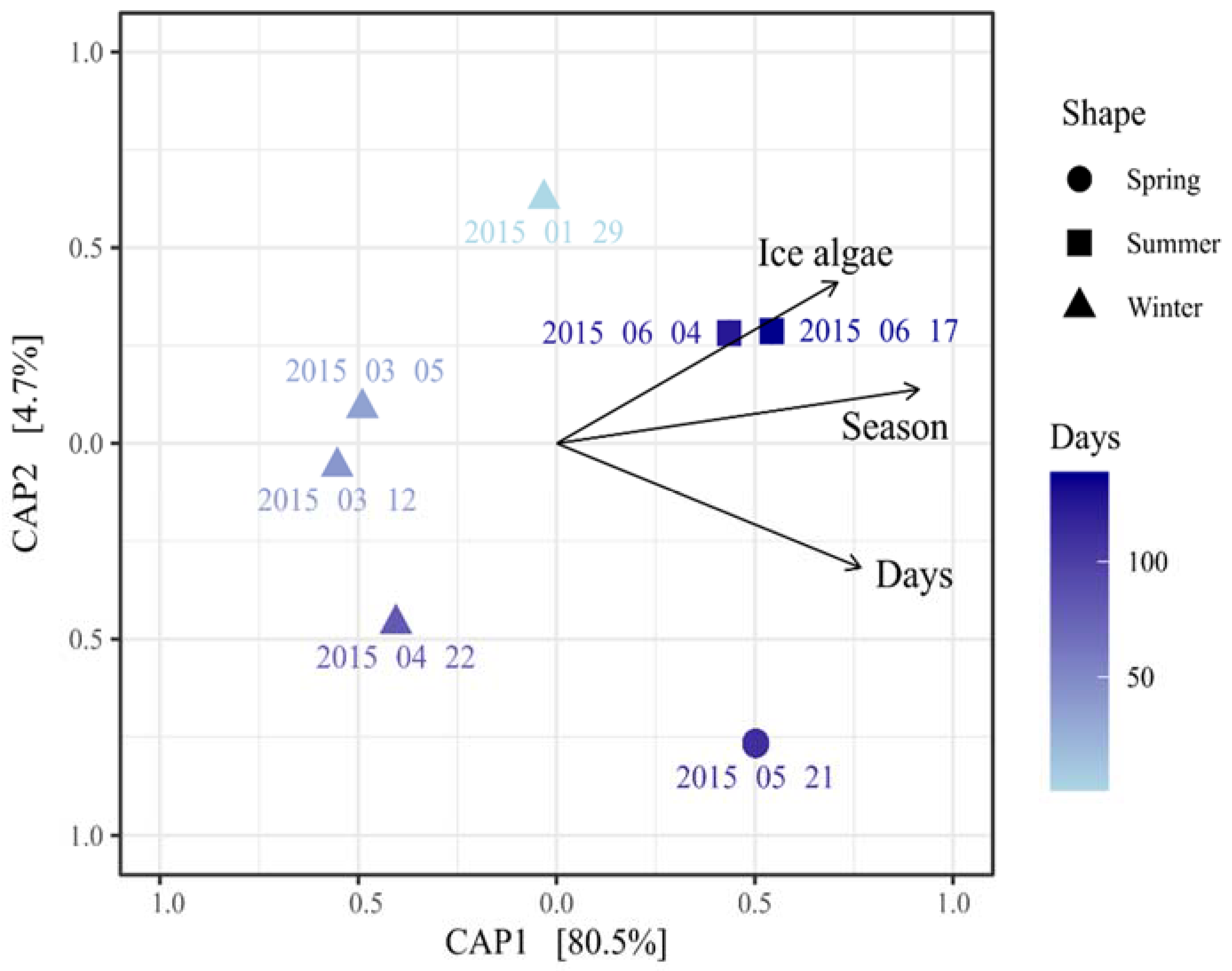
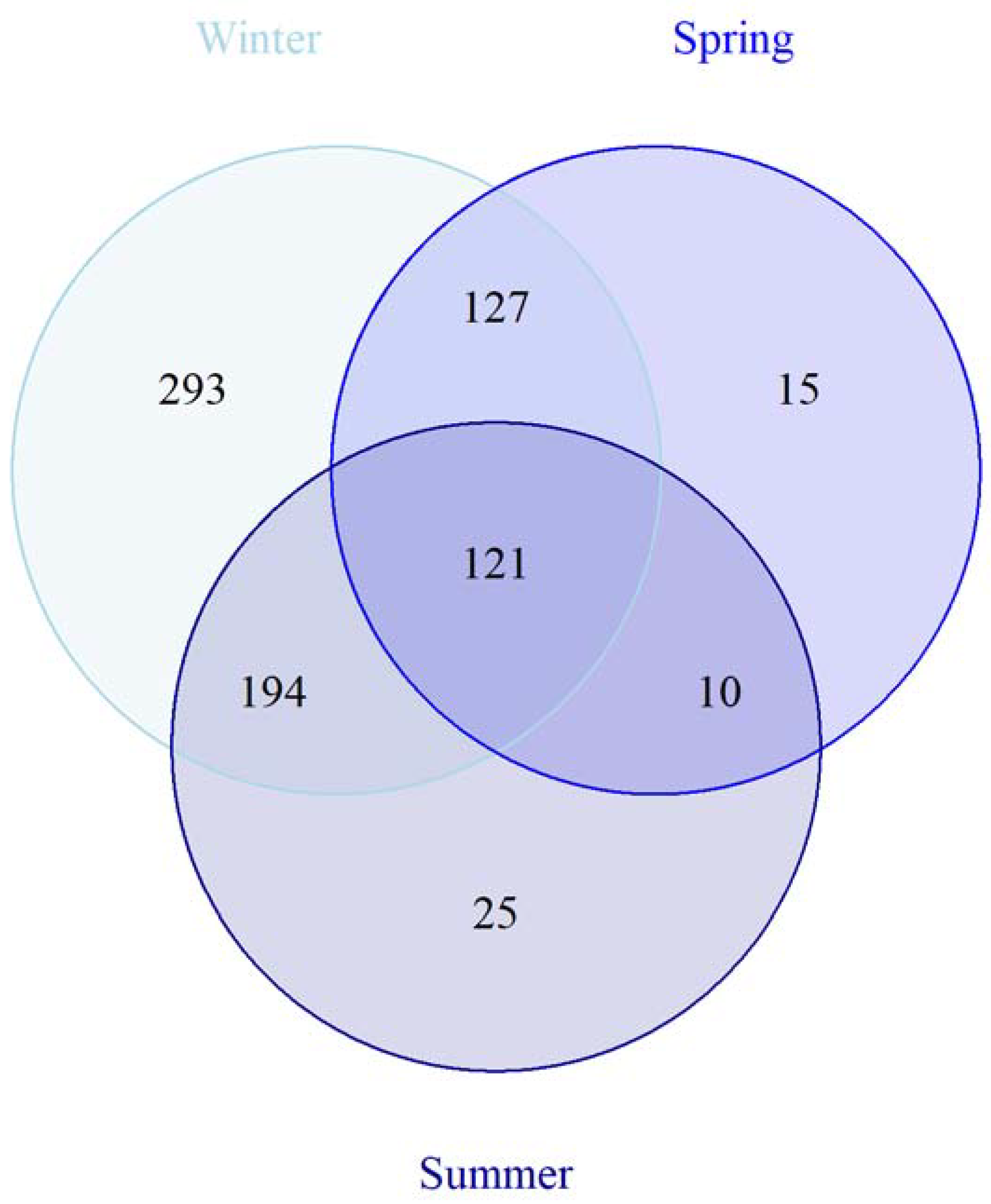
3.3. Seasonal Variation
3.4. Bacterial and Archaeal Activity Estimations
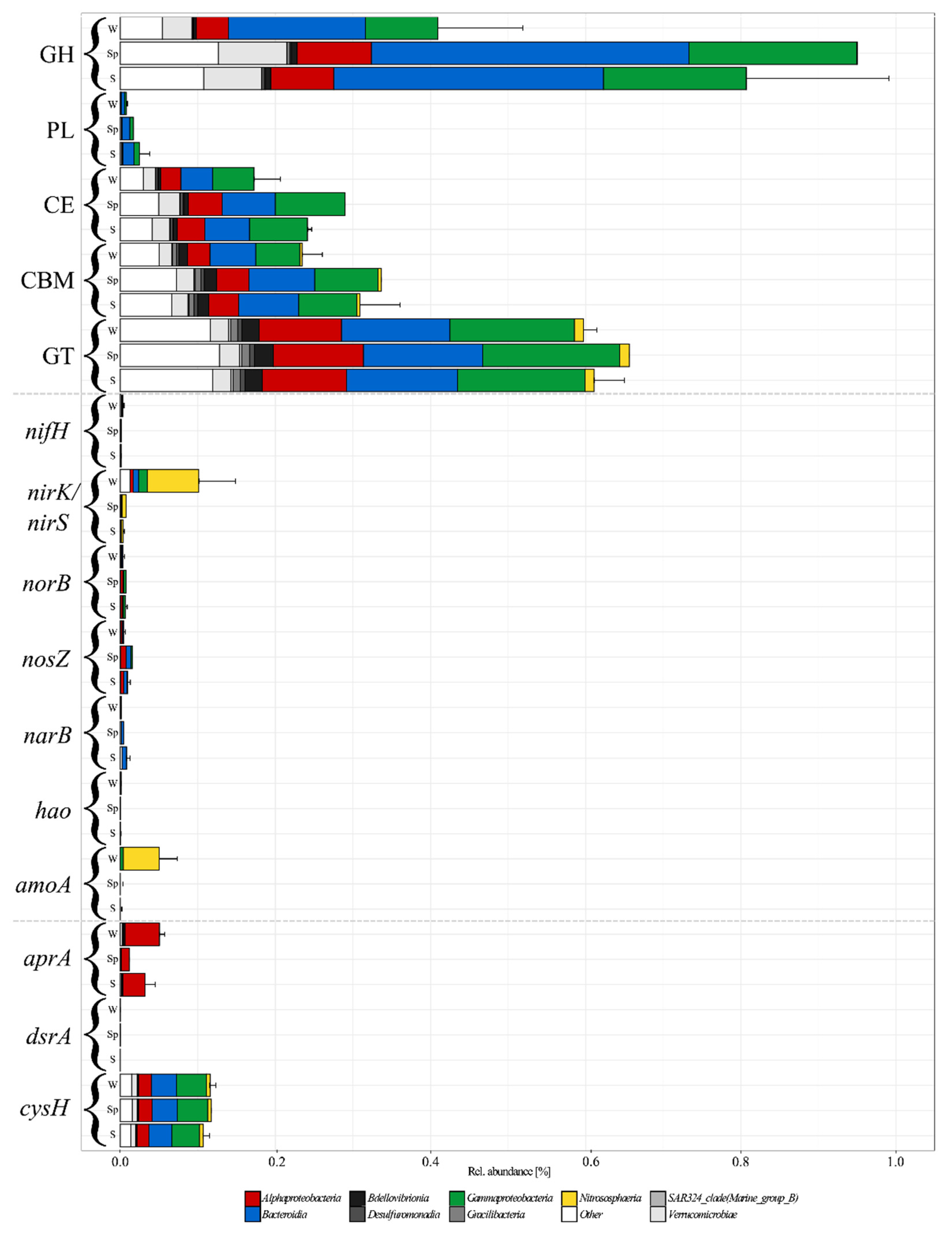
3.5. Functional Approximation Based on PiCRUST2
4. Discussion
4.1. The Winter Community
4.2. A Carbon-Based Succession in Spring/Early Summer
4.3. CAZyme Abundance Support Succession Based on Carbon Sources
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meier, W.N.; Hovelsrud, G.K.; van Oort, B.E.H.; Key, J.R.; Kovacs, K.M.; Michel, C.; Haas, C.; Granskog, M.A.; Gerland, S.; Perovich, D.K.; et al. Arctic Sea Ice in Transformation: A Review of Recent Observed Changes and Impacts on Biology and Human Activity. Rev. Geophys. 2014, 52, 185–217. [Google Scholar] [CrossRef]
- Maslanik, J.; Stroeve, J.; Fowler, C.; Emery, W. Distribution and Trends in Arctic Sea Ice Age through Spring 2011. Geophys. Res. Lett. 2011, 38, L13502. [Google Scholar] [CrossRef]
- Lindsay, R.; Schweiger, A. Arctic Sea Ice Thickness Loss Determined Using Subsurface, Aircraft, and Satellite Observations. Cryosphere 2015, 9, 269–283. [Google Scholar] [CrossRef]
- Kwok, R. Arctic Sea Ice Thickness, Volume, and Multiyear Ice Coverage: Losses and Coupled Variability (1958–2018). Environ. Res. Lett. 2018, 13, 105005. [Google Scholar] [CrossRef]
- Granskog, M.A.; Fer, I.; Rinke, A.; Steen, H. Atmosphere-Ice-Ocean-Ecosystem Processes in a Thinner Arctic Sea Ice Regime: The Norwegian Young Sea ICE (N-ICE2015) Expedition. J. Geophys. Res. Oceans 2018, 123, 1586–1594. [Google Scholar] [CrossRef]
- Assmy, P.; Fernández-Méndez, M.; Duarte, P.; Meyer, A.; Randelhoff, A.; Mundy, C.J.; Olsen, L.M.; Kauko, H.M.; Bailey, A.; Chierici, M.; et al. Leads in Arctic Pack Ice Enable Early Phytoplankton Blooms below Snow-Covered Sea Ice. Sci. Rep. 2017, 7, 40850. [Google Scholar] [CrossRef]
- De Sousa, A.G.G.; Tomasino, M.P.; Duarte, P.; Fernández-Méndez, M.; Assmy, P.; Ribeiro, H.; Surkont, J.; Leite, R.B.; Pereira-Leal, J.B.; Torgo, L.; et al. Diversity and Composition of Pelagic Prokaryotic and Protist Communities in a Thin Arctic Sea-Ice Regime. Microb. Ecol. 2019, 78, 388–408. [Google Scholar] [CrossRef]
- Teeling, H.; Fuchs, B.M.; Bennke, C.M.; Krüger, K.; Chafee, M.; Kappelmann, L.; Reintjes, G.; Waldmann, J.; Quast, C.; Glöckner, F.O.; et al. Recurring Patterns in Bacterioplankton Dynamics during Coastal Spring Algae Blooms. eLife 2016, 5, e11888. [Google Scholar] [CrossRef]
- Christman, G.D.; Cottrell, M.T.; Popp, B.N.; Gier, E.; Kirchman, D.L. Abundance, Diversity, and Activity of Ammonia-Oxidizing Prokaryotes in the Coastal Arctic Ocean in Summer and Winter. Appl. Environ. Microbiol. 2011, 77, 2026–2034. [Google Scholar] [CrossRef]
- Merbt, S.N.; Stahl, D.A.; Casamayor, E.O.; Martí, E.; Nicol, G.W.; Prosser, J.I. Differential Photoinhibition of Bacterial and Archaeal Ammonia Oxidation. FEMS Microbiol. Lett. 2012, 327, 41–46. [Google Scholar] [CrossRef]
