Strategies for the Identification and Assessment of Bacterial Strains with Specific Probiotic Traits
Abstract
1. Introduction
2. Isolation and Identification of Potential Probiotic Strains: Traditional and New Approaches
2.1. Repetitive Element Palindromic–Polymerase Chain Reaction (REP)
2.2. Random Amplified Polymorphic DNA (RAPD)
2.3. Omics Approaches for Screening Potentially Probiotic Candidate Strains
2.4. Whole-Genome Shotgun Metagenomic Sequencing (WGS)
2.5. Metabolomics
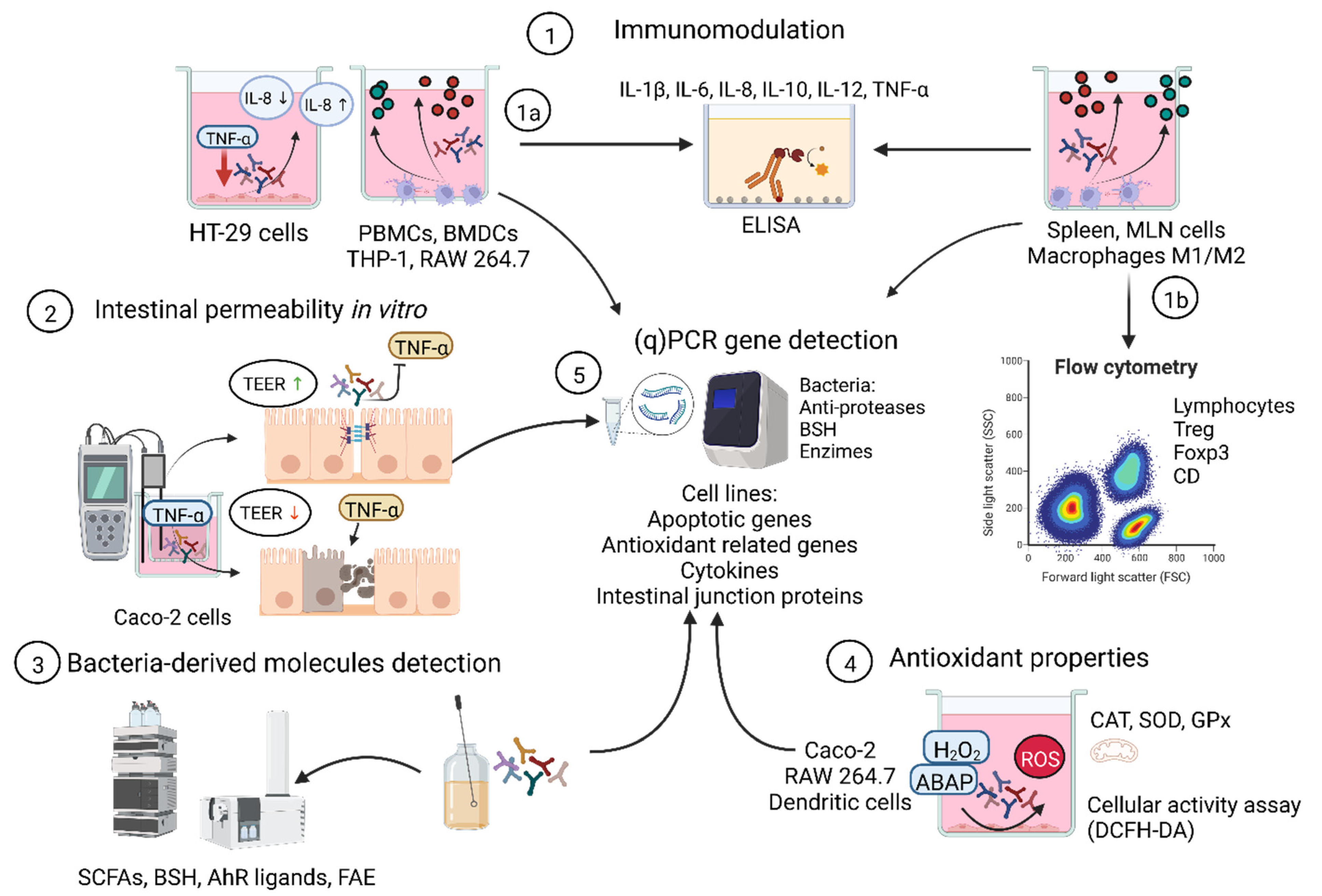
3. Target Screening to Select Bacterial Strains with Probiotic Properties
3.1. Inflammatory Bowel Diseases (IBD) and Irritable Bowel Syndrome (IBS)
3.2. Food Allergies (FA)
3.3. Metabolic Syndrome (MBS)
4. Discussion and Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ozen, M.; Dinleyici, E.C. The history of probiotics: The untold story. Benef. Microbes 2015, 6, 159–165. [Google Scholar] [CrossRef] [PubMed]
- Shoaie, S.; Karlsson, F.; Mardinoglu, A.; Nookaew, I.; Bordel, S.; Nielsen, J. Understanding the interactions between bacteria in the human gut through metabolic modeling. Sci. Rep. 2013, 3, 2532. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef]
- de Simone, C. The Unregulated Probiotic Market. Clin. Gastroenterol. Hepatol. 2019, 17, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Caselli, M.; Cassol, F.; Calò, G.; Holton, J.; Zuliani, G.; Gasbarrini, A. Actual concept of “probiotics”: Is it more functional to science or business? World J. Gastroenterol. 2013, 19, 1527–1540. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-A.; Bayo, J.; Cha, J.; Choi, Y.J.; Jung, M.Y.; Kim, D.-H.; Kim, Y. Investigating the probiotic characteristics of four microbial strains with potential application in feed industry. PLoS ONE 2019, 14, e0218922. [Google Scholar] [CrossRef] [PubMed]
- Konings, W.N.; Kok, J.; Kuipers, O.P.; Poolman, B. Lactic acid bacteria: The bugs of the new millennium. Curr. Opin. Microbiol. 2000, 3, 276–282. [Google Scholar] [CrossRef][Green Version]
- Tabssum, F.; Ahmad, Q.U.; Qazi, J.I. DNA sequenced based bacterial taxonomy should entail decisive phenotypic remarks: Towards a balanced approach. J. Basic Microbiol. 2018, 58, 918–927. [Google Scholar] [CrossRef]
- Gevers, D.; Huys, G.; Swings, J. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 2001, 205, 31–36. [Google Scholar] [CrossRef]
- Van Hoorde, K.; Vandamme, P.; Huys, G. Molecular identification and typing of lactic acid bacteria associated with the production of two artisanal raw milk cheeses. Dairy Sci. Technol. 2008, 88, 445–455. [Google Scholar] [CrossRef][Green Version]
- Williams, J.G.K.; Kubelik, A.R.; Livak, K.J.; Rafalski, J.A.; Tingey, S.V. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 1990, 18, 6531–6535. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Cabello, A.; Calero-Delgado, B.; Rodríguez-Gómez, F.; Garrido-Fernández, A.; Jiménez-Díaz, R.; Arroyo-López, F.N. Biodiversity and multifunctional features of lactic acid bacteria isolated from table olive biofilms. Front. Microbiol. 2019, 10, 836. [Google Scholar] [CrossRef] [PubMed]
- Benítez-Cabello, A.; Calero-Delgado, B.; Rodríguez-Gómez, F.; Bautista-Gallego, J.; Garrido-Fernández, A.; Jiménez-Díaz, R.; Arroyo-López, F.N. The use of multifunctional yeast-lactobacilli starter cultures improves fermentation performance of Spanish-style green table olives. Food Microbiol. 2020, 91, 103497. [Google Scholar] [CrossRef] [PubMed]
- Nami, Y.; Haghshenas, B.; Yari Khosroushahi, A. Molecular Identification and Probiotic Potential Characterization of Lactic Acid Bacteria Isolated from Human Vaginal Microbiota. Adv. Pharm. Bull. 2018, 8, 683–695. [Google Scholar] [CrossRef] [PubMed]
- Guantario, B.; Zinno, P.; Schifano, E.; Roselli, M.; Perozzi, G.; Palleschi, C.; Uccelletti, D.; Devirgiliis, C. In vitro and in vivo selection of potentially probiotic lactobacilli from Nocellara del Belice table olives. Front. Microbiol. 2018, 9, 595. [Google Scholar] [CrossRef]
- Guarcello, R.; De Angelis, M.; Settanni, L.; Formiglio, S.; Gaglio, R.; Minervini, F.; Moschetti, G.; Gobbetti, M. Selection of amine-oxidizing dairy lactic acid bacteria and identification of the enzyme and gene involved in the decrease of biogenic amines. Appl. Environ. Microbiol. 2016, 82, 6870–6880. [Google Scholar] [CrossRef]
- Zheng, J.; Wittouck, S.; Salvetti, E.; Franz, C.M.A.P.; Harris, H.; Mattarelli, P.; O’Toole, P.W.; Pot, B.; Vandamme, P.; Walter, J. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020, 70, 2782–2858. [Google Scholar] [CrossRef]
- O’Connell, T.M. The application of metabolomics to probiotic and prebiotic interventions in human clinical studies. Metabolites 2020, 10, 120. [Google Scholar] [CrossRef]
