1. Introduction
Antimicrobials are a very valuable tool—or even weapon—in human medicine, but also in the treatment of livestock, and substances overlap broadly between both fields of use. Thus, similar resistances might be selected and transferred from livestock to humans via consumption. In 2020, penicillins (278 t), tetracyclines (148 t), and sulfonamides (65 t) were used most frequently in livestock in Germany; the quantities dispensed for 3rd- and 4th-generation cephalosporins were 1.0 t and 0.3 t, respectively (
https://www.bvl.bund.de/SharedDocs/Bilder/09_Presse/01_Bilder_Pressemitteilungen/Tabelle_%20Antibiotika-Abgabemengen_2011-2020_Print.html, accessed on 19 July 2022). However, the individual quantities of active ingredients cannot be assigned to individual animal species because the majority of active ingredients are approved for use in different animal species. Similar to antibiotic use, antibiotic resistance is also monitored. Numerous monitoring programs are being performed or have been implemented in Europe, but such projects are not designed to track the origin of the resistant strains, which are mostly sampled from animal products by random inspections or from diseased humans or animals [
1]. In the last decades, a variety of studies has investigated antimicrobial resistance in primary production—mainly pigs, chickens, and cattle. While a vast amount of studies deal with the prevalence of resistant isolates within a total lot of isolates, considerably fewer studies relate the prevalence of resistant isolates to animals or primary products thereof, and even less do so for sample numbers above 200. Therefore, even if there is basically ample information on the current antimicrobial resistance situation worldwide and the problems associated with it in both human and veterinary medicine, very little is known about the actual transmission routes. From farm to fork, many sources of entry are conceivable along the entire production chain. The primary task for food control is to avoid the spread of pathogenic bacteria that are listed in (EC) No 2073/2005. However, antimicrobial resistance is not primarily a problem associated with virulent subtypes; on the contrary, antimicrobial treatment might even be contraindicated in virulent strains, such as EHEC [
2]. Anyhow, antimicrobial resistance might cause health risks independent of the virulence features of a strain, since carriers might form a reservoir for genetic transfer in the gut. It has been shown that, after oral ingestion of resistant bacteria via pork, bacteria survived the gastrointestinal passage and were detectable in the feces for up to 14 days after ingestion [
3,
4]. As early as in the 1960s, Williams Smith described the transient colonization of the human digestive tract with resistant
E. coli strains of animal origin after oral ingestion [
5]. Later studies also confirmed that the human microbiota is entered by resistant bacteria through the oral ingestion of bacteria-contaminated animal foods [
3,
6]. The reservoir function of such bacteria is the reason why, irrespective of virulence, commensal
E. coli are included in the German implementation of Directive 2003/99/EC.
No matter whether meat samples are taken at the slaughterhouse or after processing, samplings show that pork can be contaminated with phenotypically resistant
E. coli [
7,
8,
9,
10]. However, only very few tracking studies actually exist [
11], due to the complexity of studies that have to be carried out over relatively long periods of time, additionally requiring the consent of those responsible at various stages of food production (e.g., farms and slaughterhouse). In a valuable study, Burow et al. [
12] followed piglets from birth until the end of fattening, but no meat samples were included. In order to trace antimicrobial-resistant bacteria from pigs to pork along the production chain, we performed a prospective cohort study with pigs in farms and at slaughter in order to track along different production steps.
2. Materials and Methods
2.1. Study Farms
The study was conducted in six pig farms, with each farm assigned an identification code (A6, A21, E5, H18, J20, and M13). Two farms (H18 and J20) were organic pig farms, the others operated in a conventional production mode. Farms categorized as “organic” fulfilled at least the legal regulations of (EU) No 834/2007, now repealed by (EU) 2018/848. Two of the pig farms also had piglet production in addition to fattening (E5 and M13); these two farms did not purchase any fattening pigs. The other farms purchased animals from one source.
Table 1 gives an overview of the individual farms and their characteristics.
2.2. Antimicrobial Use
Antimicrobial use data were collected from the compulsory documents related to veterinary application and dispensing, as well as by involving the herd books. The documents contain information on the number and type of animals treated, type and quantity of the administered drug, diagnosis, type and duration of application, dose per animal and day, as well as waiting periods. The antimicrobial treatment was prescribed and decided by the attending veterinarian of the respective farms. The antimicrobial treatments were therefore not influenced or controlled by the authors and only recorded retrospectively.