- Olsen, L.M.; Laney, S.R.; Duarte, P.; Kauko, H.M.; Fernández-Méndez, M.; Mundy, C.J.; Rösel, A.; Meyer, A.; Itkin, P.; Cohen, L.; et al. The Seeding of Ice Algal Blooms in Arctic Pack Ice: The Multiyear Ice Seed Repository Hypothesis. J. Geophys. Res. Biogeosci. 2017, 122, 1529–1548. [Google Scholar] [CrossRef]
- Brinkmeyer, R.; Knittel, K.; Jürgens, J.; Weyland, H.; Amann, R.; Helmke, E. Diversity and Structure of Bacterial Communities in Arctic versus Antarctic Pack Ice. Appl. Environ. Microbiol. 2003, 69, 6610–6619. [Google Scholar] [CrossRef]
- Bowman, J.S.; Rasmussen, S.; Blom, N.; Deming, J.W.; Rysgaard, S.; Sicheritz-Ponten, T. Microbial Community Structure of Arctic Multiyear Sea Ice and Surface Seawater by 454 Sequencing of the 16S RNA Gene. ISME J. 2012, 6, 11–20. [Google Scholar] [CrossRef]
- Cowie, R.O.M.; Williams, G.J.; Maas, E.W.; Voyles, K.M.; Ryan, K.G. Antarctic Sea-Ice Microbial Communities Show Distinct Patterns of Zonation in Response to Algal-Derived Substrates. Aquat. Microb. Ecol. 2014, 73, 123–134. [Google Scholar] [CrossRef][Green Version]
- Han, D.; Kang, I.; Ha, H.K.; Kim, H.C.; Kim, O.-S.; Lee, B.Y.; Cho, J.-C.; Hur, H.-G.; Lee, Y.K. Bacterial Communities of Surface Mixed Layer in the Pacific Sector of the Western Arctic Ocean during Sea-Ice Melting. PLoS ONE 2014, 9, e86887. [Google Scholar] [CrossRef] [PubMed]
- Boetius, A.; Anesio, A.M.; Deming, J.W.; Mikucki, J.A.; Rapp, J.Z. Microbial Ecology of the Cryosphere: Sea Ice and Glacial Habitats. Nat. Rev. Micro. 2015, 13, 677–690. [Google Scholar] [CrossRef]
- Könneke, M.; Bernhard, A.E.; de la Torre, J.R.; Walker, C.B.; Waterbury, J.B.; Stahl, D.A. Isolation of an Autotrophic Ammonia-Oxidizing Marine Archaeon. Nature 2005, 437, 543–546. [Google Scholar] [CrossRef]
- Collins, R.E.; Rocap, G.; Deming, J.W. Persistence of Bacterial and Archaeal Communities in Sea Ice through an Arctic Winter. Environ. Microbiol. 2010, 12, 1828–1841. [Google Scholar] [CrossRef]
- Eronen-Rasimus, E.; Piiparinen, J.; Karkman, A.; Lyra, C.; Gerland, S.; Kaartokallio, H. Bacterial Communities in Arctic First-Year Drift Ice during the Winter/Spring Transition. Environ. Microbiol. Rep. 2016, 8, 527–535. [Google Scholar] [CrossRef]
- Eronen-Rasimus, E.; Luhtanen, A.-M.; Rintala, J.-M.; Delille, B.; Dieckmann, G.; Karkman, A.; Tison, J.-L. An Active Bacterial Community Linked to High Chl- a Concentrations in Antarctic Winter-Pack Ice and Evidence for the Development of an Anaerobic Sea-Ice Bacterial Community. ISME J. 2017, 11, 2345–2355. [Google Scholar] [CrossRef]
- Fernández-Gómez, B.; Díez, B.; Polz, M.F.; Arroyo, J.I.; Alfaro, F.D.; Marchandon, G.; Sanhueza, C.; Farías, L.; Trefault, N.; Marquet, P.A.; et al. Bacterial Community Structure in a Sympagic Habitat Expanding with Global Warming: Brackish Ice Brine at 85–90 °N. ISME J. 2019, 13, 316–333. [Google Scholar] [CrossRef] [PubMed]