- Klindworth, A.; Pruesse, E.; Schweer, T.; Peplies, J.; Quast, C.; Horn, M.; Glöckner, F.O. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013, 41, e1. [Google Scholar] [CrossRef]
- Gardes, M.; Bruns, T.D. ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 1993, 2, 113–118. [Google Scholar] [CrossRef]
- Toju, H.; Tanabe, A.S.; Yamamoto, S.; Sato, H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS ONE 2012, 7, e40863. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.-H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE database for molecular identification of fungi: Handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 2018, 47, D259–D264. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Bagchi, S.; Chaterji, S.; Gerlach, W.; Grama, A.; Harrison, T.; Paczian, T.; Trimble, W.L.; Wilke, A. MG-RAST version 4—lessons learned from a decade of low-budget ultra-high-throughput metagenome analysis. Brief. Bioinform. 2019, 20, 1151–1159. [Google Scholar] [CrossRef]
- Team, R.C. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.r-project.org/index.html (accessed on 8 June 2022).
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome helper: A custom and streamlined workflow for microbiome research. MSystems 2017, 2, 127. [Google Scholar] [CrossRef]
- Kim, C.-S.; Cha, L.; Sim, M.; Jung, S.; Chun, W.Y.; Baik, H.W.; Shin, D.-M. Probiotic supplementation improves cognitive function and mood with changes in gut microbiota in community-dwelling older adults: A randomized, double-blind, placebo-controlled, multicenter trial. J. Gerontol. Ser. A 2021, 76, 32–40. [Google Scholar] [CrossRef]
- Xie, H.; Guo, R.; Zhong, H.; Feng, Q.; Lan, Z.; Qin, B.; Ward, K.J.; Jackson, M.A.; Xia, Y.; Chen, X. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst. 2016, 3, 572–584.e3. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [PubMed]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef]
- Seol, D.; Jhang, S.Y.; Kim, H.; Kim, S.-Y.; Kwak, H.-S.; Kim, S.H.; Lee, W.; Park, S.; Kim, H.; Cho, S. Accurate and strict identification of probiotic species based on coverage of whole-metagenome shotgun sequencing data. Front. Microbiol. 2019, 10, 1683. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Zhao, F.; Hou, Q.; Huang, W.; Liu, Y.; Zhang, H.; Sun, Z. Metagenomic analysis revealed beneficial effects of probiotics in improving the composition and function of the gut microbiota in dogs with diarrhoea. Food Funct. 2019, 10, 2618–2629. [Google Scholar] [CrossRef]
- Chen, H.; Xia, Y.; Zhu, S.; Yang, J.; Yao, J.; Di, J.; Liang, Y.; Gao, R.; Wu, W.; Yang, Y. Lactobacillus plantarum LP-Onlly alters the gut flora and attenuates colitis by inducing microbiome alteration in interleukin-10 knockout mice. Mol. Med. Rep. 2017, 16, 5979–5985. [Google Scholar] [CrossRef]
- Jangid, A.; Fukuda, S.; Suzuki, Y.; Taylor, T.D.; Ohno, H.; Prakash, T. Shotgun metagenomic sequencing revealed the prebiotic potential of a grain-based diet in mice. Sci. Rep. 2022, 12, 6748. [Google Scholar] [CrossRef] [PubMed]
- Deleu, S.; Machiels, K.; Raes, J.; Verbeke, K.; Vermeire, S. Short chain fatty acids and its producing organisms: An overlooked therapy for IBD? EBioMedicine 2021, 66, 103293. [Google Scholar] [CrossRef] [PubMed]
- Markowiak-Kopeć, P.; Śliżewska, K. The effect of probiotics on the production of short-chain fatty acids by human intestinal microbiome. Nutrients 2020, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- Moya-Perez, A.; Perez-Villalba, A.; Benitez-Paez, A.; Campillo, I.; Sanz, Y. Bifidobacterium CECT 7765 modulates early stress-induced immune, neuroendocrine and behavioral alterations in mice. Brain Behav. Immun. 2017, 65, 43–56. [Google Scholar] [CrossRef]
- Calderón-Pérez, L.; Gosalbes, M.J.; Yuste, S.; Valls, R.M.; Pedret, A.; Llauradó, E.; Jimenez-Hernandez, N.; Artacho, A.; Pla-Pagà, L.; Companys, J. Gut metagenomic and short chain fatty acids signature in hypertension: A cross-sectional study. Sci. Rep. 2020, 10, 6436. [Google Scholar] [CrossRef]
- Bersanelli, M.; Mosca, E.; Remondini, D.; Giampieri, E.; Sala, C.; Castellani, G.; Milanesi, L. Methods for the integration of multi-omics data: Mathematical aspects. BMC Bioinform. 2016, 17, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, J.A.; Villumsen, K.R.; Ernst, M.; Hansen, M.; Forberg, T.; Gopalakrishnan, S.; Gilbert, M.T.P.; Bojesen, A.M.; Kristiansen, K.; Limborg, M.T. A multi-omics approach unravels metagenomic and metabolic alterations of a probiotic and synbiotic additive in rainbow trout (Oncorhynchus mykiss). Microbiome 2022, 10, 21. [Google Scholar] [CrossRef]
- Kiousi, D.E.; Rathosi, M.; Tsifintaris, M.; Chondrou, P.; Galanis, A. Pro-biomics: Omics technologies to unravel the role of probiotics in health and disease. Adv. Nutr. 2021, 12, 1802–1820. [Google Scholar] [CrossRef] [PubMed]