2.3. Sampling Procedure
Pigs were sampled at three different slaughterhouses, since the organically reared pigs were slaughtered at two separate slaughterhouses. Samples at slaughter were taken at the end of the regular fattening period (100–115 days in conventional farming, 126–135 in organic farming). Samples of intestinal content and meat samples were taken during the regular slaughter process from 10 randomly selected animals (sentinel) per fattening run (n = 1–4 per farm, depending on the time of study entry). Intestines were transferred to metal surface and rectum content was transferred to sterile tubes by manual manipulation of the rectum. The cecum was ligated, resected by use of a metal scissor, and transferred to sample bags. In the lab, the cecum was opened using a scalpel and content was transferred to a sterile tube. Meats samples were taken from neck and belly with the help of a sterile punch applicator (approx. 0.5 × 0.5 cm). In addition, feces samples had been taken before slaughter, from the same sentinel animals on farm during the fattening period during spontaneous defecation. The start of fecal sampling varied by farm: In E5, sampling started at the time of weaning; in M13, sampling started when the pigs entered the early growing-finishing phase (and pen) with approx. 25 kg. In the farms that purchased pigs, sampling started at the time of stalling. The sentinel animals were sampled monthly for the entire duration of the fattening period. In addition, 5 of the 10 sentinel animals were initially randomly selected and then sampled at each antimicrobial treatment—at the beginning, during, and after the end of the treatment (day 0, 2, 4, and 10). On day 0, 4, and 10, also the farmers provided stool samples by self-sampling.
2.4. Sample Processing and Escherichia coli-Isolation and Identification
The feces samples were suspended in NaCl solution (1 g in 9 mL) and a dilution series was prepared. Punch samples of the meat were swirled with 10 mL NaCl solution for 15 min at 200 rpm, and a dilution series was prepared from the suspension obtained. From those dilution series, 0.1 mL was spatted onto Fluorocult agar (Fluorocult ECD-Agar, Merck, Darmstadt, Germany) and incubated for 24 h at 44 °C. Colony material was also streaked to Gassner agar (Gassner-Agar, Merck, Darmstadt, Germany) and ESBL agar (CHROMagar ESBL, Mast Diagnostica, Reinfeld, Germany). In order to select for diversity, isolates were described morphologically. From feces samples, four isolates where picked that differed in colony morphology. From meat samples, all colonies were picked for further investigations. All isolates were confirmed by biochemical standard tests partly included in the cultivation media (fluorescence from conversion of 4-methylumbelliferyl-b-D-glucuronide; acid production from lactose, negative oxidase test, positive indole reaction). Questionable isolates were further confirmed by means of a miniaturized identification method, using modified conventional and chromogenic substrates (BBL Crystal Enteric/Nonfermenter ID Kit, Becton Dickinson, Franklin Lakes, NJ, USA). Isolates with conspicuous resistance profiles (in particular resistance to 3rd + 4th generation cephalosporins including ESBL phenotype) were additionally identified by MALDI-TOF MS (Matrix-assisted Laser Desorption/Ionisation-Time-of-Flight Mass Spectrometry, Bruker, Billerica, MA, USA).
Isolates were cryopreserved at −80 °C.
2.5. Antimicrobial Susceptibility Testing
The testing of phenotypic antimicrobial resistance was performed by microdilution following EUCAST (
https://eucast.org/ast_of_bacteria/mic_determination/, accessed on 19 July 2022). The cryopreserved
E. coli isolates were spread out on sheep blood agar and incubated at 37 °C for 24 h. The reference strain
E. coli DSM 1103 was included as a performance control and was repeatedly tested together with each tested lot of study isolates. For testing, a bacterial suspension with a turbidity level according to McFarland 0.5 was prepared. Fifty microliters of the suspension were added to two test tubes, each containing 13 mL Müller-Hinton broth, and mixed briefly. The cell density of this suspension was approximately 5 × 10
5 cfu/mL. For testing, commercial, but client-adapted 96-well microtiter plates pre-coated with antibiotics were used. Each well of the plate was filled with 100 μL suspension with the aid of the Micronaut
® Sprint automatic dispenser. The plate was covered with foil, shaken for five minutes, and then incubated for 18–20 h at 37 °C. Turbidity was visually evaluated. Based on listed breakpoints according to EUCAST, isolates were classified into susceptible, intermediate, or resistant. For three of the investigated antimicrobials, no EUCAST breakpoint was defined for
E. coli. In those cases, epidemiological cut-off values (ECOFF; EUCAST) were used to classify the microdilution results. Breakpoints and ECOFFS are shown in
Table 2.
