The Study of Soil Bacterial Diversity and the Influence of Soil Physicochemical Factors in Meltwater Region of Ny-Ålesund, Arctic
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling Site Description and Sample Collection
2.2. Soil Physicochemical Property Analyses
2.3. DNA Extraction, PCR Amplification, and Sequencing
2.4. High-Throughput Sequencing and Analysis
3. Results
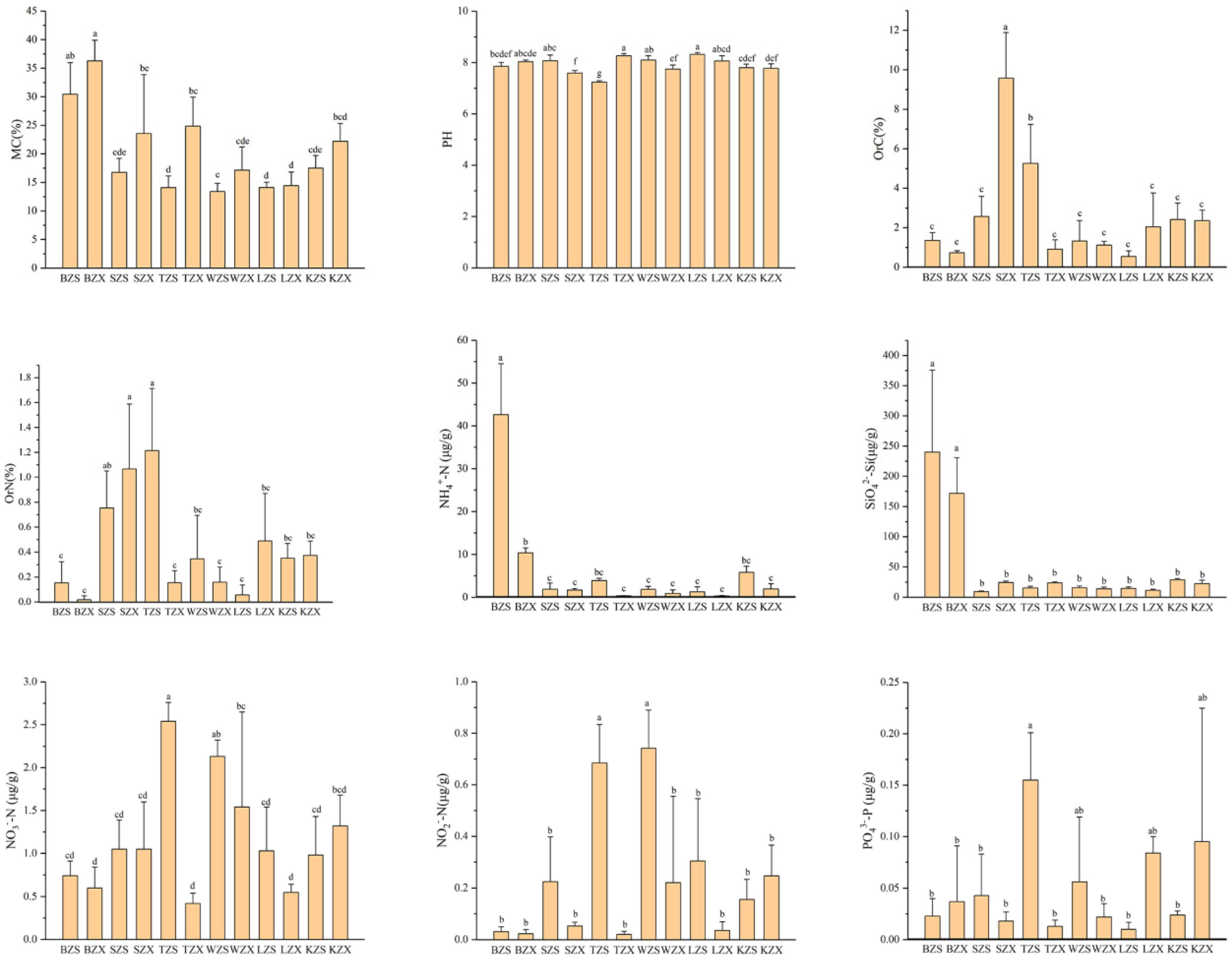
3.1. Physicochemical Properties of Soils
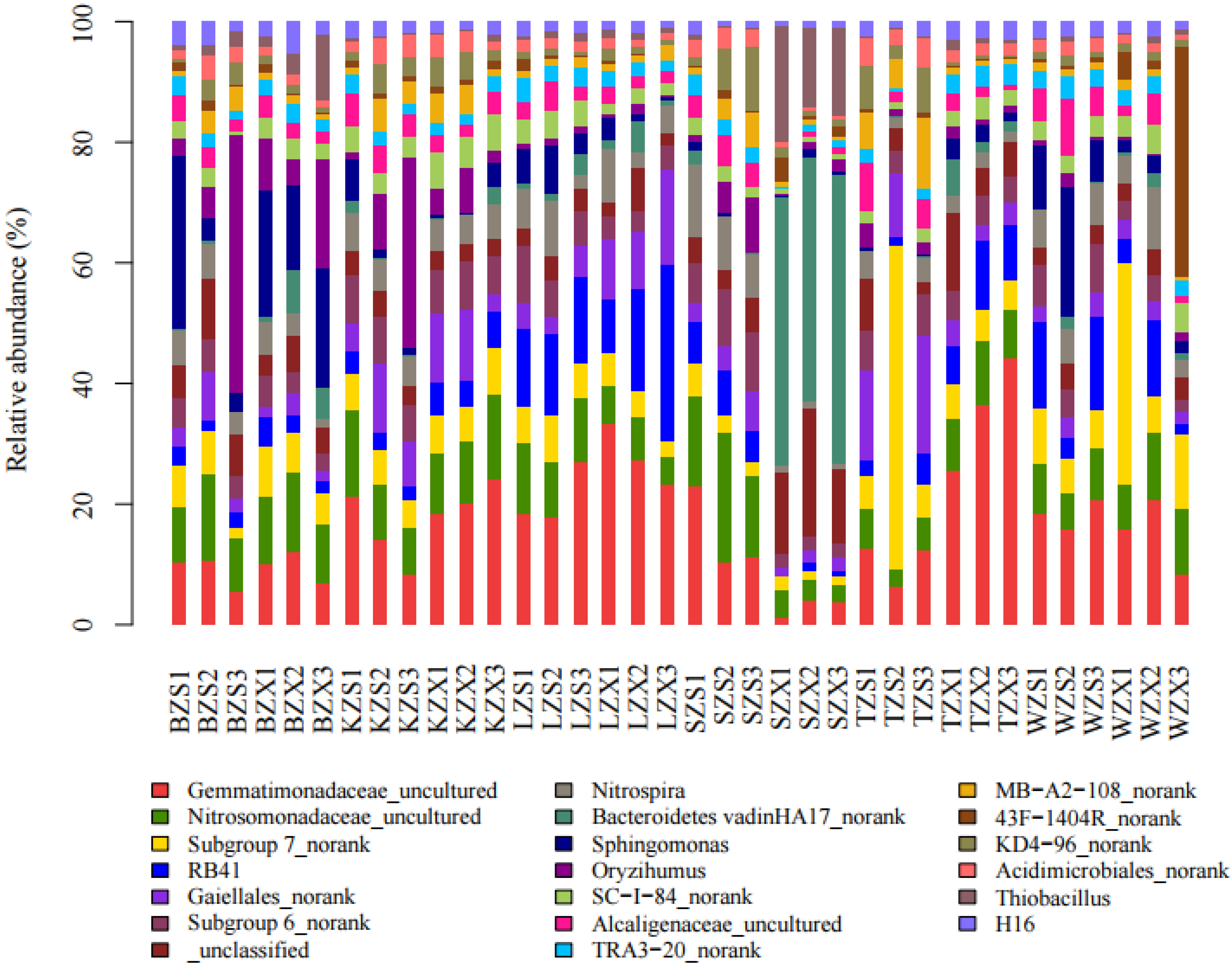
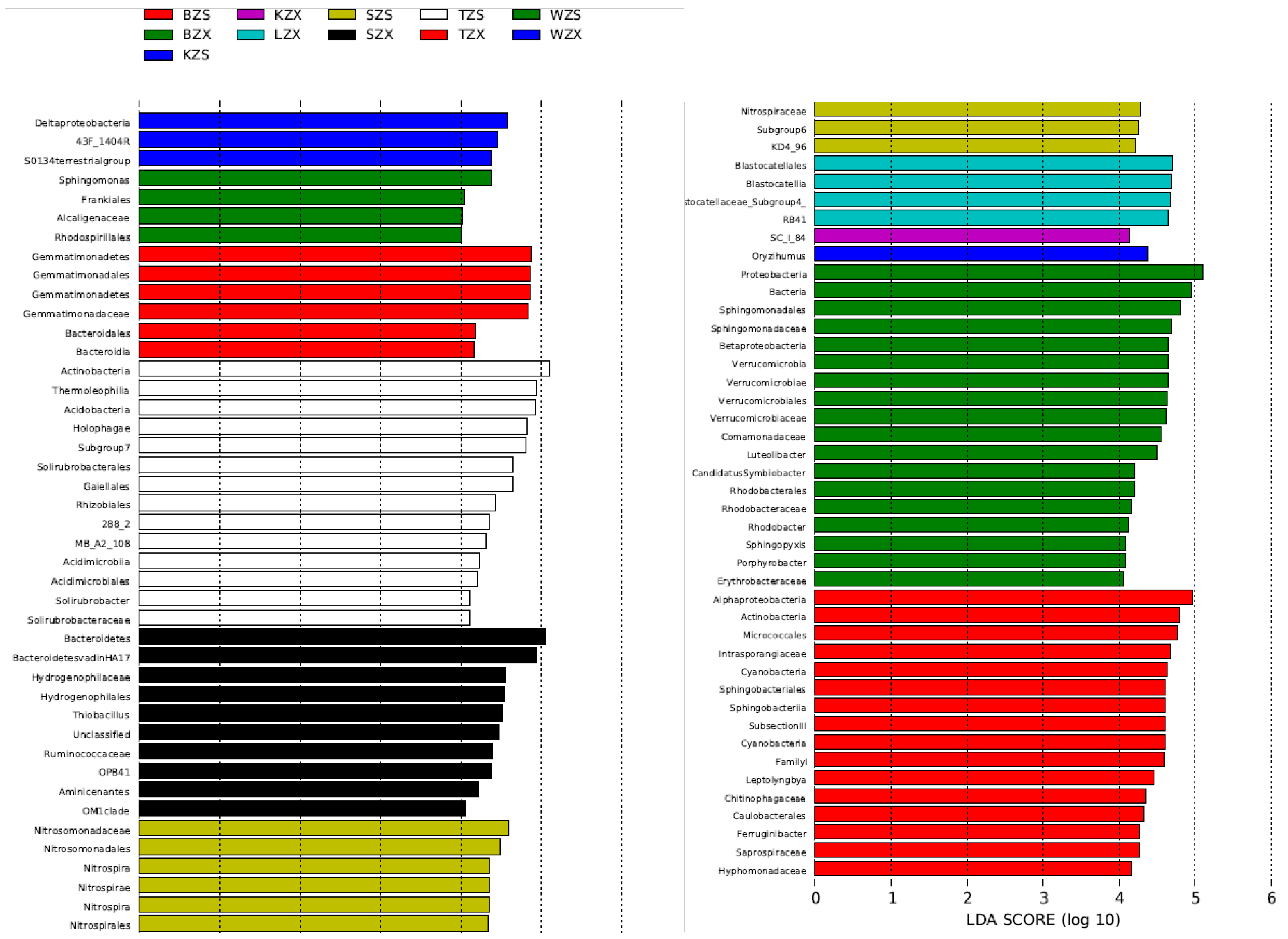
3.2. Bacterial Diversity and Community Composition
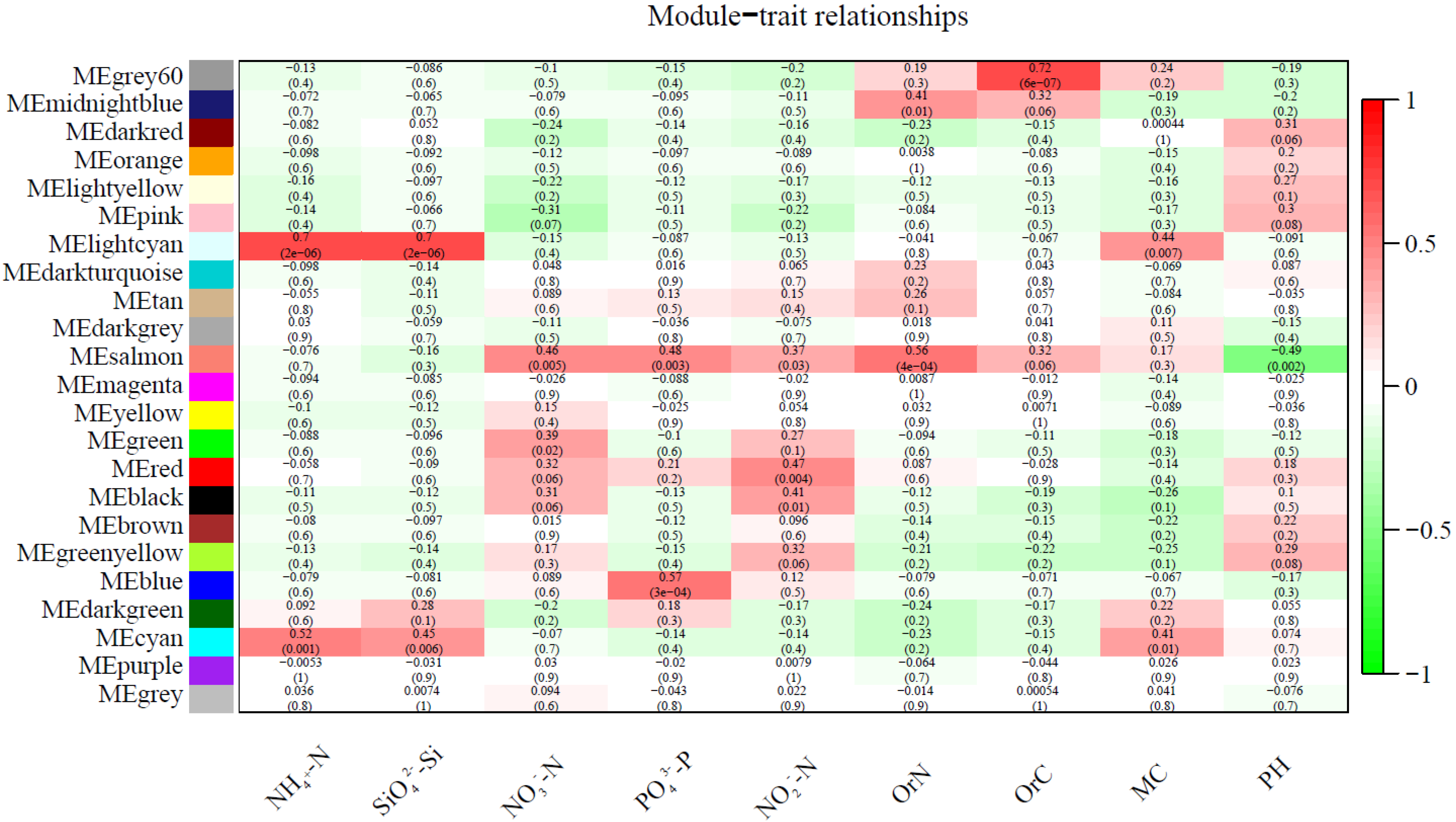
3.3. Correlation between Soil Physicochemical Factors and Bacterial Community Structure
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kerr, R.A. Global warming: Arctic summer sea ice could vanish soon but not suddenly. Science 2009, 323, 1655. [Google Scholar] [CrossRef][Green Version]
- Lutz, S.; Anesio, A.M.; Raiswell, R.; Edwards, A.; Newton, R.; Gill, F.; Benning, L.G. The biogeography of red snow microbiomes and their role in melting arctic glaciers. Nat. Commun. 2016, 7, 11968. [Google Scholar] [CrossRef]
- Rühland, K.M.; Paterson, A.M.; Keller, W.; Michelutti, N.; Smol, J.P. Global warming triggers the loss of a key Arctic refugium. Proc. R. Soc. B Boil. Sci. 2013, 280, 20131887. [Google Scholar] [CrossRef]
- Yi, S.; Wischnewski, K.; Langer, M.; Muster, S.; Boike, J. Freeze/thaw processes in complex permafrost landscapes of northern Siberia simulated using the TEM ecosystem model: Impact of thermokarst ponds and lakes. Geosci. Model Dev. 2014, 7, 1671–1689. [Google Scholar] [CrossRef]
