Transcriptomic Analysis Reveals the Role of tmRNA on Biofilm Formation in Bacillus subtilis
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Construction of Strains
2.3. Biofilm Formation Assays
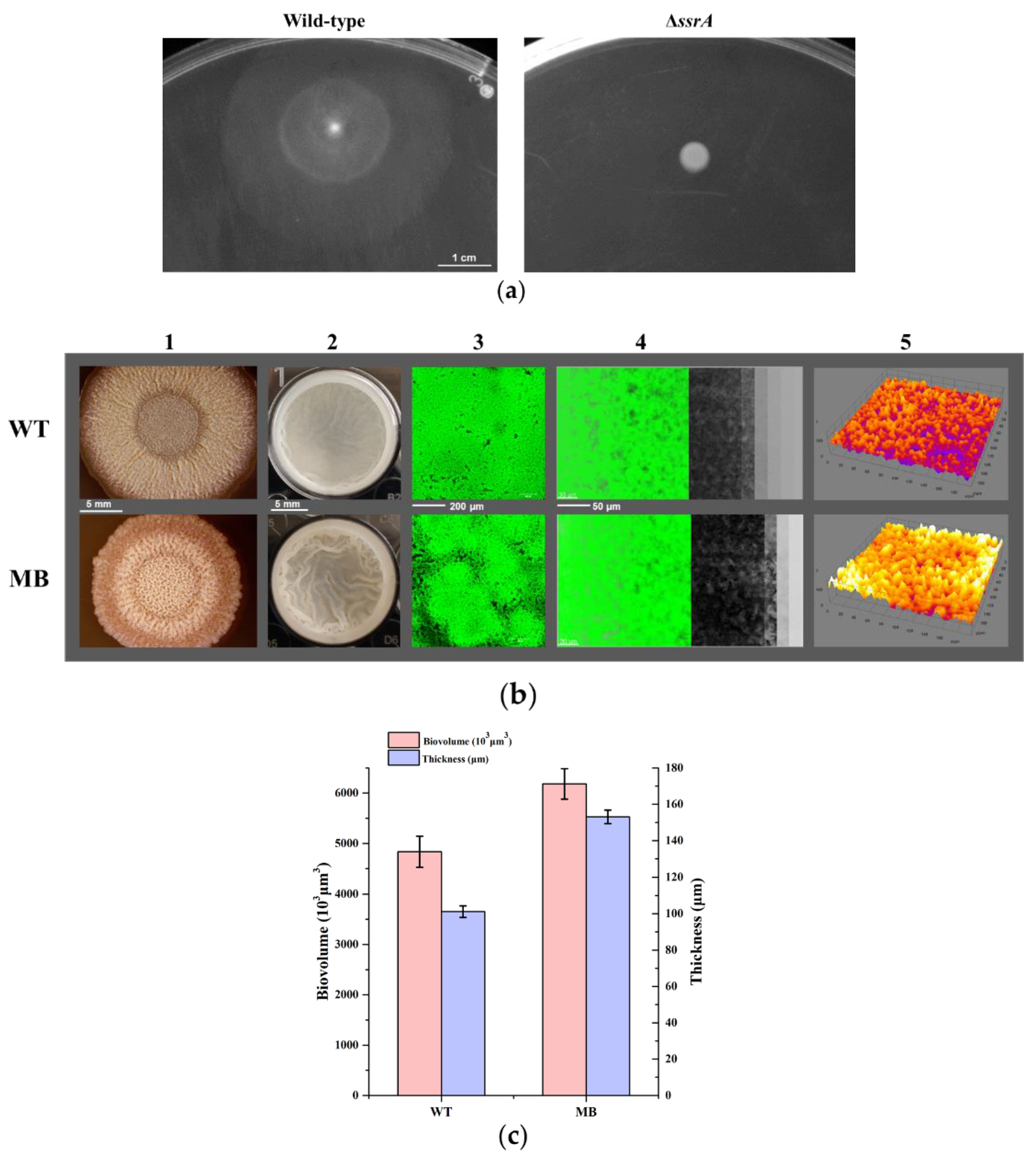
2.4. Swarming Motility Assay
2.5. Isolation of the Swarming Suppressors of ssrA in B. subtilis
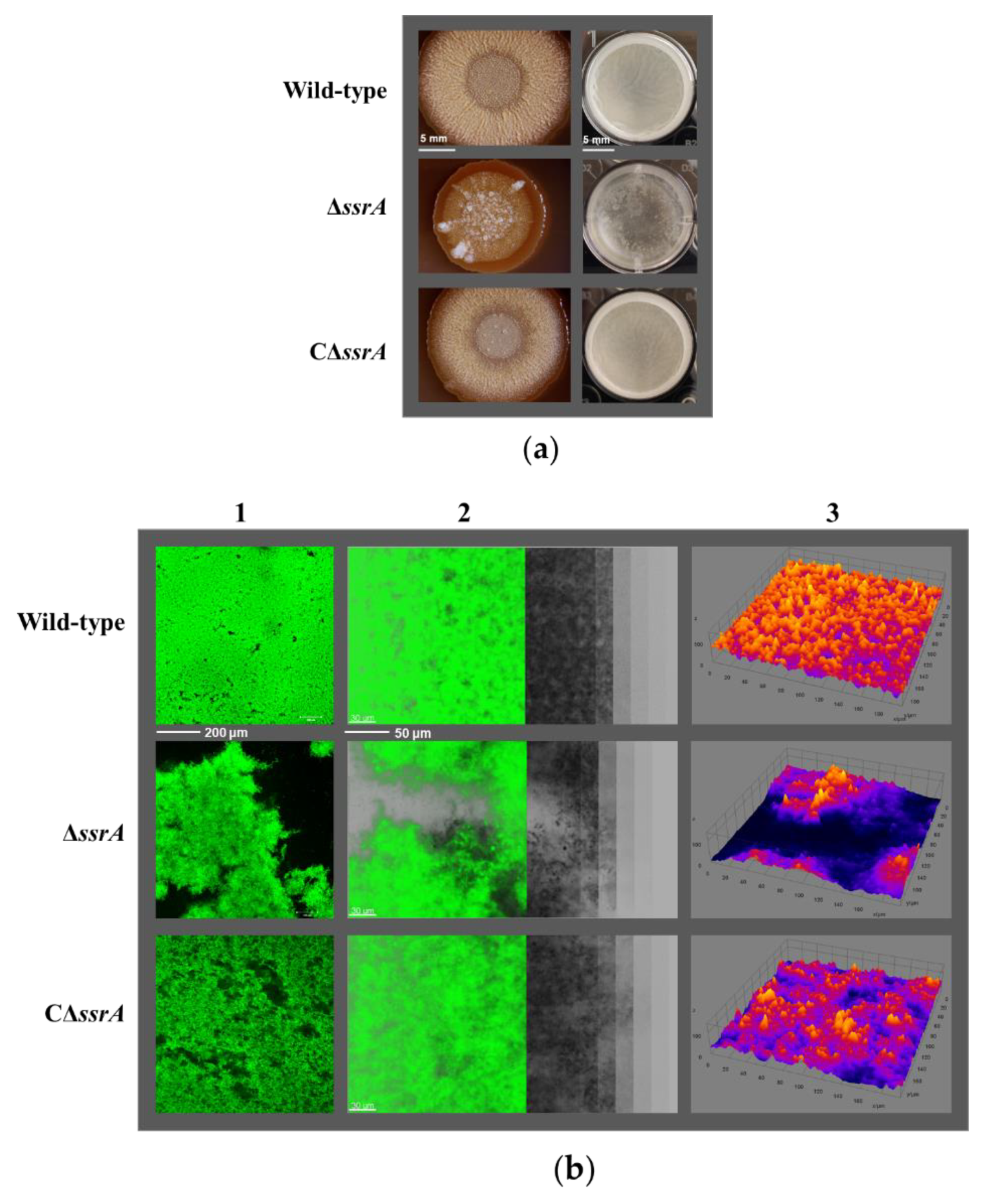
2.6. Confocal Laser Scanning Microscopy
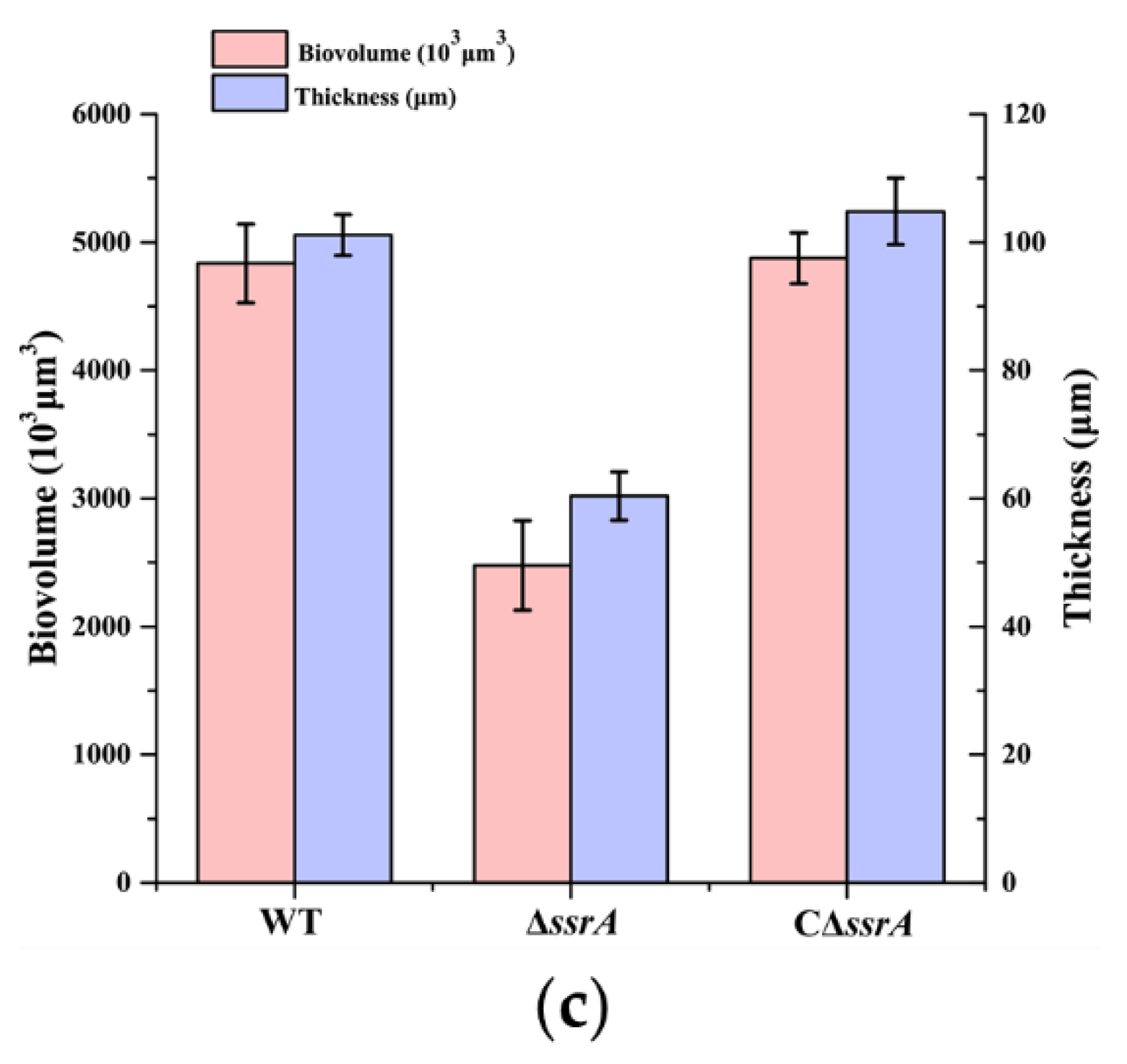
2.7. Quantification of the Average Thickness and Biovolume of Biofilms
2.8. RNA Isolation
2.9. Transcriptome Analysis
2.10. Sporulation Assay
2.11. Extraction of Extracellular Polysaccharide (EPS)
3. Results
3.1. tmRNA Positively Affects Biofilm Formation in B. subtilis
3.2. Isolation of a Suppressor of ssrA That Cured the TM Biofilm-Formation Defect
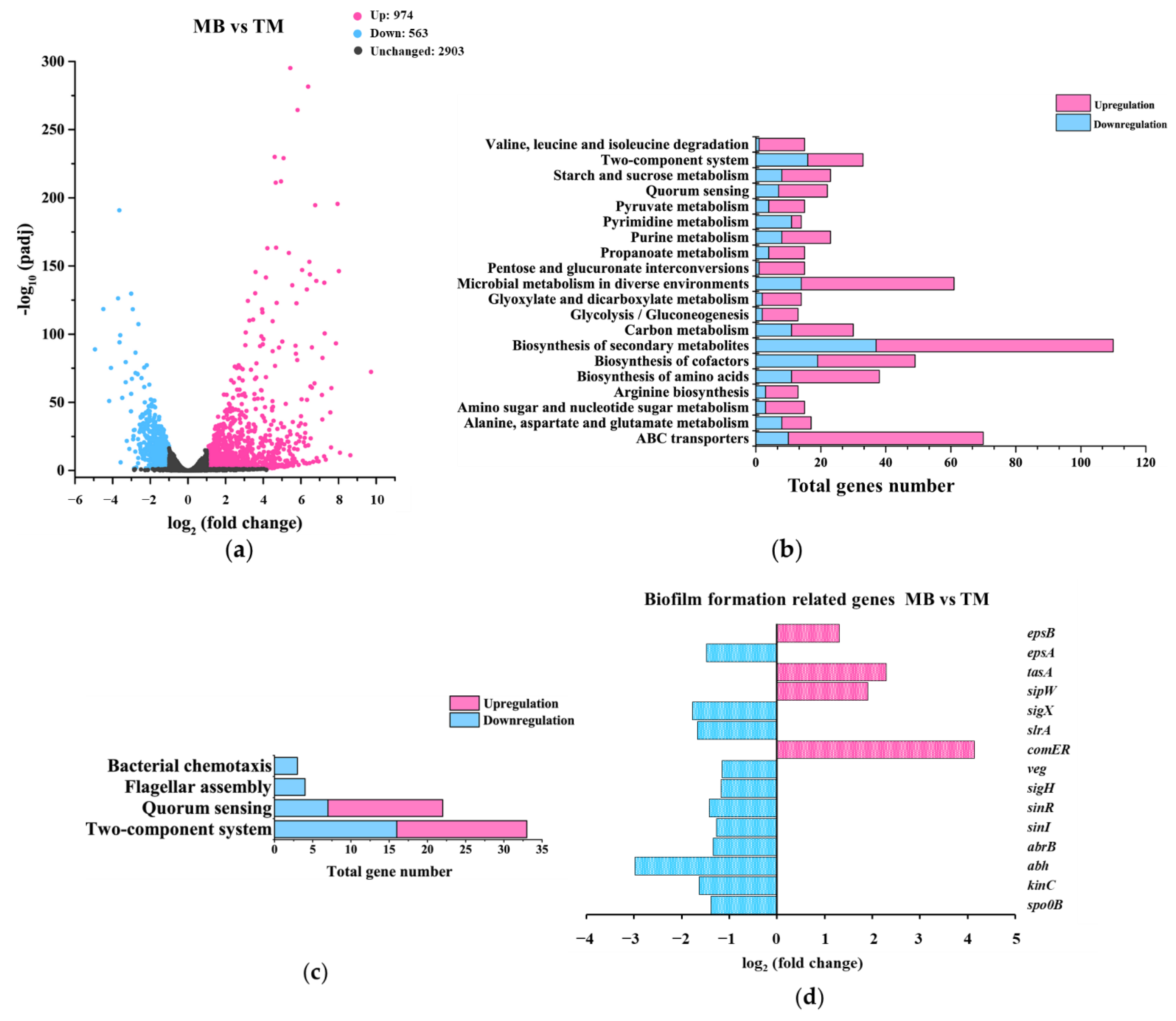
3.3. Transcriptome Analysis among the WT, TM, and MB Strain
3.3.1. Gene-Expression Differences between TM and WT
3.3.2. Gene-Expression Differences between MB and TM
3.3.3. Gene-Expression Differences between MB and WT
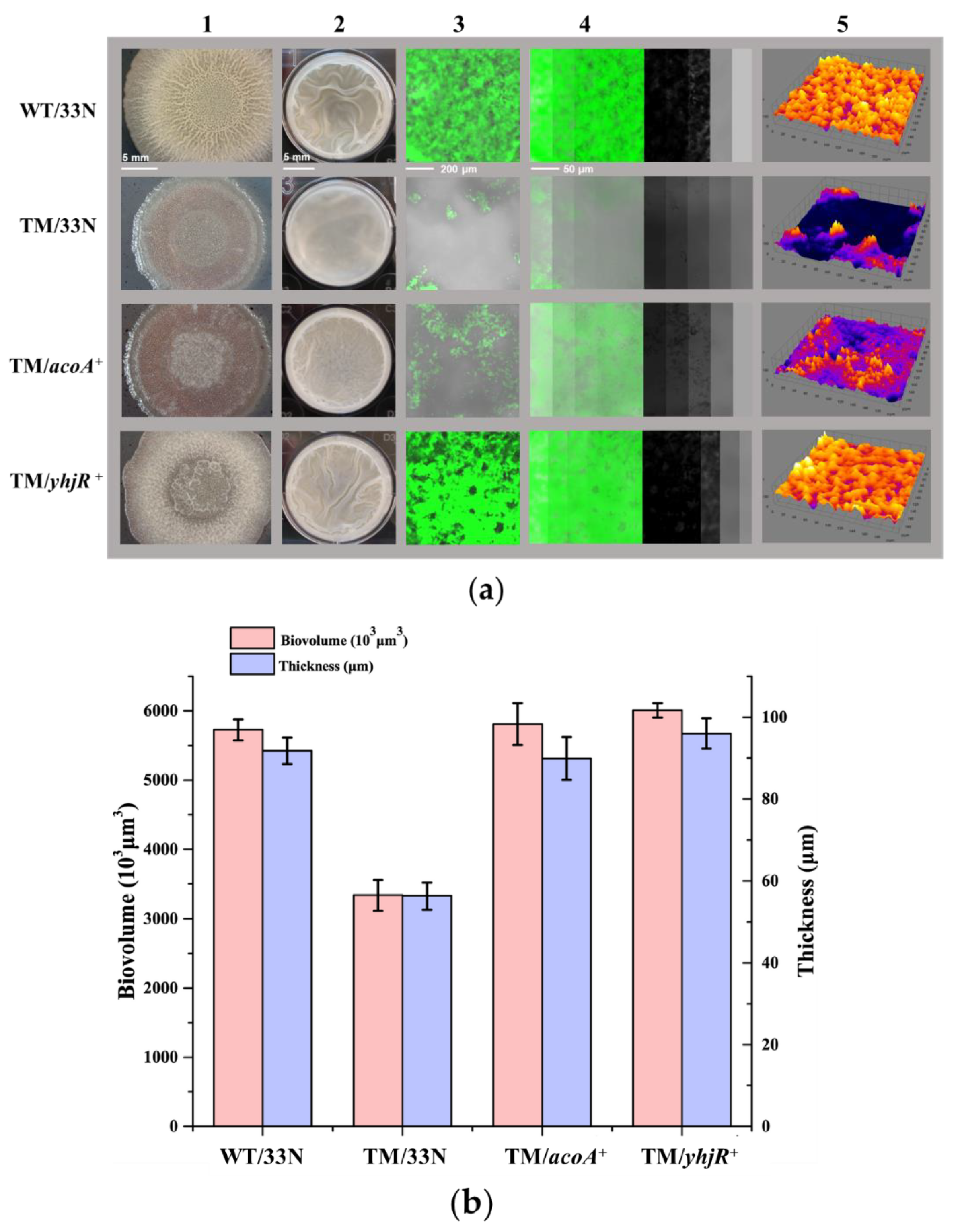
3.4. Elucidating the Roles of Known Biofilm-Formation Factors in the tmRNA Regulation of Biofilm in B. subtilis
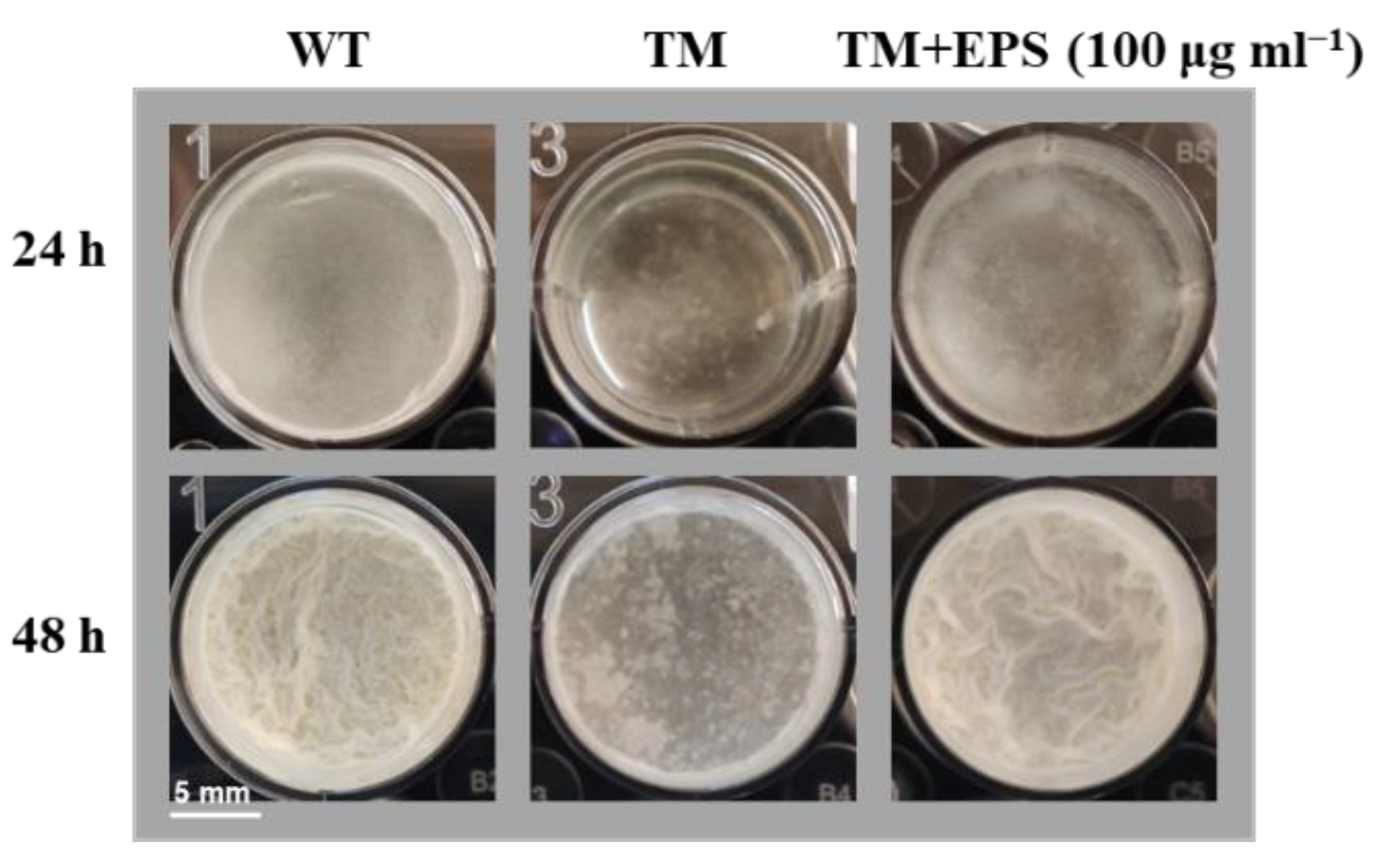
3.5. Initial Characterization of the Unknown Pathways Involved in the tmRNA Regulation of Biofilm Formation in B. subtilis
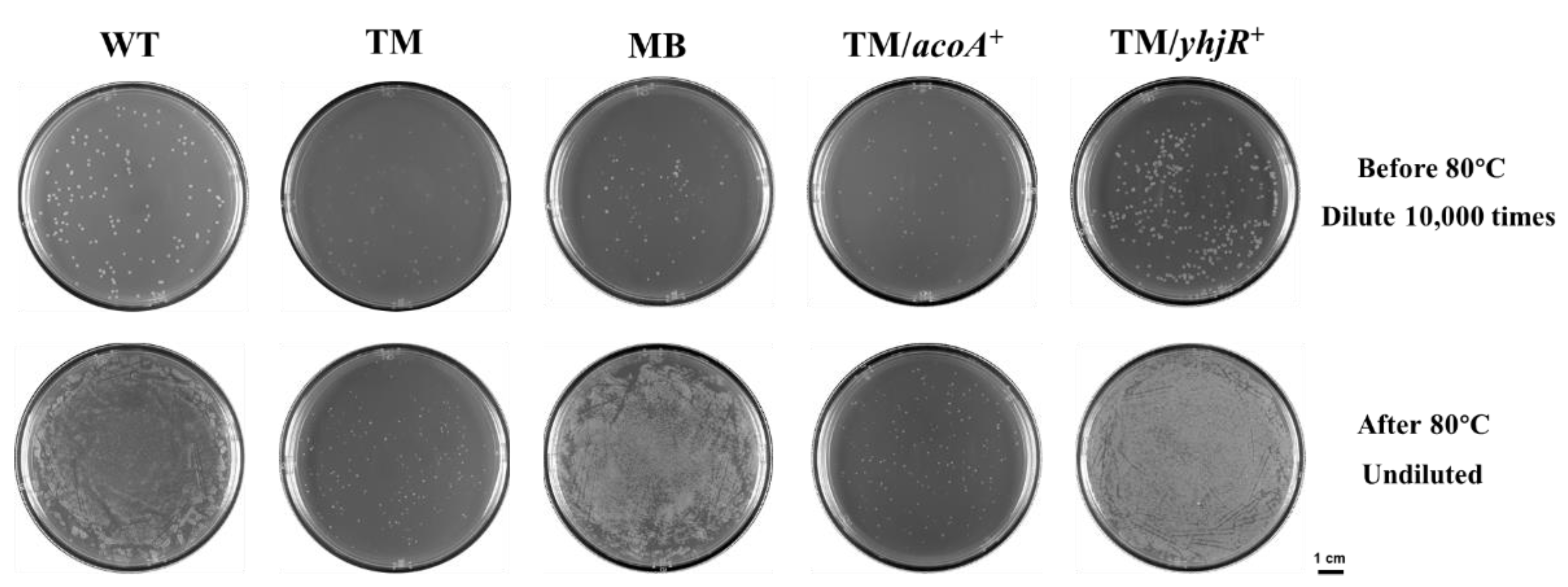
3.6. Sporulation in WT, TM, and TM Derivates
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Flemming, H.C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Høiby, N.; Ciofu, O.; Johansen, H.K.; Song, Z.J.; Moser, C.; Jensen, P.; Molin, S.; Givskov, M.; Tolker-Nielsen, T.; Bjarnsholt, T. The clinical impact of bacterial biofilms. Int. J. Oral. Sci. 2011, 3, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Erable, B.; Duţeanu, N.M.; Ghangrekar, M.M.; Dumas, C.; Scott, K. Application of electro-active biofilms. Biofouling 2010, 26, 57–71. [Google Scholar] [CrossRef]