- Nishida, A.; Inoue, R.; Inatomi, O.; Bamba, S.; Naito, Y.; Andoh, A. Gut microbiota in the pathogenesis of inflammatory bowel disease. Clin. J. Gastroenterol. 2018, 11, 1–10. [Google Scholar] [CrossRef]
- Lacy, B.E.; Pimentel, M.; Brenner, D.M.; Chey, W.D.; Keefer, L.A.; Long, M.D.; Moshiree, B. ACG Clinical Guideline: Management of Irritable Bowel Syndrome. Am. J. Gastroenterol. 2021, 116, 17–44. [Google Scholar] [CrossRef]
- Salinas, E.; Reyes-Pavón, D.; Cortes-Perez, N.G.; Torres-Maravilla, E.; Bitzer-Quintero, O.K.; Langella, P.; Bermúdez-Humarán, L.G. Bioactive Compounds in Food as a Current Therapeutic Approach to Maintain a Healthy Intestinal Epithelium. Microorganisms 2021, 9, 1634. [Google Scholar] [CrossRef]
- Vanderpool, C.; Yan, F.; Polk, B.D. Mechanisms of probiotic action: Implications for therapeutic applications in inflammatory bowel diseases. Inflamm. Bowel Dis. 2008, 14, 1585–1596. [Google Scholar] [CrossRef] [PubMed]
- Vich Vila, A.; Imhann, F.; Collij, V.; Jankipersadsing, S.A.; Gurry, T.; Mujagic, Z.; Kurilshikov, A.; Bonder, M.J.; Jiang, X.; Tigchelaar, E.F.; et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci. Transl. Med. 2018, 10, eaap8914. [Google Scholar] [CrossRef]
- Deng, Z.; Luo, X.M.; Liu, J.; Wang, H. Quorum Sensing, Biofilm, and Intestinal Mucosal Barrier: Involvement the Role of Probiotic. Front. Cell Infect. Microbiol. 2020, 10, 538077. [Google Scholar] [CrossRef]
- Chamignon, C.; Guéneau, V.; Medina, S.; Deschamps, J.; Gil-Izquierdo, A.; Briandet, R.; Mousset, P.Y.; Langella, P.; Lafay, S.; Bermúdez-Humarán, L.G. Evaluation of the Probiotic Properties and the Capacity to Form Biofilms of Various Lactobacillus Strains. Microorganisms 2020, 8, 1053. [Google Scholar] [CrossRef] [PubMed]
- Cuffaro, B.; Assohoun, A.L.W.; Boutillier, D.; Súkeníková, L.; Desramaut, J.; Boudebbouze, S.; Salomé-Desnoulez, S.; Hrdý, J.; Waligora-Dupriet, A.-J.; Maguin, E.; et al. In Vitro Characterization of Gut Microbiota-Derived Commensal Strains: Selection of Parabacteroides distasonis Strains Alleviating TNBS-Induced Colitis in Mice. Cells 2020, 9, 2104. [Google Scholar] [CrossRef]
- Gonsalkorale, W.M.; Perrey, C.; Pravica, V.; Whorwell, P.J.; Hutchinson, I.V. Interleukin 10 genotypes in irritable bowel syndrome: Evidence for an inflammatory component? Gut 2003, 52, 91–93. [Google Scholar] [CrossRef]
- Krawiec, P.; Pawłowska-Kamieniak, A.; Pac-Kożuchowska, E. Interleukin 10 and interleukin 10 receptor in paediatric inflammatory bowel disease: From bench to bedside lesson. J. Inflamm. 2021, 18, 13. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, S.; Heinig, T.; Thiele, H.G.; Raedler, A. Immunoregulatory role of interleukin 10 in patients with inflammatory bowel disease. Gastroenterology 1995, 108, 1434–1444. [Google Scholar] [CrossRef]
- Campbell, L.M.; Maxwell, P.J.; Waugh, D.J.J. Rationale and Means to Target Pro-Inflammatory Interleukin-8 (CXCL8) Signaling in Cancer. Pharmaceuticals 2013, 6, 929–959. [Google Scholar] [CrossRef] [PubMed]
- Kechaou, N.; Chain, F.; Gratadoux, J.J.; Blugeon, S.; Bertho, N.; Chevalier, C.; Le Goffic, R.; Courau, S.; Molimard, P.; Chatel, J.M.; et al. Identification of one novel candidate probiotic Lactobacillus plantarum strain active against influenza virus infection in mice by a large-scale screening. Appl. Environ. Microbiol. 2013, 79, 1491–1499. [Google Scholar] [CrossRef]
- Gad, M.; Ravn, P.; Søborg, D.A.; Lund-Jensen, K.; Ouwehand, A.C.; Jensen, S.S. Regulation of the IL-10/IL-12 axis in human dendritic cells with probiotic bacteria. FEMS Immunol. Med. Microbiol. 2011, 63, 93–107. [Google Scholar] [CrossRef]
- Engevik, M.A.; Ruan, W.; Esparza, M.; Fultz, R.; Shi, Z.; Engevik, K.A.; Engevik, A.C.; Ihekweazu, F.D.; Visuthranukul, C.; Venable, S.; et al. Immunomodulation of dendritic cells by Lactobacillus reuteri surface components and metabolites. Physiol. Rep. 2021, 9, e14719. [Google Scholar] [CrossRef]
- Kim, W.; Lee, E.J.; Bae, I.H.; Myoung, K.; Kim, S.T.; Park, P.J.; Lee, K.H.; Pham, A.V.Q.; Ko, J.; Oh, S.H.; et al. Lactobacillus plantarum-derived extracellular vesicles induce anti-inflammatory M2 macrophage polarization in vitro. J. Extracell. Vesicles 2020, 9, 1793514. [Google Scholar] [CrossRef]
- Qu, S.; Fan, L.; Qi, Y.; Xu, C.; Hu, Y.; Chen, S.; Liu, W.; Liu, W.; Si, J. Akkermansia muciniphila Alleviates Dextran Sulfate Sodium (DSS)-Induced Acute Colitis by NLRP3 Activation. Microbiol. Spectr. 2021, 9, e0073021. [Google Scholar] [CrossRef] [PubMed]
- Hu, R.; Lin, H.; Li, J.; Zhao, Y.; Wang, M.; Sun, X.; Min, Y.; Gao, Y.; Yang, M. Probiotic Escherichia coli Nissle 1917-derived outer membrane vesicles enhance immunomodulation and antimicrobial activity in RAW264.7 macrophages. BMC Microbiol. 2020, 20, 268. [Google Scholar] [CrossRef] [PubMed]