- Smith, L.C.; Sheng, Y.; Macdonald, G.M.; Hinzman, L.D. Disappearing Arctic Lakes. Science 2005, 308, 1429. [Google Scholar] [CrossRef]
- Adrian, R.; Reilly, C.M.; Zagareze, H.; Baines, S.B.; Hessen, D.O.; Keller, W.; Livingstone, D.M.; Sommaruga, R.; Straile, D.; Van Donk, E.; et al. Lakes as sentinels of climate change. Limnol. Oceanogr. 2009, 54, 2283–2297. [Google Scholar] [CrossRef]
- Inglese, C.N.; Christiansen, C.T.; Lamhonwah, D.; Moniz, K.; Montross, S.N.; Lamoureux, S.; Lafreniere, M.; Grogan, P.; Walker, V.K. Examination of soil microbial communities after permafrost thaw subsequent to an active layer detach-ment in the high Arctic. Arct. Antarct. Alp. Res. 2017, 49, 455–472. [Google Scholar] [CrossRef]
- Poosakkannu, A.; Nissinen, R.; Männistö, M.; Kytöviita, M.M. Microbial community composition but not diver-sity changes along succession in arctic sand dunes. Environ. Microbiol. 2017, 19, 698–709. [Google Scholar] [CrossRef]
- Rohart, F.; Gautier, B.; Singh, A.; Lê Cao, K.-A. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Lipson, D.A.; Raab, T.K.; Parker, M.; Kelley, S.T.; Brislawn, C.J.; Jansson, J. Changes in microbial communities along redox gradients in polygonized Arctic wet tundra soils. Environ. Microbiol. Rep. 2015, 7, 649–657. [Google Scholar] [CrossRef]
- Maccario, L.; Vogel, T.M.; Larose, C. Potential drivers of microbial community structure and function in Arctic spring snow. Front. Microbiol. 2014, 5, 413. [Google Scholar] [CrossRef]
- Kumar, N.; Shah, V.; Walker, V.K. Perturbation of an arctic soil microbial community by metal nanoparti-cles. J. Hazard. Mater. 2011, 190, 816–822. [Google Scholar] [CrossRef]