- Martin, K.J.; Nerenberg, R. The membrane biofilm reactor (MBfR) for water and wastewater treatment: Principles, applications, and recent developments. Bioresour. Technol. 2012, 122, 83–94. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, F.; Chai, Y.; Liu, H.; Kolter, R.; Losick, R.; Guo, J.H. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 2013, 15, 848–864. [Google Scholar] [CrossRef]
- Chen, Y.; Cao, S.; Chai, Y.; Clardy, J.; Kolter, R.; Guo, J.H.; Losick, R. A Bacillus subtilis sensor kinase involved in triggering biofilm formation on the roots of tomato plants. Mol. Microbiol. 2012, 85, 418–430. [Google Scholar] [CrossRef]
- Arnaouteli, S.; Bamford, N.C.; Stanley-Wall, N.R.; Kovács, Á.T. Bacillus subtilis biofilm formation and social interactions. Nat. Rev. Microbiol. 2021, 19, 600–614. [Google Scholar] [CrossRef]
- Ostrowski, A.; Mehert, A.; Prescott, A.; Kiley, T.B.; Stanley-Wall, N.R. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J. Bacteriol. 2011, 193, 4821–4831. [Google Scholar] [CrossRef]
- Marvasi, M.; Visscher, P.T.; Casillas Martinez, L. Exopolymeric substances (EPS) from Bacillus subtilis: Polymers and genes encoding their synthesis. FEMS Microbiol. Lett. 2010, 313, 1–9. [Google Scholar] [CrossRef]
- Kobayashi, K. Bacillus subtilis pellicle formation proceeds through genetically defined morphological changes. J. Bacteriol. 2007, 189, 4920–4931. [Google Scholar] [CrossRef]
- Mielich-Süss, B.; Lopez, D. Molecular mechanisms involved in Bacillus subtilis biofilm formation. Environ. Microbiol. 2015, 17, 555–565. [Google Scholar] [CrossRef] [PubMed]