2.6. Detection of Resistance Genes
DNA was extracted from pure cultures using a slightly modified form of the Chelex 100 DNA extraction method (Yang et al., 2008). The total volume of the PCR reaction was 25 µL, containing 23 µL of a commercially available master mix (LightCycler 480 SYBR Green I Master, Roche, Germany) composed of 18.25 µL nuclease-free water, 2.5 µL PCR buffer, 1.5 µL MgCl
2 (25 µM), 0.5 µL dNTP mix (10 µM), and 0.25 µL Taq-Polymerase (5 U/µL), to which 0.5 µL primer FW, 0.5 µL primer RV (25 µM, each), and 1 µL of template-DNA were added. An overview of the relevant resistance genes investigated can be found in
Table 3. PCR was performed in a thermocycler (T3000, Biometra, Jena, Germany) and included the amplification conditions listed in
Table 4.
2.7. ERIC-PCR
In order to derive taxonomic relationships, an ERIC- (Enterobacterial Repetitive Intergenic Consensus) PCR was performed on
E. coli isolates from four of the six farms (E5, A21, A6 and J20). The total volume of ERIC-PCR was 25 µL per sample, using 1 µL of the template DNA, adjusted to 15.38 ng per µL. A commercial master mix (GoTaqGreen Master Mix, Promega, Madison, WI, USA), which contains two dyes, was used to perform the ERIC-PCR under the conditions listed in
Table 5. The target gene, fragment size, and annealing temperature of the primers used for ERIC-PCR can be found in
Table 6.
The evaluation of the band patterns generated in the ERIC-PCR was carried out using the software GelCompar II (Applied Maths, Sint-Martens-Latem, Belgium). Digital images of the band patterns of the agarose gels were imported into the software program, identification numbers were assigned, and finally the cluster-analysis was carried out. Gels were aligned to each other by help of a DNA ladder and one E. coli strain was run on all gels in order to assess reproducibility.
4. Discussion
Contamination rates of meat with
E. coli were between 5.6 and 11.1% at the slaughterhouses of organically produced pigs, while between 16.7% and 36.8% of the samples were positive at the slaughterhouses where conventionally produced pigs were slaughtered. That difference might be related to differences in the slaughter procedure, which, however, could not be substantiated due to the lack of detailed information. When comparing results to a study of Schwaiger et al. [
10], even higher prevalences of 72% were found.
Escherichia coli isolates at slaughter were phenotypically resistant—or had MIC-values above the ECOFF—against/for up to 19 different substances (out of 26 tested). Resistance or reduced AMS affected, at least in single cases, all substances except meropenem. Although the spectrum of tested substances was not completely comparable, the core resistance pattern of porcine
E. coli in this study—AMP-AMC-PIP-DOX—was comparable to the core pattern found by Schwaiger et al. [
10].
No obvious correlation could be detected between antimicrobial use and resistance. To give one example, beta-lactam resistance was more pronounced in A21, which did not use beta-lactams, compared to A6, which did. However, we saw a certain correlation with overall 1-year treatment frequency, which was assessed in another part of the study (data not shown). Overall, resistance was most pronounced in isolates from A21 and the fattening sector of A21 had a 1-year treatment frequency of 6.38, which was above the third quartile of 50 farms from South Germany (6.35; median 1.0). Compared to that, the fattening sector of M13 had a 1-year treatment frequency of 1.12 (0.62 in suckling piglets, 4.99 in sows), and E5 had a 1-year treatment frequency of only 0.06 in fattening pigs, but 1.69 in suckling piglets and 8.7 in sows. H18 and J20—the two organic farms—had a 1-year treatment frequency of 0.0 and 0.04, respectively. Unfortunately, no treatment frequency could be assessed for farm A6, since the farm left that part of the study prematurely. Treatment frequency was repeatedly described as an important enhancer of selection (e.g., Cobey et al., 2017 [
21]) and is the subject of continuous monitoring in German fattening pigs now (Flor et al., 2022 [
22]).