- Canion, A.; Overholt, W.A.; Kostka, J.E.; Huettel, M.; Lavik, G.; Kuypers, M.M. Temperature response of denitri-fication and anaerobic ammonium oxidation rates and microbial community structure in Arctic fjord sediments. Environ. Microbiol. 2014, 16, 3331–3344. [Google Scholar] [CrossRef]
- Zhang, T.; Wang, N.F.; Liu, H.Y.; Zhang, Y.Q.; Yu, L.Y. Soil pH is a key determinant of soil fungal community composition in the Ny-Ålesund Region, Svalbard (High Arctic). Front. Microbiol. 2016, 7, 227. [Google Scholar] [CrossRef]
- Wang, N.F.; Zhang, T.; Yang, X.; Wang, S.; Yu, Y.; Dong, L.L.; Guo, Y.D.; Ma, Y.X.; Zang, J.Y. Diversity and composition of bacterial community in soils and lake sediments from an arctic lake area. Front. Microbiol. 2016, 7, 1170. [Google Scholar] [CrossRef]
- Venkatachalam, S.; Kannan, V.M.; Saritha, V.N.; Loganathachetti, D.S.; Mohan, M.; Krishnan, K.P. Bacterial diversity and community structure along the glacier foreland of Midtre Lovénbreen, Svalbard, Arctic. Ecol. Indic. 2021, 126, 107704. [Google Scholar] [CrossRef]
- Sun, W.; Yan, M.; Ai, S.; Zhu, G.; Wang, Z.; Liu, L.; Xu, Y.; Ren, J. Ice Temperature Characteristics of the Austre Lovénbreen Glacier in NY-lesund, Arctic Region. J. Wuhan Univ. Inf. Sci. Ed. 2016, 41, 79–85. [Google Scholar]
- Ding, Y. Study on Changes of Glacial Landforms in Ny-Ålesund, Arctic; Shandong Normal University: Jinan, China, 2016. [Google Scholar]
- Hu, L.; Shi, X.; Yu, Z.; Lin, T.; Wang, H.; Ma, D.; Guo, Z.; Yang, Z. Distribution of sedimentary organic matter in estua-rine–inner shelf regions of the East China Sea: Implications for hydrodynamic forces and anthropogenic impact. Mar. Chem. 2012, 142, 29–40. [Google Scholar] [CrossRef]
- Liu, J.; Zang, J.; Zhao, C.; Yu, Z.; Xu, B.; Li, J.; Ran, X. Phosphorus speciation, transformation, and preservation in the coastal area of Rushan Bay. Sci. Total Environ. 2016, 565, 258–270. [Google Scholar] [CrossRef]