- Hamon, M.A.; Lazazzera, B.A. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 2001, 42, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Dubnau, E.J.; Carabetta, V.J.; Tanner, A.W.; Miras, M.; Diethmaier, C.; Dubnau, D. A protein complex supports the production of Spo0A-P and plays additional roles for biofilms and the K-state in Bacillus subtilis. Mol. Microbiol. 2016, 101, 606–624. [Google Scholar] [CrossRef] [PubMed]
- Banse, A.V.; Chastanet, A.; Rahn-Lee, L.; Hobbs, E.C.; Losick, R. Parallel pathways of repression and antirepression governing the transition to stationary phase in Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2008, 105, 15547–15552. [Google Scholar] [CrossRef] [PubMed]
- Chumsakul, O.; Takahashi, H.; Oshima, T.; Hishimoto, T.; Kanaya, S.; Ogasawara, N.; Ishikawa, S. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2011, 39, 414–428. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Oshima, T.; Ogasawara, N.; Ishikawa, S. Functional analysis of the protein Veg, which stimulates biofilm formation in Bacillus subtilis. J. Bacteriol. 2013, 195, 1697–1705. [Google Scholar] [CrossRef]
- Sanchez-Vizuete, P.; Le Coq, D.; Bridier, A.; Herry, J.M.; Aymerich, S.; Briandet, R. Identification of ypqP as a new Bacillus subtilis biofilm determinant that mediates the protection of Staphylococcus aureus against antimicrobial agents in mixed-species communities. Appl. Environ. Microbiol. 2015, 81, 109–118. [Google Scholar] [CrossRef]
- Yan, F.; Yu, Y.; Wang, L.; Luo, Y.; Guo, J.H.; Chai, Y. The comER gene plays an important role in biofilm formation and sporulation in both Bacillus subtilis and Bacillus cereus. Front. Microbiol. 2016, 7, 1025. [Google Scholar] [CrossRef]
