There is growing interest in antibiotic resistance (AR) and multidrug resistance (MDR) in the agricultural sector, as they are public health concerns especially for both clinicians and veterinarians. The use of antibiotics for the treatment of animal infections and prophylactic use at sub-therapeutic dose to enhance animal growth could lead to MDR and transmission to humans. About 148 mg of antibiotics per kilogram of chicken is estimated to be consumed by humans, and the quantity is projected to continue in an upward direction [
27]. The application of animal waste as organic manure is an indirect environmental mechanism of AR transmission to humans as these anthropogenic contaminants could potentially enhance antibiotic resistance genes of the microorganisms in the ecosystem [
28]. Nevertheless, microorganisms such as
Pseudomonas aeruginosa,
Stenotrophomonas maltophilia, and
Enterococci possess intrinsic capability to develop antibiotic resistance without encountering any antimicrobial contaminant ([
29,
30]). Recent studies report the prevalence of MDR (resistance to three or more antibiotics) isolates of
Salmonella,
Listeria, and
Campylobacter, important zoonotic pathogens on poultry farms without historical or exogenous sources of antibiotics [
31]. However, the drivers of MDR from the environment (soil) or animals themselves (feces) are yet to be identified. We utilized an ensemble approach combining machine learning and deep learning to identify preharvest management practices that are predictive of MDR in poultry pathogens and determine the feature values that are associated with low MDR.
3.1. Data Analysis
Table 1 is an overview of the number of samples in our dataset that exhibit varying degrees of antibiotic resistance.
Only 27% of fecal samples had
Salmonella that were resistant to three or more antibiotics (MDR), while 37% of
Salmonella isolates were MDR in soil samples.
Salmonella MDR included antibiotics that inhibit protein synthesis (tetracycline, streptomycin) and antibiotics that target cell wall synthesis (ampicillin, augmentin, cefoxitin, ceftriaxone, ceftiofur) among the antibiotics tested in this study. High prevalence (frequency of PCR-based detection) of the
tetA and
aadA1 genes responsible for
E. coli resistance to protein-synthesis-targeting tetracycline and streptomycin (72 and 33% prevalence rate, respectively) have been previously reported in poultry broiler chicken [
32]. The same study found a similar pattern of resistance to protein-targeting antibiotics in layers. The horizontal transfer of resistance genes between
E. coli and other organisms such as
Salmonella,
Listeria, and
Campylobacter in the poultry production environment could contribute to the development of AR in these zoonotic poultry pathogens.
Salmonella isolates resistant to a combination of tetracyline, streptomycin, and ampicillin, as observed in this study, have been reported in human, animal, and environmental samples [
33]. Known mechanisms of resistance to these antibiotics involve bacterial plasmid and chromosomal DNA ([
34,
35]). Our results also show that 97.8% of the
Salmonella isolates with MDR are of the Kentucky serotype and are of lesser public health concerns in humans, consistent with an earlier observation [
9]. Only 1.1% of MDR
Salmonella isolates belonging to the Braenderup/Cholareasuis strains are of significant importance to human health [
36], while the remaining 1.1% represent ungrouped serotypes.
Our results show that only 5% of
Campylobacter isolates were MDR in feces and soil. This dataset is highly imbalanced from the perspective of machine learning, making it difficult to build a generalized predictive model. The MDR
Campylobacter identified in this study include
C. jejuni (43%),
C. coli (43%), and a mixed culture of
C. jejuni and
C. coli (14%). C_MDR included antibiotics that inhibit protein synthesis (azithromycin, gentamicin, clindamycin, erythromycin, florfenicol, telithromycin, tetracycline) and antibiotics that target DNA replication (ciprofloxacin and nalidixic acid). Similar to our observation, Aksomaitiene and colleagues reported
Campylobacter resistance to tetracycline and ciprofloxacin in broiler chickens, as well as in humans, wild birds, and cattle [
37]. Noreen et al. reported a high level of MDR in livestock-associated
Campylobacter isolates compared to
Campylobacter isolates from non-livestock sources such as water and wildlife [
38]. Furthermore, their work identified
Camplobacter MDR to antibiotics such as erythromycin, ciprofloxacin, nalidixic acid, tetracycline, and gentamicin, as observed in this study. Mutations in
tetO,
aphA, and
aadE genes that could alter antibiotic binding sites, the presence of super efflux pump
RE-CmeABC, and the carriage of
pTet plasmid that could confer resistance could be the mechanisms for the development of
Campylobacter MDR ([
39,
40]).