- Magoä, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar]
- Michelsen, C.F.; Pedas, P.; Glaring, M.A.; Schjoerring, J.K.; Stougaard, P. Bacterial diversity in Greenlandic soils as affected by potato cropping and inorganic versus organic fertilization. Polar Biol. 2013, 37, 61–71. [Google Scholar] [CrossRef]
- Jeanette, B.; Brandt, K.K.; Al-Soud, W.A.; Holm, P.E.; Hansen, L.H.; Srensen, S.R.J.; Ole, N. Selection for Cutolerant bacterial communities with altered composition, but unaltered richness, via long-term Cu exposure. Appl. Environ. Microbiol. 2012, 78, 7438–7446. [Google Scholar]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 2011, 21, 494–504. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA se-quences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Tierney, L. The R Statistical Computing Environment; Springer: New York, NY, USA, 2012; pp. 1–21. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Wagner, H. Vegan: Community Ecology Package; 2016. [Google Scholar]
- Peeters, K.; Hodgson, D.A.; Convey, P.; Willems, A. Cultiurable diversity of heterotrophic bacteria in Forlidas Pond (Pensacola Mountains) and Lundström Lake (Shackleton Range), Antarctica. Microb. Ecol. 2011, 62, 399–413. [Google Scholar] [CrossRef]
- Zhang, W.; Bahadur, A.; Sajjad, W.; Zhang, G.; Nasir, F.; Zhang, B.; Wu, X.; Liu, G.; Chen, T. Bacterial Diversity and Community Composition Distribution in Cold-Desert Habitats of Qinghai–Tibet Plateau, China. Microorganisms 2021, 9, 262. [Google Scholar] [CrossRef]