- Špacapan, M.; Danevčič, T.; Štefanic, P.; Porter, M.; Stanley-Wall, N.R.; Mandic-Mulec, I. The ComX quorum sensing peptide of Bacillus subtilis affects biofilm formation negatively and sporulation positively. Microorganisms. 2020, 8, 1131. [Google Scholar] [CrossRef]
- Keiler, K.C. Mechanisms of ribosome rescue in bacteria. Nat. Rev. Microbiol. 2015, 13, 285–297. [Google Scholar] [CrossRef] [PubMed]
- Ushida, C.; Himeno, H.; Watanabe, T.; Muto, A. tRNA-like structures in 10Sa RNAs of Mycoplasma capricolum and Bacillus subtilis. Nucleic Acids Res. 1994, 22, 3392–3396. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wolfgang, M.C.; Withey, J.; Koomey, M.; Friedman, D.I. Charged tmRNA but not tmRNA-mediated proteolysis is essential for Neisseria gonorrhoeae viability. EMBO J. 2000, 19, 1098–1107. [Google Scholar] [CrossRef] [PubMed]
- Ramadoss, N.S.; Zhou, X.; Keiler, K.C. tmRNA is essential in Shigella flexneri. PLoS ONE 2013, 8, e57537. [Google Scholar] [CrossRef][Green Version]
- Li, J.; Ji, L.; Shi, W.; Xie, J.; Zhang, Y. Trans-translation mediates tolerance to multiple antibiotics and stresses in Escherichia coli. J. Antimicrob. Chemother. 2013, 68, 2477–2481. [Google Scholar] [CrossRef]
- Okan, N.A.; Bliska, J.B.; Karzai, A.W. A Role for the SmpB-SsrA system in Yersinia pseudotuberculosis pathogenesis. PLoS Pathog. 2006, 2, e6. [Google Scholar] [CrossRef]
- Keiler, K.C.; Shapiro, L. tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. J. Bacteriol. 2003, 185, 1825–1830. [Google Scholar] [CrossRef]
- Liu, X.; Tang, K.; Zhang, D.; Li, Y.; Liu, Z.; Yao, J.; Wood, T.K.; Wang, X. Symbiosis of a P2-family phage and deep-sea Shewanella putrefaciens. Environ. Microbiol. 2019, 21, 4212–4232. [Google Scholar] [CrossRef]