The MDRs observed in this study for
Listeria in feces and soil samples are 72% and 85%, respectively.
Listeria MDR predominantly involves antimicrobials that inhibit protein synthesis such as tetracycline, streptomycin, daptomycin, lincomycin, erythromycin, streptogramins, and tigecycline. Resistance to ciprofloxacin that inhibits DNA replication was also observed. MDR isolates of
Listeria include
L. innocua (78.3%),
L. welshimeri (14.1%),
L. monocytogenes (7.1%), and uncharacterized (0.5%). These results are consistent with the report of Okorie-Kanu and colleagues that identified
L. innocua as the predominant species in chicken [
41]. Although
L. innocua is generally known to be non-pathogenic but genetically closely related to pathogenic
L. monocytogenes, atypical hemolytic
L. innocua has been reported to be virulent, albeit to a lower extent compared to
L. monocytogenes [
42]. A recent report indicates that
L. innocua is capable of causing disease in farm animals [
43]. Similar to the observation in this study,
Listeria MDR to ciprofloxacin, tetracycline, and erythromycin has been previously reported in environmental samples [
44]. Similar to
Salmonella and
Campylobacter MDR, mechanisms of
Listeria MDR could involve both chromosomal and plasmid DNA [
45].
MDR rates reported here corroborate the reported rates of 36.0%, 1.4%, and 63.9% in
Salmonella,
Campylobacter, and
Listeria, respectively, in a survey of six pastured poultry farms [
31].
3.2. Predictive Analysis
Our study examined preharvest feces and soil as separate models and a common model (combined soil and feces) for MDR classification in
Salmonella,
Campylobacter, and
Listeria. These models aim to estimate the likelihood of pastured poultry farms developing MDR as a function of the farm management practices. Prior to building a prediction model based on the data, we standardize/normalize and oversample the data as described in
Section 2.4.
Standardization: The goal of normalization is to convert numeric values in a dataset to a standardized scale while maintaining the differences in range. We compared the classification performance of four different normalization methods (unit normalization, robust-scale standardization, quantile transformation, and standard scale normalization) with our data. Our preliminary analysis showed that quantile transformations, unit normalization, and robust-scale normalization were effective in the classification of MDR in
Salmonella,
Campylobacter, and
Listeria, respectively.
(a) Unit Normalization: This method normalizes every sample, shrinking/stretching the input feature vector (x) to a unit sphere. This ensures that the vector scales to the unit norm without regard to the distribution of the samples (Equation (
1)):
(b) Robust-Scale Standardization: This scaler centers and scales each feature independently using the quantile range (IQR: Interquartile Range) to reduce the influence of outliers in the feature set. Instead of considering the mean to standardize the feature, the method uses the median that is less significant to the outliers in scaling.
(c) Quantile Transformation: A quantile function provides an approximation of the quantile positions of actual values by inversely calculating the cumulative distribution function. In quantile transformations [
46], imbalanced distributions of data with outliers are converted to uniform distributions. This non-linear transformation smooths out the relation between observations by removing the linear correlation between the input variables. It is a popular and effective way to improve prediction with complex and noisy inputs.
(d) Standard-Scale Normalization: Standardization is beneficial when the distribution of feature values follows a Gaussian distribution. It is a method for transforming feature values by subtracting from the mean and dividing by the standard deviation. Alternatively, this process is referred to as z-score standardization.
Oversampling: The percentage of negative samples outweighs the percentage of positive samples in our data. Oversampling is a technique often used to balance such skewed distributions. It maintains class balance by adding new points to the minority class rather than removing them from the majority class. We tested two popular oversampling strategies to balance the distribution during data processing. In our preliminary evaluations, random sampling proved effective for
Salmonella MDR classification, while SMOTE sampling was effective for prediction of MDR in
Campylobacter and
Listeria.