- Tian, T.; Wang, Z.; Zhang, J. Pathomechanisms of Oxidative Stress in Inflammatory Bowel Disease and Potential Antioxidant Therapies. Oxidative Med. Cell. Longev. 2017, 2017, 4535194. [Google Scholar] [CrossRef]
- Wu, Y.; Wang, B.; Xu, H.; Tang, L.; Li, Y.; Gong, L.; Wang, Y.; Li, W. Probiotic Bacillus Attenuates Oxidative Stress- Induced Intestinal Injury via p38-Mediated Autophagy. Front. Microbiol. 2019, 10, 2185. [Google Scholar] [CrossRef] [PubMed]
- Prete, R.; Garcia-Gonzalez, N.; Di Mattia, C.D.; Corsetti, A.; Battista, N. Food-borne Lactiplantibacillus plantarum protect normal intestinal cells against inflammation by modulating reactive oxygen species and IL-23/IL-17 axis. Sci. Rep. 2020, 10, 16340. [Google Scholar] [CrossRef] [PubMed]
- Bhushan, B.; Sakhare, S.M.; Narayan, K.S.; Kumari, M.; Mishra, V.; Dicks, L.M.T. Characterization of Riboflavin-Producing Strains of Lactobacillus plantarum as Potential Probiotic Candidate through in vitro Assessment and Principal Component Analysis. Probiotics Antimicrob. Proteins 2021, 13, 453–467. [Google Scholar] [CrossRef]
- Hernández-Delgado, N.C.; Torres-Maravilla, E.; Mayorga-Reyes, L.; Martín, R.; Langella, P.; Pérez-Pastén-Borja, R.; Sánchez-Pardo, M.E.; Bermúdez-Humarán, L.G. Antioxidant and Anti-Inflammatory Properties of Probiotic Candidate Strains Isolated during Fermentation of Agave (Agave angustifolia Haw). Microorganisms 2021, 9, 1063. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fang, Z.; Zhai, Q.; Cui, S.; Zhao, J.; Zhang, H.; Chen, W.; Lu, W. Supernatants of Bifidobacterium longum and Lactobacillus plantarum Strains Exhibited Antioxidative Effects on A7R5 Cells. Microorganisms 2021, 9, 452. [Google Scholar] [CrossRef]
- Mu, G.; Li, H.; Tuo, Y.; Gao, Y.; Zhang, Y. Antioxidative effect of Lactobacillus plantarum Y44 on 2,2′-azobis(2-methylpropionamidine) dihydrochloride (ABAP)-damaged Caco-2 cells. J. Dairy Sci. 2019, 102, 6863–6875. [Google Scholar] [CrossRef] [PubMed]
- Vergnolle, N. Protease inhibition as new therapeutic strategy for GI diseases. Gut 2016, 65, 1215–1224. [Google Scholar] [CrossRef] [PubMed]
- Van Spaendonk, H.; Ceuleers, H.; Smet, A.; Berg, M.; Joossens, J.; Van der Veken, P.; Francque, S.M.; Lambeir, A.-M.; De Man, J.G.; De Meester, I.; et al. The Effect of a Novel Serine Protease Inhibitor on Inflammation and Intestinal Permeability in a Murine Colitis Transfer Model. Front. Pharmacol. 2021, 12, 1601. [Google Scholar] [CrossRef] [PubMed]
- Motta, J.-P.; Bermúdez-Humarán, L.G.; Deraison, C.; Martin, L.; Rolland, C.; Rousset, P.; Boue, J.; Dietrich, G.; Chapman, K.; Kharrat, P.; et al. Food-Grade Bacteria Expressing Elafin Protect Against Inflammation and Restore Colon Homeostasis. Sci. Transl. Med. 2012, 4, 158ra144. [Google Scholar] [CrossRef] [PubMed]
- McCarville, J.L.; Dong, J.; Caminero, A.; Bermudez-Brito, M.; Jury, J.; Murray, J.A.; Duboux, S.; Steinmann, M.; Delley, M.; Tangyu, M.; et al. A Commensal Bifidobacterium longum Strain Prevents Gluten-Related Immunopathology in Mice through Expression of a Serine Protease Inhibitor. Appl. Environ. Microbiol. 2017, 83, e01323-17. [Google Scholar] [CrossRef]
- Vahora, I.S.; Tsouklidis, N.; Kumar, R.; Soni, R.; Khan, S. How Serotonin Level Fluctuation Affects the Effectiveness of Treatment in Irritable Bowel Syndrome. Cureus 2020, 12, e9871. [Google Scholar] [CrossRef] [PubMed]
- Bhattarai, Y.; Williams, B.B.; Battaglioli, E.J.; Whitaker, W.R.; Till, L.; Grover, M.; Linden, D.R.; Akiba, Y.; Kandimalla, K.K.; Zachos, N.C.; et al. Gut Microbiota-Produced Tryptamine Activates an Epithelial G-Protein-Coupled Receptor to Increase Colonic Secretion. Cell Host Microbe 2018, 23, 775–785.e5. [Google Scholar] [CrossRef] [PubMed]
- Yano, J.M.; Yu, K.; Donaldson, G.P.; Shastri, G.G.; Ann, P.; Ma, L.; Nagler, C.R.; Ismagilov, R.F.; Mazmanian, S.K.; Hsiao, E.Y. Indigenous bacteria from the gut microbiota regulate host serotonin biosynthesis. Cell 2015, 161, 264–276. [Google Scholar] [CrossRef] [PubMed]
- Natividad, J.M.; Agus, A.; Planchais, J.; Lamas, B.; Jarry, A.C.; Martin, R.; Michel, M.-L.; Chong-Nguyen, C.; Roussel, R.; Straube, M.; et al. Impaired Aryl Hydrocarbon Receptor Ligand Production by the Gut Microbiota Is a Key Factor in Metabolic Syndrome. Cell Metab. 2018, 28, 737–749.e4. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Hu, X.; Bao, L.; Wu, K.; Feng, L.; Qiu, M.; Hao, H.; Fu, Y.; Zhang, N. Aryl hydrocarbon receptor activation by Lactobacillus reuteri tryptophan metabolism alleviates Escherichia coli-induced mastitis in mice. PLOS Pathog. 2021, 17, e1009774. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, S.; Toshimitsu, T.; Matsuoka, S.; Maruyama, A.; Oh-Oka, K.; Takamura, T.; Nakamura, Y.; Ishimaru, K.; Fujii-Kuriyama, Y.; Ikegami, S.; et al. Identification of a probiotic bacteria-derived activator of the aryl hydrocarbon receptor that inhibits colitis. Immunol. Cell Biol. 2014, 92, 460–465. [Google Scholar] [CrossRef] [PubMed]