- Muto, A.; Fujihara, A.; Ito, K.I.; Matsuno, J.; Ushida, C.; Himeno, H. Requirement of transfer-messenger RNA for the growth of Bacillus subtilis under stresses. Genes Cells 2000, 5, 627–635. [Google Scholar] [CrossRef]
- Shin, J.H.; Price, C.W. The SsrA-SmpB ribosome rescue system is important for growth of Bacillus subtilis at low and high temperatures. J. Bacteriol. 2007, 189, 3729–3737. [Google Scholar] [CrossRef]
- Abe, T.; Sakaki, K.; Fujihara, A.; Ujiie, H.; Ushida, C.; Himeno, H.; Sato, T.; Muto, A. tmRNA-dependent trans-translation is required for sporulation in Bacillus subtilis. Mol. Microbiol. 2008, 69, 1491–1498. [Google Scholar] [CrossRef] [PubMed]
- Gallegos-Monterrosa, R.; Mhatre, E.; Kovács, Á.T. Specific Bacillus subtilis 168 variants form biofilms on nutrient-rich medium. Microbiology 2016, 162, 1922–1932. [Google Scholar] [CrossRef] [PubMed]
- Branda, S.S.; González-Pastor, J.E.; Ben-Yehuda, S.; Losick, R.; Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2001, 98, 11621–11626. [Google Scholar] [CrossRef] [PubMed]
- Altenbuchner, J. Editing of the Bacillus subtilis Genome by the CRISPR-Cas9 System. Appl. Environ. Microbiol. 2016, 82, 5421–5427. [Google Scholar] [CrossRef] [PubMed]
- Rhee, M.S.; Kim, J.W.; Qian, Y.; Ingram, L.O.; Shanmugam, K.T. Development of plasmid vector and electroporation condition for gene transfer in sporogenic lactic acid bacterium, Bacillus coagulans. Plasmid 2007, 58, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Heydorn, A.; Nielsen, A.T.; Hentzer, M.; Sternberg, C.; Givskov, M.; Ersbøll, B.K.; Molin, S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology 2000, 146, 2395–2407. [Google Scholar] [CrossRef]