(a) Random Oversampling: In random oversampling [
25], an increase in sample size is achieved by selecting minority class examples in random order and including them in the training set. This sampling technique iterates until a majority sample equals a minority sample.
(b) Synthetic Minority Oversampling Technique: SMOTE [
47] is based on the selection of a random sample from the minority class and one of its nearest neighbors, followed by the generation of a new synthetic sample within that range. Oversampling uses the nearest neighbor method in place of adding random duplicate samples to the minority class.
Deep Neural Network Learning We compared the performance of three different deep learning architectures to find an efficient method of detecting MDR for
Salmonella,
Campylobacter, and
Listeria. In preliminary analyses, a generative adversarial network (GAN) proved more effective than Auto-Encoder for
Listeria MDR, while a multi-layer perceptron (MLP) provided the best results for
Campylobacter MDR. Additionally,
Salmonella MDR classification was best performed with the Auto-Encoder design.
(a) Multi-Layer Perceptron: An MLP [
48] is a type of feed-forward artificial neural network that can distinguish data that cannot be linearly separated. These multi-layered networks consist of hidden nodes with a non-linear activation function that are connected with specific weights to the next layer of nodes. At the learning stage, connection weights are adjusted based on the amount of error in the output using a backpropagation function.
(b) Generative Adversarial Network: A GAN [
49] is a deep learning method for generating models from data using supervised learning techniques. Generative modeling involves discovering and understanding regularities and patterns in data. Rather than treating the problem as an unsupervised problem, GAN treats it as a supervised problem with two submodels: a generator model and a discriminator model. The generator model attempts to generate new samples from the negatives, while the discriminator model tries to determine what is positive and what is negative. Using backpropagation, we train the generator and discriminator models together.
(c) Auto-Encoder: Autoencoder [
50] is a stacked neural network layer system comprising an encoder layer, a latent or representative layer, a decoder layer, and an output layer. The latent layer will embed data without labels in a meaningful manner, and the output layer will attempt to recreate the original input. With the backpropagation algorithm, the networks are learned by minimizing the reconstruction error, which is the difference between the original and the reconstructed inputs.
For MDR classification using the above models, the prediction the confusion matrix, which is used to compare the model performance, is shown in
Table 2. The ground truth has been presented as the actual value and the model’s prediction has been presented as the predicted value in this table. When the actual value and predicted value are both positive (+), this is known as a true-positive prediction in the confusion matrix (also known as the error matrix). False-positive predictions occur when the actual value is negative (−) and the predicted value is positive (+). A true-negative prediction is one in which both the actual and the predicted results are negative (−), while a false-negative prediction is one in which the actual is positive (+) and the prediction is negative (−). Precision is the measure of the proportion of correct positive predictions among the positive predictions. Similarly, recall measures the proportion of actual positives identified. Specificity refers to the number of correctly identified negative samples. Furthermore, the F1-Score is the harmonic mean of precision and recall. The purpose of these metrics is to evaluate the predictive ability of a model.