- Hinsa-Leasure, S.M.; Bhavaraju, L.; Rodrigues, J.L.; Bakermans, C.; Gilichinsky, D.A.; Tiedje, J.M. Characterization of a bacterial community from a Northeast Siberian seacoast permafrost sample. FEMS Microbiol. Ecol. 2010, 74, 103–113. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Shu, H.Y.; Lin, X.R.; Zhou, Q.X.; Bramryd, T.; Shu, W.S.; Huang, L.N. Microbial community structure and function in sediments from e-waste contaminat-ed rivers at Guiyu area of China. Environ. Pollut. 2018, 235, 171–179. [Google Scholar] [CrossRef]
- Peng, F.J.; Diepens, N.J.; Pan, C.G.; Ying, G.G.; Salvito, D.; Selck, H.; Van den Brink, P.J. Response of sediment bacterial community to triclosan in subtropical fresh-water benthic microcosms. Environ. Pollut. 2019, 248, 676–683. [Google Scholar] [CrossRef]
- Zhang, M.; Wu, Z.; Sun, Q.; Ding, Y.; Ding, Z.; Sun, L. The spatial and seasonal variations of bacterial community structure and in-fluencing factors in river sediments. J. Environ. Manag. 2019, 248, 109293. [Google Scholar] [CrossRef]
- Oyugi, J.O.; Qiu, H.; Safronetz, D. Global warming and the emergence of ancient pathogens in Canada’s arctic regions. Med. Hypotheses 2007, 68, 709. [Google Scholar] [CrossRef]
- Shi, Y.; Grogan, P.; Sun, H.; Xiong, J.; Yang, Y.; Zhou, J.; Chu, H. Multi-scale variability analysis reveals the im-portance of spatial distance in shaping Arctic soil microbial functional communities. Soil Biol. Biochem. 2015, 86, 126–134. [Google Scholar] [CrossRef]
- Wang, J.; Jia, H. Metagenome-wide association studies: Fine-mining the microbiome. Nat. Rev. Microbiol. 2016, 14, 508–522. [Google Scholar] [CrossRef]
- Wang, Y.; Han, W.; Zhu, Q.; Lin, L.; Wang, N.; Zhang, B. Effects of soluble inorganic ni-trogen salts on the diversity of rhizosphere soil bacteria in New Ohlson, Arctic. Adv. Polar Sci. 2021, 33, 395–413. [Google Scholar]