- Torstensson, A.; Margolin, A.R.; Showalter, G.M.; Smith, W.O., Jr.; Shadwick, E.H.; Carpenter, S.D.; Bolinesi, F.; Deming, J.W. Sea-Ice Microbial Communities in the Central Arctic Ocean: Limited Responses to Short-Term PCO2 Perturbations. Limnol. Oceanogr. 2021, 66, S383–S400. [Google Scholar] [CrossRef]
- Cohen, L.; Hudson, S.R.; Walden, V.P.; Graham, R.M.; Granskog, M.A. Meteorological Conditions in a Thinner Arctic Sea Ice Regime from Winter to Summer during the Norwegian Young Sea Ice Expedition (N-ICE2015). J. Geophys. Res. Atmos. 2017, 122, 7235–7259. [Google Scholar] [CrossRef]
- Assmy, P.; Dodd, P.A.; Duarte, P.; Dujardin, J.; Elliott, A.; Fernández-Méndez, M.; Fransson, A.; Granskog, M.A.; Hendry, K.; Hodgson, R.; et al. N-ICE2015 Sea Ice Biogeochemistry; Norwegian Polar Institute: Tromsø, Norway, 2017. [Google Scholar] [CrossRef]
- Marie, D.; Brussaard, C.P.D.; Thyrhaug, R.; Bratbak, G.; Vaulot, D. Enumeration of Marine Viruses in Culture and Natural Samples by Flow Cytometry. Appl. Environ. Microbiol. 1999, 65, 45–52. [Google Scholar] [CrossRef]
- Wilson, B.; Müller, O.; Nordmann, E.-L.; Seuthe, L.; Bratbak, G.; Øvreås, L. Changes in Marine Prokaryote Composition with Season and Depth Over an Arctic Polar Year. Front. Mar. Sci. 2017, 4, 95. [Google Scholar] [CrossRef]
- Ovreås, L.; Forney, L.; Daae, F.L.; Torsvik, V. Distribution of Bacterioplankton in Meromictic Lake Saelenvannet, as Determined by Denaturing Gradient Gel Electrophoresis of PCR-Amplified Gene Fragments Coding for 16S RRNA. Appl. Environ. Microbiol. 1997, 63, 3367–3373. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor Revision to V4 Region SSU RRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Meth. 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucl. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Katoh, K.; Misawa, K.; Kuma, K.; Miyata, T. MAFFT: A Novel Method for Rapid Multiple Sequence Alignment Based on Fast Fourier Transform. Nucl. Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef] [PubMed]
- Cáceres, M.D.; Legendre, P. Associations between Species and Groups of Sites: Indices and Statistical Inference. Ecology 2009, 90, 3566–3574. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Boutros, P.C. VennDiagram: A Package for the Generation of Highly-Customizable Venn and Euler Diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes, R Package Version 1.1-2; 2014. Available online: https://rdrr.io/cran/RColorBrewer/man/ColorBrewer.html (accessed on 29 June 2022).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Wickham, H. Forcats: Tools for Working with Categorical Variables (Factors), R Package Version 0.5.0.; 2020. Available online: https://forcats.tidyverse.org/reference/forcats-package.html (accessed on 29 June 2022).