In order to avoid overfitting in our models, the scores are averaged from four-fold stratified cross-validation. The imbalanced dataset could be responsible for the high number of false negatives shown in the table for
Listeria MDR (
Table 2). Overall, our learning models are able to predict MDR with a greater than 86% F1-Score confidence. Considering the imbalance in the dataset, the prediction scores are reasonable (
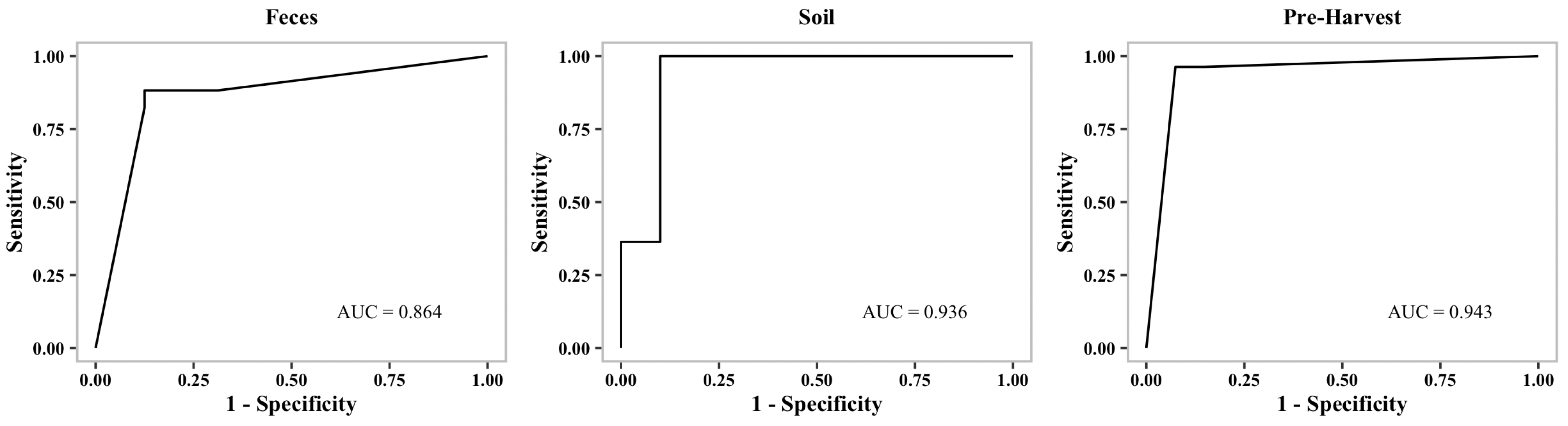
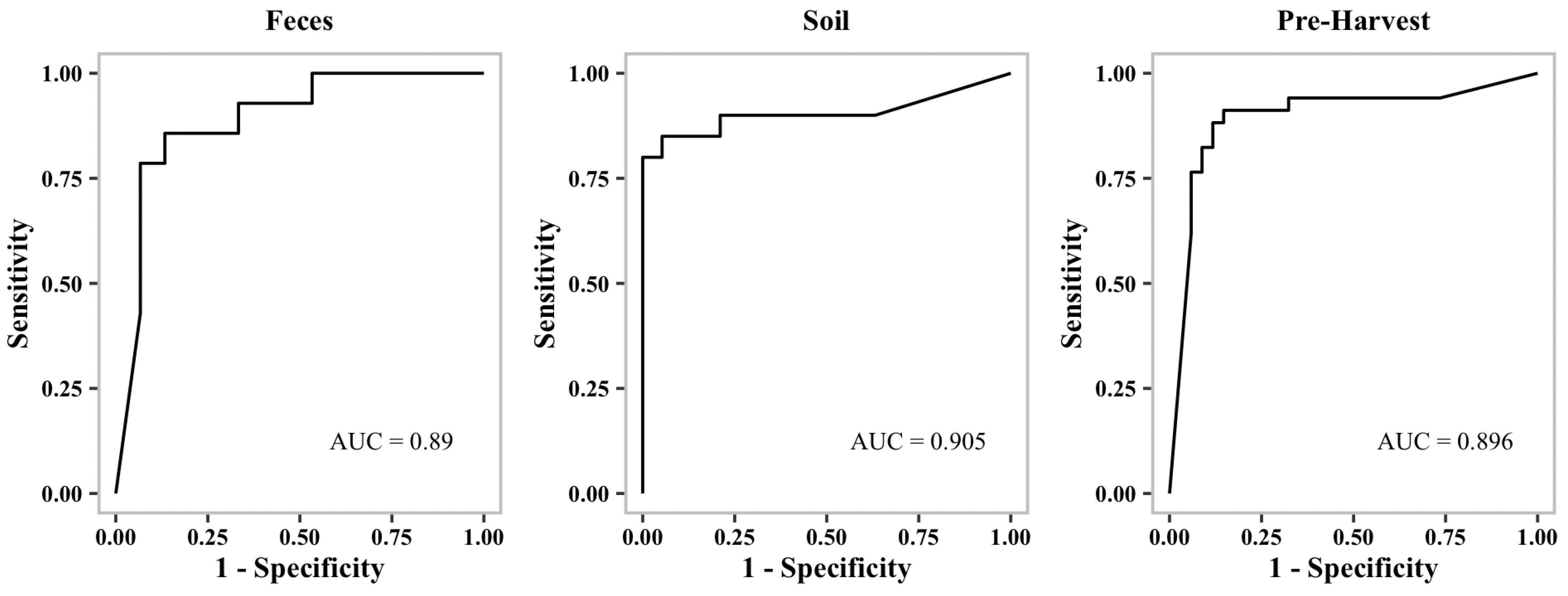
Table 2). Additionally, we generated receiver operating characteristic curves (ROC curves) to show the performance of our binary classification models at different thresholds.