- Chinthrajah, R.S.; Hernandez, J.D.; Boyd, S.D.; Galli, S.J.; Nadeau, K.C. Molecular and cellular mechanisms of food allergy and food tolerance. J. Allergy Clin. Immunol. 2016, 137, 984–997. [Google Scholar] [CrossRef]
- Cao, S.; Feehley, T.J.; Nagler, C.R. The role of commensal bacteria in the regulation of sensitization to food allergens. FEBS Lett. 2014, 588, 4258–4266. [Google Scholar] [CrossRef] [PubMed]
- Sampson, H.A.; O’Mahony, L.; Burks, A.W.; Plaut, M.; Lack, G.; Akdis, C.A. Mechanisms of food allergy. J. Allergy Clin. Immunol. 2018, 141, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Tan-Lim, C.S.C.; Esteban-Ipac, N.A.R. Probiotics as treatment for food allergies among pediatric patients: A meta-analysis. World Allergy Organ. J. 2018, 11, 25. [Google Scholar] [CrossRef]
- Santos, S.C.D.; Konstantyner, T.; Cocco, R.R. Effects of probiotics in the treatment of food hypersensitivity in children: A systematic review. Allergol. Et Immunopathol. 2020, 48, 95–104. [Google Scholar] [CrossRef]
- Lopez-Santamarina, A.; Gonzalez, E.G.; Lamas, A.; Mondragon, A.d.C.; Regal, P.; Miranda, J.M. Probiotics as a Possible Strategy for the Prevention and Treatment of Allergies. A Narrative Review. Foods 2021, 10, 701. [Google Scholar] [CrossRef] [PubMed]
- Ghavami, S.B.; Yadegar, A.; Aghdaei, H.A.; Sorrentino, D.; Farmani, M.; Mir, A.S.; Azimirad, M.; Balaii, H.; Shahrokh, S.; Zali, M.R. Immunomodulation and Generation of Tolerogenic Dendritic Cells by Probiotic Bacteria in Patients with Inflammatory Bowel Disease. Int. J. Mol. Sci. 2020, 21, 6266. [Google Scholar] [CrossRef] [PubMed]
- You, J.; Dong, H.; Mann, E.R.; Knight, S.C.; Yaqoob, P. Probiotic modulation of dendritic cell function is influenced by ageing. Immunobiology 2014, 219, 138–148. [Google Scholar] [CrossRef]
- Ruiz, L.; Delgado, S.; Ruas-Madiedo, P.; Sánchez, B.; Margolles, A. Bifidobacteria and Their Molecular Communication with the Immune System. Front. Microbiol. 2017, 8, 2345. [Google Scholar] [CrossRef]
- Konieczna, P.; Groeger, D.; Ziegler, M.; Frei, R.; Ferstl, R.; Shanahan, F.; Quigley, E.M.; Kiely, B.; Akdis, C.A.; O’Mahony, L. Bifidobacterium infantis 35,624 administration induces Foxp3 T regulatory cells in human peripheral blood: Potential role for myeloid and plasmacytoid dendritic cells. Gut 2012, 61, 354–366. [Google Scholar] [CrossRef]
- Fu, L.; Song, J.; Wang, C.; Fu, S.; Wang, Y. Bifidobacterium infantis Potentially Alleviates Shrimp Tropomyosin-Induced Allergy by Tolerogenic Dendritic Cell-Dependent Induction of Regulatory T Cells and Alterations in Gut Microbiota. Front. Immunol. 2017, 8, 1536. [Google Scholar] [CrossRef]
- Santos, S.S.; Miranda, V.C.; Trindade, L.M.; Cardoso, V.N.; Reis, D.C.; Cassali, G.D.; Nicoli, J.R.; Cara, D.C.; Martins, F.S. Bifidobacterium longum subsp. longum 51A Attenuates Signs of Inflammation in a Murine Model of Food Allergy. Probiotics Antimicrob. Proteins 2021. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.-K.; Lee, C.-G.; So, J.-S.; Chae, C.-S.; Hwang, J.-S.; Sahoo, A.; Nam, J.H.; Rhee, J.H.; Hwang, K.-C.; Im, S.-H. Generation of regulatory dendritic cells and CD4+Foxp3+ T cells by probiotics administration suppresses immune disorders. Proc. Natl. Acad. Sci. USA 2010, 107, 2159–2164. [Google Scholar] [CrossRef]
- Shin, H.S.; Eom, J.E.; Shin, D.U.; Yeon, S.H.; Lim, S.I.; Lee, S.Y. Preventive Effects of a Probiotic Mixture in an Ovalbumin-Induced Food Allergy Model. J. Microbiol. Biotechnol. 2018, 28, 65–76. [Google Scholar] [CrossRef]
- Ma, J.; Zhang, J.; Li, Q.; Shi, Z.; Wu, H.; Zhang, H.; Tang, L.; Yi, R.; Su, H.; Sun, X. Oral administration of a mixture of probiotics protects against food allergy via induction of CD103+ dendritic cells and modulates the intestinal microbiota. J. Funct. Foods 2019, 55, 65–75. [Google Scholar] [CrossRef]
- Balmer, M.L.; Ma, E.H.; Bantug, G.R.; Grählert, J.; Pfister, S.; Glatter, T.; Jauch, A.; Dimeloe, S.; Slack, E.; Dehio, P.; et al. Memory CD8+ T Cells Require Increased Concentrations of Acetate Induced by Stress for Optimal Function. Immunity 2016, 44, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; van der Veeken, J.; deRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Wu, W.; Chen, L.; Yang, W.; Huang, X.; Ma, C.; Chen, F.; Xiao, Y.; Zhao, Y.; Ma, C.; et al. Microbiota-derived short-chain fatty acids promote Th1 cell IL-10 production to maintain intestinal homeostasis. Nat. Commun. 2018, 9, 3555. [Google Scholar] [CrossRef]
- Zhao, Y.; Chen, F.; Wu, W.; Sun, M.; Bilotta, A.J.; Yao, S.; Xiao, Y.; Huang, X.; Eaves-Pyles, T.D.; Golovko, G.; et al. GPR43 mediates microbiota metabolite SCFA regulation of antimicrobial peptide expression in intestinal epithelial cells via activation of mTOR and STAT3. Mucosal Immunol. 2018, 11, 752–762. [Google Scholar] [CrossRef]