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for Modern Phylogenetics and Evolutionary Analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J.; et al. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Kassambara, A. Ggpubr: “ggplot2” Based Publication Ready Plots, R Package Version 0.4.0. 2020. Available online: https://rpkgs.datanovia.com/ggpubr/ (accessed on 29 June 2022).
- Lin Pedersen, T. Patchwork: The Composer of Plots, R Package Version 1.1.0. 2020. Available online: https://patchwork.data-imaginist.com/ (accessed on 29 June 2022).
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package, R Package Version 2.5-7. 2020. Available online: https://github.com/vegandevs/vegan (accessed on 29 June 2022).
- Wickham, H.; Seidel, D. Scales: Scale Functions for Visualization, R Package Version 1.1.1. 2020. Available online: https://scales.r-lib.org/ (accessed on 29 June 2022).
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; ISBN 3-900051-07-0. [Google Scholar]
- Grüning, B.; Dale, R.; Sjödin, A.; Chapman, B.A.; Rowe, J.; Tomkins-Tinch, C.H.; Valieris, R.; Köster, J. Bioconda: Sustainable and Comprehensive Software Distribution for the Life Sciences. Nat. Methods 2018, 15, 475–476. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotech. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Rösel, A.; Itkin, P.; King, J.; Divine, D.; Wang, C.; Granskog, M.A.; Krumpen, T.; Gerland, S. Thin Sea Ice, Thick Snow, and Widespread Negative Freeboard Observed During N-ICE2015 North of Svalbard. J. Geophys. Res. Ocean. 2018, 123, 1156–1176. [Google Scholar] [CrossRef]
- Sun, S.; Jones, R.B.; Fodor, A.A. Inference-Based Accuracy of Metagenome Prediction Tools Varies across Sample Types and Functional Categories. Microbiome 2020, 8, 46. [Google Scholar] [CrossRef]
- Walker, C.B.; de la Torre, J.R.; Klotz, M.G.; Urakawa, H.; Pinel, N.; Arp, D.J.; Brochier-Armanet, C.; Chain, P.S.G.; Chan, P.P.; Gollabgir, A.; et al. Nitrosopumilus Maritimus Genome Reveals Unique Mechanisms for Nitrification and Autotrophy in Globally Distributed Marine Crenarchaea. Proc. Natl. Acad. Sci. USA 2010, 107, 8818–8823. [Google Scholar] [CrossRef] [PubMed]
- Grzymski, J.J.; Riesenfeld, C.S.; Williams, T.J.; Dussaq, A.M.; Ducklow, H.; Erickson, M.; Cavicchioli, R.; Murray, A.E. A Metagenomic Assessment of Winter and Summer Bacterioplankton from Antarctica Peninsula Coastal Surface Waters. ISME J. 2012, 6, 1901–1915. [Google Scholar] [CrossRef] [PubMed]
- Müller, O.; Wilson, B.; Paulsen, M.L.; Rumińska, A.; Armo, H.R.; Bratbak, G.; Øvreås, L. Spatiotemporal Dynamics of Ammonia-Oxidizing Thaumarchaeota in Distinct Arctic Water Masses. Front. Microbiol. 2018, 9, 24. [Google Scholar] [CrossRef] [PubMed]