Figure 1,
Figure 2 and
Figure 3 show representative ROC curves of MDR models. AUC-ROC curves are generally used to measure the performance of classification problems at different threshold levels. It depicts the trade-off between the True-Positive Rate (sensitivity) and the False-Positive Rate (1—specificity). ROC represents a probability curve, while AUC represents the measure of separation. It is indicative of better performance when classifiers provide curves that are close to the top-left corner. According to the ROC plots, our MDR prediction models are able to distinguish the classes fairly well. For all MDR models, the AUC provides a distinguishing metric that is higher than 86%. It is an indication of the effectiveness of our classification models.
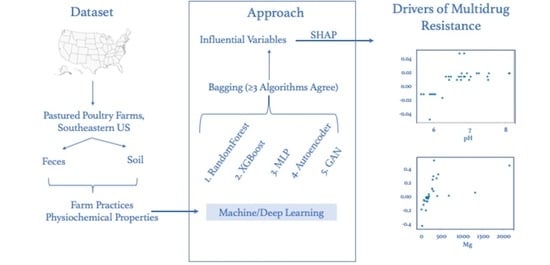
3.3. Critical Farm Feature Prediction
Our ensemble approach for selecting critical features utilized two traditional machine learning (RandomForest (RT) and eXtreme Gradient Boosting (XG)) and three deep learning (Multi-layer Perceptron (MLP), Generative Adversarial Network (GAN), and Auto-Encoder (ENC)) methods (see
Section 2.5). In the RandomForest machine learning algorithm [
51], individual decision trees that work together make up an ensemble of powerful algorithms. Using random samples from the dataset with replacement, random forest constructs several decision trees and predicts the outcome based on the majority vote. XGBoost [
52] is another powerful decision-tree-based ensemble technique that incrementally improves the performance by adding new models in sequence to fix previous models’ errors. The gradient descent optimization algorithm minimizes the weak prediction loss of current models when adding new models.
The most influential variables agreed by at least three machine learning models and their rankings in individual algorithm predictions are enumerated in the following
Table 3,
Table 4 and
Table 5. In machine learning, the approach determines the performance of the algorithm. There is no single model that is suitable for all applications. Thus, different models produce different results. To ensure reliability, majority voting is used in this study.
While Mg, P, and flock size appear to affect MDR in all pathogens,
Salmonella and
Campylobacter MDR is further affected by EC, Mn, flock age, and
Listeria, and
Campylobacter MDR appears to be affected by K, C:N, and Cu.
Salmonella MDR alone is affected by pH, Na, Ca, water source, and housing type.
Campylobacter MDR alone is affected by Zn, egg source, and brood feed and
Listeria MDR by Cr and years of farming (
Table 3,
Table 4 and
Table 5). However, as shown in
Table 1 and reported earlier, the prevalence of
Campylobacter MDR is negligible in the pastured poultry farms included in this study. Therefore, the focus will be on
Salmonella and
Listeria MDR in this manuscript.
3.4. Feature Values Associated with Low MDR Incidence
As described in
Section 2.5, we use SHAP (SHapley Additive exPlanations) [
26] to compute the effect of each feature on the model output. A model’s output with and without a specific feature is compared to determine its relative importance. When SHAP values are positive, they indicate greater importance than when SHAP values are negative. Feature values with negative SHAP values are recommended to lower the presence of multidrug resistance. We present our analysis of the most influential features in
Table 6 and
Table 7 along with value recommendations for reducing MDR.