- Rohrbeck, L.; Adori, M.; Wang, S.; He, C.; Tibbitt, C.A.; Chernyshev, M.; Sirel, M.; Ribacke, U.; Murrell, B.; Bohlooly-Y, M.; et al. GPR43 regulates marginal zone B-cell responses to foreign and endogenous antigens. Immunol. Cell Biol. 2021, 99, 234–243. [Google Scholar] [CrossRef]
- Erlich, T.H.; Sharkia, I.; Landolina, N.; Assayag, M.; Goldberger, O.; Berkman, N.; Levi-Schaffer, F.; Razin, E. Modulation of allergic responses by mitochondrial STAT3 inhibitors. Allergy 2018, 73, 2160–2171. [Google Scholar] [CrossRef]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic Acid Produced by Commensal Peptostreptococcus Species Suppresses Inflammation. Cell Host Microbe 2017, 22, 25–37.e26. [Google Scholar] [CrossRef]
- Lu, W.; Qian, L.; Fang, Z.; Wang, H.; Zhu, J.; Lee, Y.-K.; Zhao, J.; Zhang, H.; Chen, W. Probiotic strains alleviated OVA-induced food allergy in mice by regulating the gut microbiota and improving the level of indoleacrylic acid in fecal samples. Food Funct. 2022, 13, 3704–3719. [Google Scholar] [CrossRef] [PubMed]
- Oksaharju, A.; Kankainen, M.; Kekkonen, R.A.; Lindstedt, K.A.; Kovanen, P.T.; Korpela, R.; Miettinen, M. Probiotic Lactobacillus rhamnosus downregulates FCER1 and HRH4 expression in human mast cells. World J. Gastroenterol. 2011, 17, 750–759. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.-H.; Jeun, E.-J.; Hong, C.-P.; Kim, S.-H.; Jang, M.S.; Lee, E.-J.; Moon, S.J.; Yun, C.H.; Im, S.-H.; Jeong, S.-G.; et al. Extracellular vesicle–derived protein from Bifidobacterium longum alleviates food allergy through mast cell suppression. J. Allergy Clin. Immunol. 2016, 137, 507–516.e8. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Luo, Y.; Liu, Z.; Yang, P.; Gui, Y. Probiotics SOD inhibited food allergy via downregulation of STAT6-TIM4 signaling on DCs. Mol. Immunol. 2018, 103, 71–77. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; Wang, Y.; Wang, P.; Huang, Y.; Wang, F. Short-Chain Fatty Acids Manifest Stimulative and Protective Effects on Intestinal Barrier Function Through the Inhibition of NLRP3 Inflammasome and Autophagy. Cell. Physiol. Biochem. 2018, 49, 190–205. [Google Scholar] [CrossRef] [PubMed]
- Tulyeu, J.; Kumagai, H.; Jimbo, E.; Watanabe, S.; Yokoyama, K.; Cui, L.; Osaka, H.; Mieno, M.; Yamagata, T. Probiotics Prevents Sensitization to Oral Antigen and Subsequent Increases in Intestinal Tight Junction Permeability in Juvenile–Young Adult Rats. Microorganisms 2019, 7, 463. [Google Scholar] [CrossRef] [PubMed]
- Jin, B.-Y.; Li, Z.; Xia, Y.-N.; Li, L.-X.; Zhao, Z.-X.; Li, X.-Y.; Li, Y.; Li, B.; Zhou, R.-C.; Fu, S.-C.; et al. Probiotic Interventions Alleviate Food Allergy Symptoms Correlated With Cesarean Section: A Murine Model. Front. Immunol. 2021, 12, 741371. [Google Scholar] [CrossRef] [PubMed]
- Dixon, A.; Robertson, K.; Yung, A.; Que, M.; Randall, H.; Wellalagodage, D.; Cox, T.; Robertson, D.; Chi, C.; Sun, J. Efficacy of Probiotics in Patients of Cardiovascular Disease Risk: A Systematic Review and Meta-analysis. Curr. Hypertens. Rep. 2020, 22, 74. [Google Scholar] [CrossRef]
- McCracken, E.; Monaghan, M.; Sreenivasan, S. Pathophysiology of the metabolic syndrome. Clin. Dermatol. 2018, 36, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Ji, Y.; Jung, H.-Y.; Park, H.; Kang, J.; Choi, S.-H.; Shin, H.; Hyun, C.-K.; Kim, K.-T.; Holzapfel, W.H. Lactobacillus plantarum HAC01 regulates gut microbiota and adipose tissue accumulation in a diet-induced obesity murine model. Appl. Microbiol. Biotechnol. 2017, 101, 1605–1614. [Google Scholar] [CrossRef]
- Fabersani, E.; Abeijon-Mukdsi, M.C.; Ross, R.; Medina, R.; González, S.; Gauffin-Cano, P. Specific Strains of Lactic Acid Bacteria Differentially Modulate the Profile of Adipokines In Vitro. Front. Immunol. 2017, 8, 266. [Google Scholar] [CrossRef] [PubMed]
- Aoki, R.; Kamikado, K.; Suda, W.; Takii, H.; Mikami, Y.; Suganuma, N.; Hattori, M.; Koga, Y. A proliferative probiotic Bifidobacterium strain in the gut ameliorates progression of metabolic disorders via microbiota modulation and acetate elevation. Sci. Rep. 2017, 7, 43522. [Google Scholar] [CrossRef]
- Kimura, I.; Inoue, D.; Hirano, K.; Tsujimoto, G. The SCFA Receptor GPR43 and Energy Metabolism. Front. Endocrinol. 2014, 5, 85. [Google Scholar] [CrossRef]
- Lu, Y.; Fan, C.; Li, P.; Lu, Y.; Chang, X.; Qi, K. Short Chain Fatty Acids Prevent High-fat-diet-induced Obesity in Mice by Regulating G Protein-coupled Receptors and Gut Microbiota. Sci. Rep. 2016, 6, 37589. [Google Scholar] [CrossRef] [PubMed]
- Queipo-Ortuño, M.I.; Seoane, L.M.; Murri, M.; Pardo, M.; Gomez-Zumaquero, J.M.; Cardona, F.; Casanueva, F.; Tinahones, F.J. Gut Microbiota Composition in Male Rat Models under Different Nutritional Status and Physical Activity and Its Association with Serum Leptin and Ghrelin Levels. PLoS ONE 2013, 8, e65465. [Google Scholar] [CrossRef] [PubMed]