- Luecker, S.; Nowka, B.; Rattei, T.; Spieck, E.; Daims, H. The Genome of Nitrospina Gracilis Illuminates the Metabolism and Evolution of the Major Marine Nitrite Oxidizer. Front. Microbiol. 2013, 4, 27. [Google Scholar] [CrossRef] [PubMed]
- Reji, L.; Tolar, B.B.; Smith, J.M.; Chavez, F.P.; Francis, C.A. Differential Co-Occurrence Relationships Shaping Ecotype Diversification within Thaumarchaeota Populations in the Coastal Ocean Water Column. ISME J. 2019, 13, 1144–1158. [Google Scholar] [CrossRef]
- Wietz, M.; Bienhold, C.; Metfies, K.; Torres-Valdés, S.; von Appen, W.-J.; Salter, I.; Boetius, A. The Polar Night Shift: Seasonal Dynamics and Drivers of Arctic Ocean Microbiomes Revealed by Autonomous Sampling. ISME Commun. 2021, 1, 76. [Google Scholar] [CrossRef]
- Mori, J.F.; Chen, L.-X.; Jessen, G.L.; Rudderham, S.B.; McBeth, J.M.; Lindsay, M.B.J.; Slater, G.F.; Banfield, J.F.; Warren, L.A. Putative Mixotrophic Nitrifying-Denitrifying Gammaproteobacteria Implicated in Nitrogen Cycling Within the Ammonia/Oxygen Transition Zone of an Oil Sands Pit Lake. Front. Microbiol. 2019, 10, 2435. [Google Scholar] [CrossRef]
- Liu, Y.; Blain, S.; Crispi, O.; Rembauville, M.; Obernosterer, I. Seasonal Dynamics of Prokaryotes and Their Associations with Diatoms in the Southern Ocean as Revealed by an Autonomous Sampler. Environ. Microbiol. 2020, 22, 3968–3984. [Google Scholar] [CrossRef]
- Murray, A.E.; Preston, C.M.; Massana, R.; Taylor, L.T.; Blakis, A.; Wu, K.; DeLong, E.F. Seasonal and Spatial Variability of Bacterial and Archaeal Assemblages in the Coastal Waters near Anvers Island, Antarctica. Appl. Environ. Microbiol. 1998, 64, 2585–2595. [Google Scholar] [CrossRef]
- Mincer, T.J.; Church, M.J.; Taylor, L.T.; Preston, C.; Karl, D.M.; DeLong, E.F. Quantitative Distribution of Presumptive Archaeal and Bacterial Nitrifiers in Monterey Bay and the North Pacific Subtropical Gyre. Environ. Microbiol. 2007, 9, 1162–1175. [Google Scholar] [CrossRef]
- Smith, J.M.; Chavez, F.P.; Francis, C.A. Ammonium Uptake by Phytoplankton Regulates Nitrification in the Sunlit Ocean. PLoS ONE 2014, 9, e108173. [Google Scholar] [CrossRef]
- Church, M.J.; DeLong, E.F.; Ducklow, H.W.; Karner, M.B.; Preston, C.M.; Karl, D.M. Abundance and Distribution of Planktonic Archaea and Bacteria in the Waters West of the Antarctic Peninsula. Limnol. Oceanogr. 2003, 48, 1893–1902. [Google Scholar] [CrossRef]
- Herfort, L.; Schouten, S.; Abbas, B.; Veldhuis, M.J.W.; Coolen, M.J.L.; Wuchter, C.; Boon, J.P.; Herndl, G.J.; Sinninghe Damsté, J.S. Variations in Spatial and Temporal Distribution of Archaea in the North Sea in Relation to Environmental Variables. FEMS Microbiol. Ecol. 2007, 62, 242–257. [Google Scholar] [CrossRef][Green Version]
- Morris, R.M.; Rappe, M.S.; Connon, S.A.; Vergin, K.L.; Siebold, W.A.; Carlson, C.A.; Giovannoni, S.J. SAR11 Clade Dominates Ocean Surface Bacterioplankton Communities. Nature 2002, 420, 806–810. [Google Scholar] [CrossRef]