- Arnaouteli, S.; Matoz-Fernandez, D.A.; Porter, M.; Kalamara, M.; Abbott, J.; MacPhee, C.E.; Davidson, F.A.; Stanley-Wall, N.R. Pulcherrimin formation controls growth arrest of the Bacillus subtilis biofilm. Proc. Natl. Acad. Sci. USA 2019, 116, 13553–13562. [Google Scholar] [CrossRef]
- Guttenplan, S.B.; Kearns, D.B. Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 2013, 37, 849–871. [Google Scholar] [CrossRef]
- Hummels, K.R.; Kearns, D.B. Suppressor mutations in ribosomal proteins and FliY restore Bacillus subtilis swarming motility in the absence of EF-P. PLoS Genet. 2019, 15, e1008179. [Google Scholar] [CrossRef]
- Pisithkul, T.; Schroeder, J.W.; Trujillo, E.A.; Yeesin, P.; Stevenson, D.M.; Chaiamarit, T.; Coon, J.J.; Wang, J.D.; Amador-Noguez, D. Metabolic Remodeling during Biofilm Development of Bacillus subtilis. mBio 2019, 10, e00623-19. [Google Scholar] [CrossRef]
- Marciniak, B.C.; Pabijaniak, M.; de Jong, A.; Dűhring, R.; Seidel, G.; Hillen, W.; Kuipers, O.P. High- and low-affinity cre boxes for CcpA binding in Bacillus subtilis revealed by genome-wide analysis. BMC Genom. 2012, 13, 401. [Google Scholar] [CrossRef] [PubMed]
- Arrieta-Ortiz, M.L.; Hafemeister, C.; Bate, A.R.; Chu, T.; Greenfield, A.; Shuster, B.; Barry, S.N.; Gallitto, M.; Liu, B.; Kacmarczyk, T.; et al. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol. Syst. Biol. 2015, 11, 839. [Google Scholar] [CrossRef] [PubMed]
- Behravan, J.; Chirakkal, H.; Masson, A.; Moir, A. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J. Bacteriol. 2000, 182, 1987–1994. [Google Scholar] [CrossRef] [PubMed]
- Steil, L.; Serrano, M.; Henriques, A.O.; Völker, U. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 2005, 151, 399–420. [Google Scholar] [CrossRef]