The spectrum of clinical resistances in isolates at slaughter was much broader in farm E5, compared to farm A6. Farm E5 combined several practices that are known to favor the selection of antimicrobial resistance: First, and as a routine, piglets were treated very early in life, with the very first additional feeding offered when they still fed on milk. Early exposure to antimicrobials had been shown to be linked with delayed maturation of microbiomes and lowered alpha-diversity [
23], which depicts selection. Second, a broad-spectrum antibiotic (chlortetracycline) was applied, while broad spectrum antibiotics are known to aggravate selection [
24]. Third, two antibiotics were combined when treating the piglets, which is also known to increase selection [
25], at least when done without appropriate diagnostics, and apart from antibiotics which exploit synergistic pathways like sulfonamides plus trimethoprim. Fourth, a broad variety of different antibiotics was used in the farm (14 substances from 9 classes)—a practice also proven to select AMR [
26]. Anyhow, unfavorable treatments were carried out in farm A6 as well, where pigs of one lot were treated consecutively with four different antibiotics during a total time of 23 days.
In farm E5, all fattening pigs were bred on the farm. This does not necessarily favor antimicrobial resistance—on the contrary, purchasing piglets from elsewhere is suspected to negatively affect the health state of pigs and thus to increase the need for treatment. However, it helps resistant isolates, once selected, to permanently establish, especially if antibiotic selection is continued as a prophylactic routine practice. To be clear, routine treatment is clearly discouraged by German veterinary guidelines (
https://www.bundestieraerztekammer.de/btk/downloads/antibiotika/AB_Leitlinien2015_EN.pdf; accessed on 19 July 2022). Anyhow, such routine treatment is kind of “accepted reality”—albeit with gritted teeth: Guideline 3 states that the use of antimicrobials always requires a (clinical plus minus laboratory) diagnosis, but gives further advice in case that antibiotics are used “at regular intervals for repeated or long-term use in animal groups or herds”. In that case, susceptibility testing is required. The seeming discrepancy between both passages (obligatory diagnosis/application at “regular intervals”) is best explained by cases like farm E5: Indeed, a clinical diagnosis was documented for all piglets, so this prerequisite did not hinder the farm to apply routine treatment. In such cases, the specified guideline on repeated treatment ensures susceptibility testing, at least (and might put off farmers from applying routine treatment due to diagnostic efforts). In 2018, guidelines had been made legally binding by incorporating them into national legislation (“Tierärztliche Haus-apothekenverordnung (TÄHAV)”).
One point to mention is that we used phenotypic resistance only as a marker for phenotypic similarity between isolates, not as a therapeutic forecast. Therefore, we included substances for which no clinical breakpoints exist, and our results should not be misread as clinical diagnostics. In cases where breakpoints were available (all but 3), we used EUCAST-breakpoints, which tend to be set more cautious than CLSI-breakpoints [
27] and were not specified for livestock. This approach was chosen since resistance of isolates at slaughter might, in the event of transfer, be relevant for human therapy, not for veterinary therapy.
For the same reason, we included substances that are not approved for livestock, but might be affected by co- and crossresistance, as proven before [
10].
Looking at the zoonotic relevance of our general findings, focus is surely aimed at resistance to critically important antimicrobials. Third and fourth-generation cefalosporines as well as quinolones were affected by clinical resistance. Up to 13% (in total: 6.3%) of meat samples were contaminated with
E. coli that carried
blaCTX-M. Comparing German data of 2017, the prevalence of presumptive ESBL-isolates in fattening pigs was 41.8%, while it was 4.9% in pork meat. These data are in the range of our findings for meat, and in the upper range within Northern and Western Europe [
28]. Since ESBL-
E. coli might colonize the human intestine, these bacteria might act as reservoirs or opportunistic pathogens at any time later in life, as illustrated by Hölzel et al. [
29]. However, Sharp et al. [
30] do not see major indication for meat consumption playing a role in ESBL-colonization, and vegetarians bear more risk of being colonized by antimicrobial-resistant bacteria then humans with an omnivore life style [
31]. The latter might be connected to biasing factors in the life style of vegetarians, such as having stayed abroad [
32].