- Torres-Fuentes, C.; Golubeva, A.V.; Zhdanov, A.V.; Wallace, S.; Arboleya, S.; Papkovsky, D.B.; Aidy, S.E.; Ross, P.; Roy, B.L.; Stanton, C.; et al. Short-chain fatty acids and microbiota metabolites attenuate ghrelin receptor signaling. FASEB J. 2019, 33, 13546–13559. [Google Scholar] [CrossRef] [PubMed]
- Pingitore, A.; Gonzalez-Abuin, N.; Ruz-Maldonado, I.; Huang, G.C.; Frost, G.; Persaud, S.J. Short chain fatty acids stimulate insulin secretion and reduce apoptosis in mouse and human islets in vitro: Role of free fatty acid receptor 2. Diabetes Obes. Metab. 2019, 21, 330–339. [Google Scholar] [CrossRef]
- Psichas, A.; Sleeth, M.L.; Murphy, K.G.; Brooks, L.; Bewick, G.A.; Hanyaloglu, A.C.; Ghatei, M.A.; Bloom, S.R.; Frost, G. The short chain fatty acid propionate stimulates GLP-1 and PYY secretion via free fatty acid receptor 2 in rodents. Int. J. Obes. 2015, 39, 424–429. [Google Scholar] [CrossRef]
- Xiong, Y.; Miyamoto, N.; Shibata, K.; Valasek, M.A.; Motoike, T.; Kedzierski, R.M.; Yanagisawa, M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc. Natl. Acad. Sci. USA 2004, 101, 1045–1050. [Google Scholar] [CrossRef]
- Larraufie, P.; Martin-Gallausiaux, C.; Lapaque, N.; Dore, J.; Gribble, F.M.; Reimann, F.; Blottiere, H.M. SCFAs strongly stimulate PYY production in human enteroendocrine cells. Sci. Rep. 2018, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Torres, S.; Fabersani, E.; Marquez, A.; Gauffin-Cano, P. Adipose tissue inflammation and metabolic syndrome. The proactive role of probiotics. Eur. J. Nutr. 2019, 58, 27–43. [Google Scholar] [CrossRef] [PubMed]
- Nissen, L.; di Carlo, E.; Gianotti, A. Prebiotic potential of hemp blended drinks fermented by probiotics. Food Res. Int. 2020, 131, 109029. [Google Scholar] [CrossRef] [PubMed]
- Tomaro-Duchesneau, C.; Jones, M.L.; Shah, D.; Jain, P.; Saha, S.; Prakash, S. Cholesterol Assimilation by Lactobacillus Probiotic Bacteria: An In Vitro Investigation. BioMed Res. Int. 2014, 2014, 380316. [Google Scholar] [CrossRef]
- Chand, D.; Avinash, V.S.; Yadav, Y.; Pundle, A.V.; Suresh, C.G.; Ramasamy, S. Molecular features of bile salt hydrolases and relevance in human health. Biochim. Biophys. Acta (BBA) Gen. Subj. 2017, 1861, 2981–2991. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Vazquez, R.; Azaola-Espinosa, A.; Mayorga-Reyes, L.; Reyes-Nava, L.A.; Shah, N.P.; Rivera-Espinoza, Y. Isolation, Identification and Partial Characterization of a Lactobacillus casei Strain with Bile Salt Hydrolase Activity from Pulque. Probiotics Antimicrob. Proteins 2015, 7, 242–248. [Google Scholar] [CrossRef]
- Hernández-Gómez, J.G.; López-Bonilla, A.; Trejo-Tapia, G.; Ávila-Reyes, S.V.; Jiménez-Aparicio, A.R.; Hernández-Sánchez, H. In Vitro Bile Salt Hydrolase (BSH) Activity Screening of Different Probiotic Microorganisms. Foods 2021, 10, 674. [Google Scholar] [CrossRef]
- Paiva, L.; Goldbeck, R.; dos Santos, W.; Squina, F.M. Ferulic acid and derivatives: Molecules with potential application in the pharmaceutical field. Braz. J. Pharm. Sci. 2013, 49, 395–411. [Google Scholar] [CrossRef]
- Tomaro-Duchesneau, C.; Saha, S.; Malhotra, M.; Coussa-Charley, M.; Al-Salami, H.; Jones, M.; Labbé, A. Lactobacillus fermentum NCIMB 5221 has a greater potential for the production of ferulic acid when compared to other ferulic acid esterase active Lactobacilli. Int. J. Probiotics Prebiotics 2012, 7, 23–32. [Google Scholar] [CrossRef]
- Zhang, F.; Qiu, L.; Xu, X.; Liu, Z.; Zhan, H.; Tao, X.; Shah, N.P.; Wei, H. Beneficial effects of probiotic cholesterol-lowering strain of Enterococcus faecium WEFA23 from infants on diet-induced metabolic syndrome in rats. J. Dairy Sci. 2017, 100, 1618–1628. [Google Scholar] [CrossRef]
- Huo, Y.; Lu, X.; Wang, X.; Wang, X.; Chen, L.; Guo, H.; Zhang, M.; Li, Y. Bifidobacterium animalis subsp. lactis A6 Alleviates Obesity Associated with Promoting Mitochondrial Biogenesis and Function of Adipose Tissue in Mice. Molecules 2020, 25, 1490. [Google Scholar] [CrossRef]
- Martorell, P.; Llopis, S.; González, N.; Chenoll, E.; López-Carreras, N.; Aleixandre, A.; Chen, Y.; Karoly, E.D.; Ramón, D.; Genovés, S. Probiotic Strain Bifidobacterium animalis subsp. lactis CECT 8145 Reduces Fat Content and Modulates Lipid Metabolism and Antioxidant Response in Caenorhabditis elegans. J. Agric. Food Chem. 2016, 64, 3462–3472. [Google Scholar] [CrossRef]
- Carreras, N.L.; Martorell, P.; Chenoll, E.; Genovés, S.; Ramón, D.; Aleixandre, A. Anti-obesity properties of the strain Bifidobacterium animalis subsp. lactis CECT 8145 in Zücker fatty rats. Benef. Microbes 2018, 9, 629–641. [Google Scholar] [CrossRef] [PubMed]