- Hsiao, T.L.; Revelles, O.; Chen, L.; Sauer, U.; Vitkup, D. Automatic policing of biochemical annotations using genomic correlations. Nat. Chem. Biol. 2010, 6, 34–40. [Google Scholar] [CrossRef]
- Guariglia-Oropeza, V.; Helmann, J.D. Bacillus subtilis σ(V) confers lysozyme resistance by activation of two cell wall modification pathways, peptidoglycan O-acetylation and D-alanylation of teichoic acids. J. Bacteriol. 2011, 193, 6223–6232. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Eiamphungporn, W.; Mäder, U.; Liebeke, M.; Lalk, M.; Hecker, M.; Helmann, J.D.; Antelmann, H. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: Control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR). Mol. Microbiol. 2009, 71, 876–894. [Google Scholar]
- Tanner, J.J. Structural Biology of Proline Catabolic Enzymes. Antioxid. Redox Signal. 2019, 30, 650–673. [Google Scholar] [CrossRef]
- McKenney, P.T.; Eichenberger, P. Dynamics of spore coat morphogenesis in Bacillus subtilis. Mol. Microbiol. 2012, 83, 245–260. [Google Scholar] [CrossRef]
- Pedreira, T.; Elfmann, C.; Stülke, J. The current state of SubtiWiki, the database for the model organism Bacillus subtilis. Nucleic Acids Res. 2022, 50, D875–D882. [Google Scholar] [CrossRef]
- Morinaga, T.; Kobayashi, K.; Ashida, H.; Fujita, Y.; Yoshida, K.I. Transcriptional regulation of the Bacillus subtilis asnH operon and role of the 5′-proximal long sequence triplication in RNA stabilization. Microbiology 2010, 156, 1632–1641. [Google Scholar] [CrossRef] [PubMed]
- Milton, M.E.; Draughn, G.L.; Bobay, B.G.; Stowe, S.D.; Olson, A.L.; Feldmann, E.A.; Thompson, R.J.; Myers, K.H.; Santoro, M.T.; Kearns, D.B.; et al. The solution structures and interaction of SinR and SinI: Elucidating the mechanism of action of the master regulator switch for biofilm formation in Bacillus subtilis. J. Mol. Biol. 2020, 432, 343–357. [Google Scholar] [CrossRef] [PubMed]