As limitations of our study, it has to be mentioned that we could not synchronize the different analyses, so that the isolates that underwent phenotyping and genotyping were not analyzed by amplicon typing, and isolates that were amplicon typed were not phenotyped or genotyped, vice versa. However, when we did so in another part of the study with isolates from humans and pigs that also resembled each other in ERIC-patterns, we found that they did not resemble each other in phenotypes, genotypes, and plasmid typing (data not shown). This is no surprise, since AMR features in Enterobacteriaceae are thought to be mainly mobile [
33,
34] while isolates themselves, of course, might be clonally spread—as it is very well known for pathogenic serovars of
E. coli [
35].
Genotypic and phenotypic profiles were not congruent in most cases, but rather two different features of the same organism. One exception of that rule was doxycycline “resistance” (means: MIC above ECOFF), where the most common causative genes in porcine
E. coli—
tet(A) and
tet(B) [
36]– were included. In that case, we could prove good accordance between genotypes and phenotypes: All doxycycline resistant isolates that were genotyped had at least one of the two causative genes. When matching genotypes with genotypes in six pairings between isolates from feces and pork,
tet(A) was shared in five of the six pairings where it was present, while
tet(B) was not shared between isolates of both pairs where it had occurred in one of the isolates. Given the high prevalence of
tet(A) in porcine
E. coli—Schwaiger et al. [
36] found a prevalence of 57.7% in the same geographical region—the simultaneous occurrence of
tet(A) in two isolates has limited significance as an epidemiologic link. Of other genes,
strA-
strB occurred in six pairings, but were shared in only two. One pair shared all four resistance genes that were detected, namely
tet(A),
strA-
strB, and
sul2. However, of seven detected phenotypic markers, only three were shared within that pair.
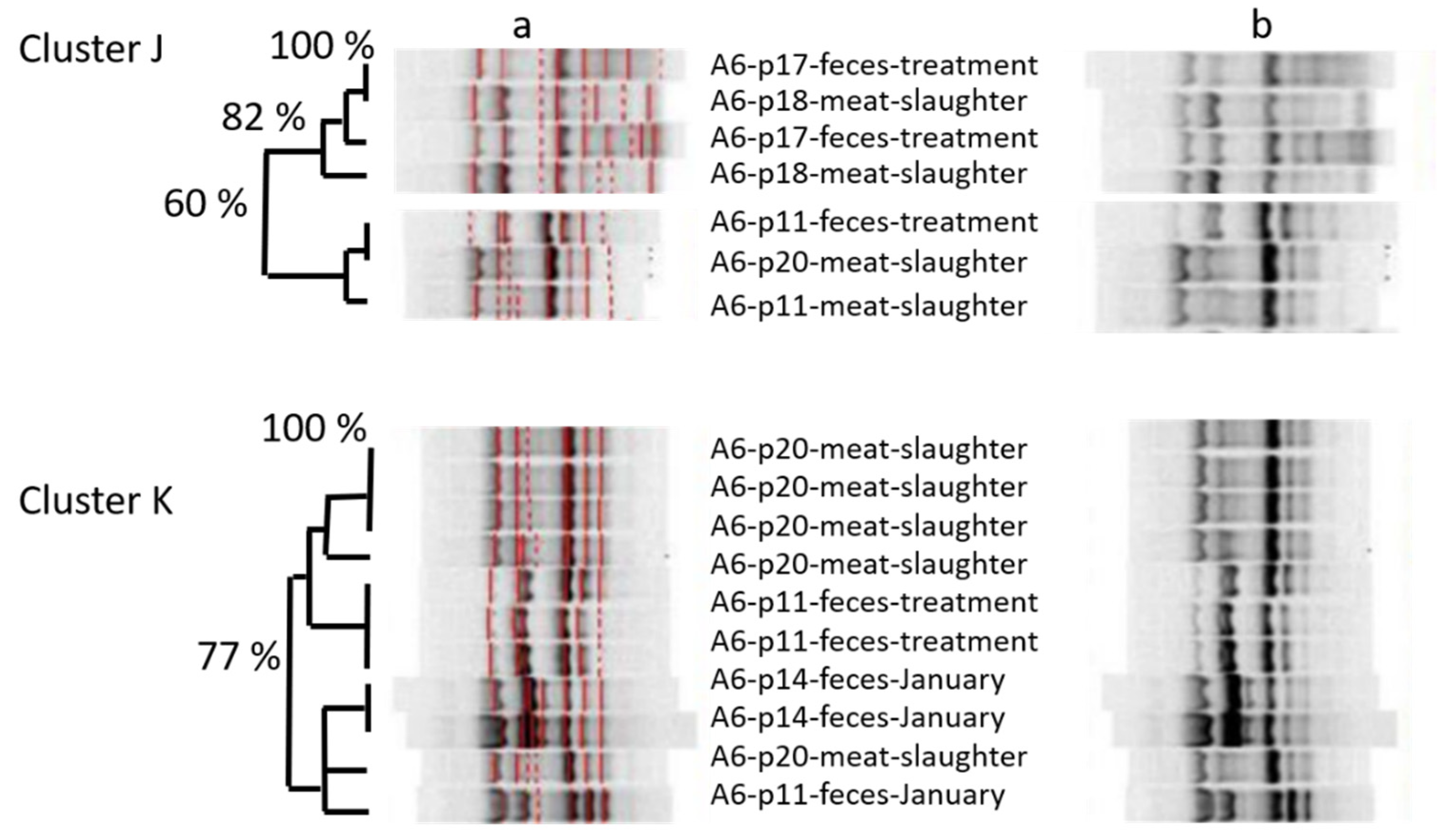
Looking at the amplicon-based cluster analysis, we saw similarities (i) between isolates of different lots at farm, (ii) between isolates from pigs on farm and at slaughter, and (iii) between isolates from pigs at slaughter and from the farmer. The GelCompare Software was allowed to include weak bands, so that the human eye would see less similarity than the software. In addition, we chose a high position tolerance for “identity” between bands, due to a low reproducibility of patterns: one strain, included as positive control in each gel, generated different band patterns with identity values as low as 63.4%, when no position tolerance was applied. However, even the visual judgment resulted in high similarity between one isolate at slaughter and two fecal isolates from the farmer, sampled months ago (