- Kuypers, M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Daims, H.; Lebedeva, E.V.; Pjevac, P.; Han, P.; Herbold, C.; Albertsen, M.; Jehmlich, N.; Palatinszky, M.; Vierheilig, J.; Bulaev, A.; et al. Complete nitrification by Nitrospira bacteria. Nature 2015, 528, 504–509. [Google Scholar] [CrossRef]
- Van Kessel, M.A.; Speth, D.R.; Albertsen, M.; Nielsen, P.H.; Op den Camp, H.J.; Kartal, B.; Jetten, M.S.; Lücker, S. Complete nitrification by a single microorganism. Nature 2015, 528, 555–559. [Google Scholar] [CrossRef]
- Sun, Y.; Yang, Q.; Bian, R.; Li, W.; Liu, Z.; Ma, Q. Analysis of bacterial diversity in the bioreactor of mineralized waste. Environ. Eng. 2016, 34, 142–146. [Google Scholar]



| Sample Code | Location Profile | Coordination | |
|---|---|---|---|
| BZS | Intertidal soil in the meltwater area near the Glacier | 78.9036° N | 12.0447° E |
| BZX | Subtidal soil in the meltwater area near the Glacier | ||
| SZS | Intertidal soil in the meltwater area near the Sand | 78.95779° N | 11.7050° E |
| SZX | Subtidal soil in the meltwater area near the Sand | ||
| TZS | Intertidal soil in the meltwater area near the Tundra | 78.9652° N | 11.6013° E |
| TZX | Subtidal soil in the meltwater area near the Tundra | ||
| WZS | Intertidal soil in the meltwater area near the Bay | 78.9664° N | 11.5792° E |
| WZX | Subtidal soil in the meltwater area near the Bay | ||
| LZS | Intertidal soil in the meltwater area near the Lake London | 78.9626° N | 12.0592° E |
| LZX | Subtidal soil in the meltwater area near the Lake London | ||
| KZS | Intertidal soil in the meltwater area near the Reservoir bay | 78.9194° N | 11.8766° E |