- Srinivasan, S.; Vladescu, I.D.; Koehler, S.A.; Wang, X.; Mani, M.; Rubinstein, S.M. Matrix production and sporulation in Bacillus subtilis biofilms localize to propagating wave fronts. Biophys. J. 2018, 114, 1490–1498. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Shao, J.; Li, B.; Yan, X.; Shen, Q.; Zhang, R. Contribution of bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl. Environ. Microbiol. 2013, 79, 808–815. [Google Scholar] [CrossRef]
- Li, H.; Han, X.; Dong, Y.; Xu, S.; Chen, C.; Feng, Y.; Cui, Q.; Li, W. Bacillaenes: Decomposition trigger point and biofilm enhancement in Bacillus. ACS Omega 2021, 6, 1093–1098. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, S.; Cao, Q.; Liu, Z.; Chen, J.; Yan, P.; Li, B.; Xu, Y. Transcriptomic Analysis Reveals the Role of tmRNA on Biofilm Formation in Bacillus subtilis. Microorganisms 2022, 10, 1338. https://doi.org/10.3390/microorganisms10071338
Xu S, Cao Q, Liu Z, Chen J, Yan P, Li B, Xu Y. Transcriptomic Analysis Reveals the Role of tmRNA on Biofilm Formation in Bacillus subtilis. Microorganisms. 2022; 10(7):1338. https://doi.org/10.3390/microorganisms10071338
Chicago/Turabian StyleXu, Shanshan, Qianqian Cao, Zengzhi Liu, Junpeng Chen, Peiguang Yan, Bingyu Li, and Ying Xu. 2022. "Transcriptomic Analysis Reveals the Role of tmRNA on Biofilm Formation in Bacillus subtilis" Microorganisms 10, no. 7: 1338. https://doi.org/10.3390/microorganisms10071338
APA StyleXu, S., Cao, Q., Liu, Z., Chen, J., Yan, P., Li, B., & Xu, Y. (2022). Transcriptomic Analysis Reveals the Role of tmRNA on Biofilm Formation in Bacillus subtilis. Microorganisms, 10(7), 1338. https://doi.org/10.3390/microorganisms10071338