SHAP dependency plots show the potential internal factors from feces that drive
Salmonella MDR, such as EC and pH, while the external factors determined from the soil include P, Mn, and Na (
Table 7). Only Mg appears to be critical for
Salmonella MDR in both feces and soil. In
Listeria, P, Cu, and Cr are correlated to MDR in feces while Mg, C:N, and years of farming are identified to be important for the MDR in soil (
Table 6). Flock size and K appear to be critical for
Listeria MDR in both soil and feces models. Although the exact mechanisms by which these farm management practices and physicochemical properties impact MDR are not fully understood, based on the current literature, we provide insights into the impact of variables on
Listeria and
Salmonella MDR.
3.4.1. Drivers of Listeria MDR in Pastured Poultry
Possible mechanisms by which preharvest variables, specifically physicochemical properties of soil, contribute to the development of MDR are discussed below.
Magnesium (Mg): The Mg ion provides a strong cohesive force that strengthens bacterial ribosomes, the protein synthesis machinery. Its presence provides stability for the cell and counteracts the action of ribosome targeting antibiotics. Diminished levels of Mg have been shown to promote the activities of antibiotics and hamper the protein synthesis in bacteria [
53]. The presence of Mg has also been reported to inhibit the transcription of genes involved in biofilm formation and promote the penetration of antibiotics [
54]. In addition, Mg itself has been shown to have antimicrobial properties [
55]. While ([
56]) reported the Mg level of conventional poultry soil to be between 285 and 463 ppm, our analysis suggests that the optimum level of Mg to prevent MDR is <300 ppm. Pastured poultry farmers could utilize gypsum, a calcium sulphate salt that could reduce soil Mg by displacing the Mg within the soil with calcium.
Phosphorus (P): The buffering effect of phosphate formed from phosphorus in the bacterial culture medium has been reported to both enhance and diminish antimicrobial activities depending on the antimicrobial agents. Phosphate promotes the resistance of
S. lactis to streptomycin but increases its sensitivity to tetracycline [
57]. Moreover, the addition of 30 to 300 ug/L phosphorus has been shown to significantly increase biofilm formation in drinking water supply [
58], which could potentially increase antibiotic resistance. Reducing the phosphorus level to below 5000 ppm as suggested by this study could potentially reduce MDR.
Potassium (K): Acesulfame potassium is an artificial sweetener found in various consumables and products such as soft drinks, jellies, beverages, and also poultry feed. The recent work of [
59] shows that with increasing concentration of acesulfame potassium, and invariably potassium, the growth of bacteria with antibiotic resistance genes (ARGs) is inhibited. Contrarily, [
60] show that uptake of K is essential for growth and antibiotic resistance in
Staphylococcus aureus. While potassium levels in the conventional poultry litter and soil are reported to be between 3000 and 13,000 ppm, our models indicate that potassium levels between 7000 and 12,000 ppm are optimal to reduce the incidence of
Listeria MDR. This supports the observation of [
59] on the detrimental effects of potassium on antibiotic resistance bacteria.
Carbon–Nitrogen Ratio (C:N): Composting is one important way to remove undesirable antibiotics that are not metabolized by animals and that enter the environment through excretion. The C:N levels positively correlate and are indicative of the presence of ARGs in the environment [
61]. Our predictive models recommend a C:N higher than 15 to mitigate MDR in
Listeria. However, the recent work of [
62] shows that a C:N of 26 is sufficient to remove ARGs from compost, and values higher than this may not be effective. Therefore, C:N values between 15 and 26 ppm are recommended to reduce MDR.
Copper (Cu): Based on Cu exposure studies with 96 microorganism isolates, Cu led to increased resistance to different clinically important antibiotics [
63]. However, a comprehensive review on the effect of the antibiotic-binding capability of Cu indicates that it could both enhance or diminish antibiotic resistance [
64]. This is not surprising, as Cu is an important cofactor for many enzymes required by bacteria. However, at elevated levels, it becomes toxic and acts as an antimicrobial and invokes adaptive response in the form of resistance from the organism [
65]. Our prediction algorithms recommend Cu levels greater than 18 ppm to combat MDR.
Chromium (Cr): Investigation into the role of Cr in
Staphylococcus aureus and
Escherichia coli has established that it acts as an antibiotic and works synergistically with conventional antimicrobial agents to induce oxidative stress in the organisms [
66]. Nevertheless, bacteria such as
Pseudomonas aeruginosa have developed effective ways of reducing and ejecting chromium from the cell, thereby making it less potent. Increasing the level of chromium to > 3 ppm as predicted by our models could help prevent MDR in
Listeria.