- Douillard, F.P.; de Vos, W.M. Biotechnology of health-promoting bacteria. Biotechnol. Adv. 2019, 37, 107369. [Google Scholar] [CrossRef] [PubMed]
- Jaswal, R.; Pathak, A.; Edwards Iii, B.; Lewis Iii, R.; Seaman, J.C.; Stothard, P.; Krivushin, K.; Blom, J.; Rupp, O.; Chauhan, A. Metagenomics-guided survey, isolation, and characterization of uranium resistant microbiota from the Savannah River Site, USA. Genes 2019, 10, 325. [Google Scholar] [CrossRef] [PubMed]
- de Castro, A.; Ruiz-Barba, J.L.; Romero, C.; Sánchez, A.H.; García, P.; Brenes, M. Formation of gas pocket defect in Spanish-style green olives by the halophile Celerinatantimonas sp. Food Control 2022, 136, 108868. [Google Scholar] [CrossRef]
- Sokol, H.; Pigneur, B.; Watterlot, L.; Lakhdari, O.; Bermúdez-Humarán, L.G.; Gratadoux, J.-J.; Blugeon, S.; Bridonneau, C.; Furet, J.-P.; Corthier, G.; et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. USA 2008, 105, 16731–16736. [Google Scholar] [CrossRef] [PubMed]
- Pittayanon, R.; Lau, J.T.; Leontiadis, G.I.; Tse, F.; Yuan, Y.; Surette, M.; Moayyedi, P. Differences in Gut Microbiota in Patients with vs. without Inflammatory Bowel Diseases: A Systematic Review. Gastroenterology 2020, 158, 930–946.e1. [Google Scholar] [CrossRef]
- Tian, P.; Zhu, H.; Zou, R.; Kong, Q.; Xu, M.; Zhao, J.; Zhang, H.; Chen, W.; Wang, G. An in vitro screening method for probiotics with antidepressant-like effect using the enterochromaffin cell model. Food Funct. 2021, 12, 646–655. [Google Scholar] [CrossRef]
- Lee, J.-E.; Lee, J.; Kim, J.H.; Cho, N.; Lee, S.H.; Park, S.B.; Koh, B.; Kang, D.; Kim, S.; Yoo, H.M. Characterization of the Anti-Cancer Activity of the Probiotic Bacterium Lactobacillus fermentum Using 2D vs. 3D Culture in Colorectal Cancer Cells. Biomolecules 2019, 9, 557. [Google Scholar] [CrossRef]
- Lenoir, M.; Martín, R.; Torres-Maravilla, E.; Chadi, S.; González-Dávila, P.; Sokol, H.; Langella, P.; Chain, F.; Bermúdez-Humarán, L.G. Butyrate mediates anti-inflammatory effects of Faecalibacterium prausnitzii in intestinal epithelial cells through Dact3. Gut Microbes 2020, 12, 1826748. [Google Scholar] [CrossRef] [PubMed]
- Kropp, C.; Le Corf, K.; Relizani, K.; Tambosco, K.; Martinez, C.; Chain, F.; Rawadi, G.; Langella, P.; Claus, S.P.; Martin, R. The Keystone commensal bacterium Christensenella minuta DSM 22607 displays anti-inflammatory properties both in vitro and in vivo. Sci. Rep. 2021, 11, 11494. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.-H.; Ngo, H.-P.-T.; Nam, H.-K.; Kim, K.-R.; Kang, L.-W.; Oh, D.-K.; Liu, S.-J. Alternative Biotransformation of Retinal to Retinoic Acid or Retinol by an Aldehyde Dehydrogenase from Bacillus cereus. Appl. Environ. Microbiol. 2016, 82, 3940–3946. [Google Scholar] [CrossRef]
- Choi, S.S.; Seo, Y.B.; Nam, S.-W.; Kim, G.-D. Enhanced Production of Astaxanthin by Co-culture of Paracoccus haeundaensis and Lactic Acid Bacteria. Front. Mar. Sci. 2021, 7, 597553. [Google Scholar] [CrossRef]
- Jiang, L.; Luo, Y.; Cao, X.; Liu, W.; Song, G.; Zhang, Z. LuxS quorum sensing system mediating Lactobacillus plantarum probiotic characteristics. Arch. Microbiol. 2021, 203, 4141–4148. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, H.; Zheng, L.; Li, J.; Hong, Y.; Liang, P.; Kwok, L.-Y.; Zuo, Y.; Zhang, W.; Zhang, H. iProbiotics: A machine learning platform for rapid identification of probiotic properties from whole-genome primary sequences. Brief. Bioinform. 2021, 23, bbab477. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres-Maravilla, E.; Reyes-Pavón, D.; Benítez-Cabello, A.; González-Vázquez, R.; Ramírez-Chamorro, L.M.; Langella, P.; Bermúdez-Humarán, L.G. Strategies for the Identification and Assessment of Bacterial Strains with Specific Probiotic Traits. Microorganisms 2022, 10, 1389. https://doi.org/10.3390/microorganisms10071389
Torres-Maravilla E, Reyes-Pavón D, Benítez-Cabello A, González-Vázquez R, Ramírez-Chamorro LM, Langella P, Bermúdez-Humarán LG. Strategies for the Identification and Assessment of Bacterial Strains with Specific Probiotic Traits. Microorganisms. 2022; 10(7):1389. https://doi.org/10.3390/microorganisms10071389
Chicago/Turabian StyleTorres-Maravilla, Edgar, Diana Reyes-Pavón, Antonio Benítez-Cabello, Raquel González-Vázquez, Luis M. Ramírez-Chamorro, Philippe Langella, and Luis G. Bermúdez-Humarán. 2022. "Strategies for the Identification and Assessment of Bacterial Strains with Specific Probiotic Traits" Microorganisms 10, no. 7: 1389. https://doi.org/10.3390/microorganisms10071389
APA StyleTorres-Maravilla, E., Reyes-Pavón, D., Benítez-Cabello, A., González-Vázquez, R., Ramírez-Chamorro, L. M., Langella, P., & Bermúdez-Humarán, L. G. (2022). Strategies for the Identification and Assessment of Bacterial Strains with Specific Probiotic Traits. Microorganisms, 10(7), 1389. https://doi.org/10.3390/microorganisms10071389