| KZX | Subtidal soil in the meltwater area near the Reservoir bay | ||
| RDA1 | RDA2 | r2 | Pr (> r) | ||
|---|---|---|---|---|---|
| MC | −0.825606 | −0.564247 | 0.3396 | 0.003 | ** |
| PH | 0.139400 | −0.990236 | 0.0512 | 0.427 | - |
| OrC | 0.806820 | −0.590797 | 0.0739 | 0.261 | - |
| OrN | 0.756620 | −0.653855 | 0.1074 | 0.138 | - |
| PO43−- | 0.634393 | 0.773011 | 0.0784 | 0.234 | - |
| NH4+-N | −0.761526 | −0.648134 | 0.3321 | 0.005 | ** |
| SiO32−-Si | −0.796176 | −0.605065 | 0.3571 | 0.004 | ** |
| NO3−-N | 0.087895 | 0.996130 | 0.0395 | 0.549 | - |
| NO2−-N | 0.715481 | 0.698633 | 0.0109 | 0.864 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, L.; Liu, J.; Yuan, J.; Wang, N. The Study of Soil Bacterial Diversity and the Influence of Soil Physicochemical Factors in Meltwater Region of Ny-Ålesund, Arctic. Microorganisms 2022, 10, 1913. https://doi.org/10.3390/microorganisms10101913
Wang L, Liu J, Yuan J, Wang N. The Study of Soil Bacterial Diversity and the Influence of Soil Physicochemical Factors in Meltwater Region of Ny-Ålesund, Arctic. Microorganisms. 2022; 10(10):1913. https://doi.org/10.3390/microorganisms10101913
Chicago/Turabian StyleWang, Long, Jie Liu, Jialin Yuan, and Nengfei Wang. 2022. "The Study of Soil Bacterial Diversity and the Influence of Soil Physicochemical Factors in Meltwater Region of Ny-Ålesund, Arctic" Microorganisms 10, no. 10: 1913. https://doi.org/10.3390/microorganisms10101913
APA StyleWang, L., Liu, J., Yuan, J., & Wang, N. (2022). The Study of Soil Bacterial Diversity and the Influence of Soil Physicochemical Factors in Meltwater Region of Ny-Ålesund, Arctic. Microorganisms, 10(10), 1913. https://doi.org/10.3390/microorganisms10101913