3.4.2. Drivers of Salmonella MDR in Pastured Poultry
Magnesium (Mg): Similar to the observation with
Listeria, Mg appears to be an important driver of MDR in
Salmonella. Mg is important for stabilizing protein synthesis machinery. It is interesting to note that both
Salmonella and
Listeria exhibit MDR to classes of antimicrobials that target protein synthesis such as aminoglycosides, macrolides, glycylcycline, and ketolides. While the median range in conventional poultry for Mg is around 374 ppm, our prediction models recommend Mg levels lower than 300 ppm to destabilize bacterial ribosomes and increase the pathogen sensitivity to the antibiotics. Furthermore, the role of Mg in modulating nitrosative stress has been established [
67]. Reduction in the Mg levels proposed in this study may lower
Salmonella viability and increase its susceptibility to antimicrobial activity.
Phosphorus (P): P is capable of modulating buffering capacity of the pathogen environment as well as the capability to form biofilms, thereby potentially altering the sensitivity to antibiotics. Therefore, reducing access of
Salmonella to phosphorus could reduce the MDR. Our model suggests P
ppm in the soil could help reduce
Salmonella MDR. In an in vivo study, dietary and systemic increases in the level of phosphorus have been reported to lower pig mortality during
Salmonella infection [
68], possibly due to phosphorus-mediated stimulation of leukocyte production and defense mechanisms against the pathogen. Therefore, while P restriction in the poultry soil where
Salmonella is found may be a good practice, supplementation of the poultry brood feed with P may enhance immune response and have a synergistic effect in further lowering presence and MDR of
Salmonella.
Electrical Conductivity (EC): Treatment of MDR methicillin-resistant
Staphylococcus aureus (MDR-MRSA) with chlorhexidine acetate nanoemulsion has been shown to have both in vivo and in vitro efficacy against the pathogen that correlated with an increase in the electrical conductivity [
69]. ECs that range from 0.97 to 10.07 S/m, i.e., 970 mS/m to 10,070 mS/m, in graphene oxide have been reported to promote wound healing against MDR-MRSA. While EC levels detected in the samples in this study are below 1000
S/cm, our models suggest that soil EC of 2000
S/cm (200 mS/m) would be sufficient to reduce MDR incidence in
Salmonella.
pH:Salmonella is equipped with mechanisms to survive a wide range of pH between 4.4 and 9.0 with an optimum pH of around 7.0 [
70]. Food safety regulation indicates acidic pH of 4.2 or lower is effective against
Salmonella [
71]. The recent work of [
72] also indicates that a pH between 4.0 and 6.0 is sufficient to inhibit biofilm formation in
Salmonella. The formation of biofilm is known to protect
Salmonella from both in vitro and in vivo actions of antibiotics [
73]. Our predictive model for soil suggests that a pH 6.5 could reduce MDR, possibly by inhibiting biofilm formation.
Manganese (Mn): Mn complex with antibiotic colistin has been reported to be effective against poultry avian pathogenic
Escherichia coli, known for being highly antibiotic resistant. Mn in the metal complex form alone ([Mn(CO)
)]Br) has antimicrobial activity, and synergistic combination with colistin further increases the killing efficiency of Mn [
74]. The recommendation here to increase the Mn levels up to 70 ppm or above in the soil compared to 5 to 7 ppm found in the conventional poultry soil [
56] could increase its availability and ultimately its antibacterial activity.
Sodium (Na): The presence of Na in the form of sodium chloride salt has been established to both increase the thermal and antibiotic resistance in multiple strains of
Salmonella [
75]. This was speculated to be a result of an increase in osmotic stress. Our predictive models suggest that decreasing the Na content of the soil to below 50 ppm could mitigate
Salmonella MDR.
Factors such as water source, housing type, flock size, and age are identified as variables that could contribute to Salmonella MDR. However, the observational study described here does not account for factors that may influence such dynamic management practices. Future experiments that control for these changing variables are warranted.