Figure 1—lower part). Since the farmer was not present at slaughter, any exchange or colonization from a third, common source must have occurred on the farm, not at slaughter. One should take into account that the discriminative power of amplicon-typing is limited (Wilson & Sharp, 2006) and no whole genome sequencing was performed yet. Anyhow, the analysis revealed plausible results by identifying clones within the four fecal isolates monthly taken from the same pig. Furthermore, MLST-analysis (
https://enterobase.warwick.ac.uk/species/ecoli/allele_st_search, accessed on 19 July 2022)—exemplarily performed in other cases where isolates form pigs and farmers resembled each other in ERIC profiles—found up to 99.9% identity of base pairs between ERIC-clones (5351 of 5356 bp), although other ERIC-clones had lower similarity in the MLST analysis (minimum 99.2% or 5407 of 5451 bp). The fact that Marshall et al. [
37] could prove the spread of labelled
E. coli from the intestine of cattle and pigs to farmers already in 1990 renders our observations plausible.
The spread of bacteria does not necessarily mean spread of antimicrobial resistance, as illustrated by the low congruence of antimicrobial resistance in feces-pork-pairs, as well as the high variability of phenotypic resistance profiles in 29
E. coli isolated from the same piece of meat. To look into those 29 isolates gives the impression of seeing recombination at work, since 14 of 16 antimicrobial resistances found in the most multiresistant isolate were also found in other isolates—in different combinations and with different frequency. No mating experiments could be performed with multiresistant strains, since no S2-facilities were available at the time and place of the study. However, we do not expect further insights from an in-vitro experiment, compared to the in-vivo indication that recombination probably had happened. All phenotypic resistances that formed part of the multiresistance profile had already been proven by others to be transferrable [
38,
39,
40,
41,
42,
43,
44,
45]. In total, 8.1% of all meat isolates were positive for
sulI plus
qacEΔ1, indicating the presence of integrons (data not shown), which confirms the reports of others [
46]. Thus, once meat is contaminated with antimicrobial-resistant pathogens, this might set humans at risk—at least the risk of introducing transferrable resistance genes into their intestines [
47,
48]. However, simple measures of kitchen hygiene, such as careful handling and cooking, might prevent that risk [
8].