- West, N.J.; Obernosterer, I.; Zemb, O.; Lebaron, P. Major Differences of Bacterial Diversity and Activity inside and Outside of a Natural Iron-Fertilized Phytoplankton Bloom in the Southern Ocean. Environ. Microbiol. 2008, 10, 738–756. [Google Scholar] [CrossRef] [PubMed]
- Schattenhofer, M.; Wulf, J.; Kostadinov, I.; Glöckner, F.O.; Zubkov, M.V.; Fuchs, B.M. Phylogenetic Characterisation of Picoplanktonic Populations with High and Low Nucleic Acid Content in the North Atlantic Ocean. Syst. Appl. Microbiol. 2011, 34, 470–475. [Google Scholar] [CrossRef]
- Thiele, S.; Fuchs, B.M.; Ramaiah, N.; Amann, R. Microbial Community Response during the Iron Fertilization Experiment LOHAFEX. Appl. Environ. Microbiol. 2012, 78, 8803–8812. [Google Scholar] [CrossRef]
- Meyer, B.; Kuever, J. Molecular Analysis of the Diversity of Sulfate-Reducing and Sulfur-Oxidizing Prokaryotes in the Environment, Using AprA as Functional Marker Gene. Appl. Environ. Microbiol. 2007, 73, 7664–7679. [Google Scholar] [CrossRef]
- Thiele, S.; Richter, M.; Balestra, C.; Glöckner, F.O.; Casotti, R. Taxonomic and Functional Diversity of a Coastal Planktonic Bacterial Community in a River-Influenced Marine Area. Mar. Genom. 2017, 32, 61–69. [Google Scholar] [CrossRef]
- Walsh, D.A.; Zaikova, E.; Howes, C.G.; Song, Y.C.; Wright, J.J.; Tringe, S.G.; Tortell, P.D.; Hallam, S.J. Metagenome of a Versatile Chemolithoautotroph from Expanding Oceanic Dead Zones. Science 2009, 326, 578–582. [Google Scholar] [CrossRef]
- Sheik, C.S.; Jain, S.; Dick, G.J. Metabolic Flexibility of Enigmatic SAR324 Revealed through Metagenomics and Metatranscriptomics. Environ. Microbiol. 2014, 16, 304–317. [Google Scholar] [CrossRef]
- Kuever, J.; Rainey, F.A.; Widdel, F. Desulfuromonas. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–7. ISBN 978-1-118-96060-8. [Google Scholar]
- Simon, M.; Glöckner, F.; Amann, R. Different Community Structure and Temperature Optima of Heterotrophic Picoplankton in Various Regions of the Southern Ocean. Aquat. Microb. Ecol. 1999, 18, 275–284. [Google Scholar] [CrossRef]
- Puddu, A.; Zoppini, A.; Fazi, S.; Rosati, M.; Amalfitano, S.; Magaletti, E. Bacterial Uptake of DOM Released from P-Limited Phytoplankton. FEMS Microbiol. Ecol. 2003, 46, 257–268. [Google Scholar] [CrossRef]
- Alderkamp, A.C.; Sintes, E.; Herndl, G.J. Abundance and Activity of Major Groups of Prokaryotic Plankton in the Coastal North Sea during Spring and Summer. Aquat. Microb. Ecol. 2006, 45, 237–246. [Google Scholar] [CrossRef]
- Gomez-Pereira, P.R.; Fuchs, B.M.; Alonso, C.; Oliver, M.J.; van Beusekom, J.E.E.; Amann, R. Distinct Flavobacterial Communities in Contrasting Water Masses of the North Atlantic Ocean. ISME J. 2010, 4, 472–487. [Google Scholar] [CrossRef]
- Teeling, H.; Fuchs, B.; Becher, D.; Klockow, C.; Gardebrecht, A.; Bennke, C.M.; Kassabgy, M.; Huang, S.; Mann, A.J.; Waldmann, J.; et al. Substrate-Controlled Succession of Marine Bacterioplankton Populations Induced by a Phytoplankton Bloom. Science 2012, 336, 608–611. [Google Scholar] [CrossRef]
- Reintjes, G.; Arnosti, C.; Fuchs, B.; Amann, R. Selfish, Sharing and Scavenging Bacteria in the Atlantic Ocean: A Biogeographical Study of Bacterial Substrate Utilisation. ISME J. 2019, 13, 1119–1132. [Google Scholar] [CrossRef]
- Reintjes, G.; Fuchs, B.M.; Scharfe, M.; Wiltshire, K.H.; Amann, R.; Arnosti, C. Short-Term Changes in Polysaccharide Utilization Mechanisms of Marine Bacterioplankton during a Spring Phytoplankton Bloom. Environ. Microbiol. 2020, 22, 1884–1900. [Google Scholar] [CrossRef]
- Allers, E.; Gómez-Consarnau, L.; Pinhassi, J.; Gasol, J.M.; Šimek, K.; Pernthaler, J. Response of Alteromonadaceae and Rhodobacteriaceae to Glucose and Phosphorus Manipulation in Marine Mesocosms. Environ. Microbiol. 2007, 9, 2417–2429. [Google Scholar] [CrossRef]
- Sakami, T.; Watanabe, T.; Kakehi, S.; Taniuchi, Y.; Kuwata, A. Spatial Variation of Bacterial Community Composition at the Expiry of Spring Phytoplankton Bloom in Sendai Bay, Japan. Gene 2016, 576, 610–617. [Google Scholar] [CrossRef]
- Luria, C.M.; Amaral-Zettler, L.A.; Ducklow, H.W.; Repeta, D.J.; Rhyne, A.L.; Rich, J.J. Seasonal Shifts in Bacterial Community Responses to Phytoplankton-Derived Dissolved Organic Matter in the Western Antarctic Peninsula. Front. Microbiol. 2017, 8, 2117. [Google Scholar] [CrossRef]
- Mönnich, J.; Tebben, J.; Bergemann, J.; Case, R.; Wohlrab, S.; Harder, T. Niche-Based Assembly of Bacterial Consortia on the Diatom Thalassiosira Rotula Is Stable and Reproducible. ISME J. 2020, 14, 1614–1625. [Google Scholar] [CrossRef]
- Park, B.S.; Lee, M.; Shin, K.; Baek, S.H. Response of the Bacterioplankton Composition to Inorganic Nutrient Loading and Phytoplankton in Southern Korean Coastal Waters: A Mesocosm Study. Mar. Ecol. 2020, 41, e12591. [Google Scholar] [CrossRef]
- Wang, J.; Lu, Y.; Nawaz, M.Z.; Xu, J. Comparative Genomics Reveals Evidence of Genome Reduction and High Extracellular Protein Degradation Potential in Kangiella. Front. Microbiol. 2018, 9, 1224. [Google Scholar] [CrossRef] [PubMed]
- Thiele, S.; Fuchs, B.M.; Amann, R.; Iversen, M.H. Colonization in the Photic Zone and Subsequent Changes during Sinking Determine Bacterial Community Composition in Marine Snow. Appl. Environ. Microbiol. 2015, 81, 1463–1471. [Google Scholar] [CrossRef] [PubMed]
- Xing, P.; Hahnke, R.L.; Unfried, F.; Markert, S.; Huang, S.; Barbeyron, T.; Harder, J.; Becher, D.; Schweder, T.; Glöckner, F.O.; et al. Niches of Two Polysaccharide-Degrading Polaribacter Isolates from the North Sea during a Spring Diatom Bloom. ISME J. 2015, 9, 1410–1422. [Google Scholar] [CrossRef] [PubMed]
- Signori, C.N.; Pellizari, V.H.; Enrich-Prast, A.; Sievert, S.M. Spatiotemporal Dynamics of Marine Bacterial and Archaeal Communities in Surface Waters off the Northern Antarctic Peninsula. Deep Sea Res. II Top. Stud. Oceanogr. 2018, 149, 150–160. [Google Scholar] [CrossRef]
- Liu, Y.; Debeljak, P.; Rembauville, M.; Blain, S.; Obernosterer, I. Diatoms Shape the Biogeography of Heterotrophic Prokaryotes in Early Spring in the Southern Ocean. Environ. Microbiol. 2019, 21, 1452–1465. [Google Scholar] [CrossRef]
- Martinez-Garcia, M.; Brazel, D.M.; Swan, B.K.; Arnosti, C.; Chain, P.S.G.; Reitenga, K.G.; Xie, G.; Poulton, N.J.; Gomez, M.L.; Masland, D.E.D.; et al. Capturing Single Cell Genomes of Active Polysaccharide Degraders: An Unexpected Contribution of Verrucomicrobia. PLoS ONE 2012, 7, e35314. [Google Scholar] [CrossRef]
- Cardman, Z.; Arnosti, C.; Durbin, A.; Ziervogel, K.; Cox, C.; Steen, A.D.; Teske, A. Verrucomicrobia Are Candidates for Polysaccharide-Degrading Bacterioplankton in an Arctic Fjord of Svalbard. Appl. Environ. Microbiol. 2014, 80, 3749–3756. [Google Scholar] [CrossRef]
- Sichert, A.; Corzett, C.H.; Schechter, M.S.; Unfried, F.; Markert, S.; Becher, D.; Fernandez-Guerra, A.; Liebeke, M.; Schweder, T.; Polz, M.F.; et al. Verrucomicrobia Use Hundreds of Enzymes to Digest the Algal Polysaccharide Fucoidan. Nat. Meth. 2020, 5, 1026–1039. [Google Scholar] [CrossRef]
- Taylor, J.D.; Cottingham, S.D.; Billinge, J.; Cunliffe, M. Seasonal Microbial Community Dynamics Correlate with Phytoplankton-Derived Polysaccharides in Surface Coastal Waters. ISME J. 2014, 8, 245–248. [Google Scholar] [CrossRef]
- Tisserand, L.; Dadaglio, L.; Intertaglia, L.; Catala, P.; Panagiotopoulos, C.; Obernosterer, I.; Joux, F. Use of Organic Exudates from Two Polar Diatoms by Bacterial Isolates from the Arctic Ocean. Philos. Trans. R. Soc. A 2020, 378, 20190356. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thiele, S.; Storesund, J.E.; Fernández-Méndez, M.; Assmy, P.; Øvreås, L. A Winter-to-Summer Transition of Bacterial and Archaeal Communities in Arctic Sea Ice. Microorganisms 2022, 10, 1618. https://doi.org/10.3390/microorganisms10081618
Thiele S, Storesund JE, Fernández-Méndez M, Assmy P, Øvreås L. A Winter-to-Summer Transition of Bacterial and Archaeal Communities in Arctic Sea Ice. Microorganisms. 2022; 10(8):1618. https://doi.org/10.3390/microorganisms10081618
Chicago/Turabian StyleThiele, Stefan, Julia E. Storesund, Mar Fernández-Méndez, Philipp Assmy, and Lise Øvreås. 2022. "A Winter-to-Summer Transition of Bacterial and Archaeal Communities in Arctic Sea Ice" Microorganisms 10, no. 8: 1618. https://doi.org/10.3390/microorganisms10081618
APA StyleThiele, S., Storesund, J. E., Fernández-Méndez, M., Assmy, P., & Øvreås, L. (2022). A Winter-to-Summer Transition of Bacterial and Archaeal Communities in Arctic Sea Ice. Microorganisms, 10(8), 1618. https://doi.org/10.3390/microorganisms10081618





