Genome-Wide Transcription Start Sites Mapping in Methylorubrum Grown with Dichloromethane and Methanol
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Growth and RNA Extraction
2.2. Construction of 5′-End-Mapping Libraries and cDNA Sequencing
2.3. Identification of 5-Ends and Discrimination between Transcription Start Sites (TSS) and Cleavage Sites
2.4. Promoter Motif Discovery
2.5. Analysis of 5′Untranslated Regions (5′UTR)
2.6. Data Deposition
3. Results and Discussion
3.1. Mapping and Annotation
3.2. Focus on TSS and Promoter Region Associated with Genes of One-Carbon Metabolism
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Durrani, T.; Clapp, R.; Harrison, R.; Shusterman, D. Solvent-based paint and varnish removers: A focused toxicologic review of existing and alternative constituents. J. Appl. Toxicol. 2020, 40, 1325–1341. [Google Scholar] [CrossRef] [PubMed]
- Hossaini, R.; Chipperfield, M.; Montzka, S.A.; Leeson, A.; Dhomse, S.S.; Pyle, J.A. The increasing threat to stratospheric ozone from dichloromethane. Nat. Commun. 2017, 8, 15962. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Fisch, A.; Gibson, C.M.; Mack, E.E.; Seger, E.S.; Campagna, S.R.; Löffler, F.E. Mineralization versus fermentation: Evidence for two distinct anaerobic bacterial degradation pathways for dichloromethane. ISME J. 2020, 14, 959–970. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.E.; Bringel, F.; Vuilleumier, S. Dichloromethane-degrading bacteria in the genomic age. Res. Microbiol. 2011, 162, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Bringel, F.; Postema, C.P.; Mangenot, S.; Bibi-Triki, S.; Chaignaud, P.; Ul Haque, M.F.; Gruffaz, C.; Hermon, L.; Louhichi, Y.; Maucourt, B.; et al. Genome sequence of the dichloromethane-degrading bacterium Hyphomicrobium sp. strain GJ21. Genome Announc. 2017, 5, e00622-17. [Google Scholar] [CrossRef]
- Vuilleumier, S.; Chistoserdova, L.; Lee, M.-C.; Bringel, F.; Lajus, A.; Zhou, Y.; Gourion, B.; Barbe, V.; Chang, J.; Cruveiller, S.; et al. Methylobacterium Genome Sequences: A reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS ONE 2009, 4, e5584. [Google Scholar] [CrossRef] [PubMed]
- Muller, E.E.L.; Hourcade, E.; Louhichi-Jelail, Y.; Hammann, P.; Vuilleumier, S.; Bringel, F. Functional genomics of dichloromethane utilization in Methylobacterium extorquens DM4. Environ. Microbiol. 2011, 13, 2518–2535. [Google Scholar] [CrossRef] [PubMed]
- Ochsner, A.M.; Sonntag, F.; Buchhaupt, M.; Schrader, J.; Vorholt, J.A. Methylobacterium extorquens: Methylotrophy and biotechnological applications. Appl. Microbiol. Biotechnol. 2015, 99, 517–534. [Google Scholar] [CrossRef] [PubMed]
- Kohler-Staub, D.; Hartmans, S.; Galli, R.; Suter, F.; Leisinger, T. Evidence for identical dichloromethane dehalogenases in different methylotrophic bacteria. Microbiology 1986, 132, 2837–2843. [Google Scholar] [CrossRef][Green Version]
- Schmid-Appert, M.; Zoller, K.; Traber, H.; Vuilleumier, S.; Leisinger, T. Association of newly discovered IS elements with the dichloromethane utilization genes of methylotrophic bacteria. Microbiology 1997, 143, 2557–2567. [Google Scholar] [CrossRef]
- Bibi-Triki, S.; Husson, G.; Maucourt, B.; Vuilleumier, S.; Carapito, C.; Bringel, F. N-terminome and proteogenomic analysis of the Methylobacterium extorquens DM4 reference strain for dichloromethane utilization. J. Proteom. 2018, 179, 131–139. [Google Scholar] [CrossRef]
- Chaignaud, P.; Maucourt, B.; Weiman, M.; Alberti, A.; Kolb, S.; Cruveiller, S.; Vuilleumier, S.; Bringel, F. Genomic and transcriptomic analysis of growth-supporting dehalogenation of chlorinated methanes in Methylobacterium. Front. Microbiol. 2017, 8, 1600. [Google Scholar] [CrossRef]
- Maucourt, B.; Vuilleumier, S.; Bringel, F. Transcriptional regulation of organohalide pollutant utilisation in bacteria. FEMS Microbiol. Rev. 2020, 44, 189–207. [Google Scholar] [CrossRef]
- Michener, J.; Neves, A.A.C.; Vuilleumier, S.; Bringel, F.; Marx, C.J. Effective use of a horizontally-transferred pathway for dichloromethane catabolism requires post–transfer refinement. eLife 2014, 3, e04279. [Google Scholar] [CrossRef]
- Anderson, D.J.; Morris, C.J.; Nunn, D.N.; Anthony, C.; Lidstrom, M.E. Nucleotide sequence of the Methylobacterium extorquens AM1 moxF and moxJ genes involved in methanol oxidation. Gene 1990, 90, 173–176. [Google Scholar] [CrossRef]
- La Roche, S.D.; Leisinger, T. Identification of dcmR, the regulatory gene governing expression of dichloromethane dehalogenase in Methylobacterium sp. strain DM4. J. Bacteriol. 1991, 173, 6714–6721. [Google Scholar] [CrossRef]
- Ramamoorthi, R.; Lidstrom, M.E. Transcriptional analysis of pqqD and study of the regulation of pyrroloquinoline quinone biosynthesis in Methylobacterium extorquens AM1. J. Bacteriol. 1995, 177, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Lidstrom, M.E. Promoters and transcripts for genes involved in methanol oxidation in Methylobacterium extorquens AM1. Microbiology 2003, 149, 1033–1040. [Google Scholar] [CrossRef]
- Kalyuzhnaya, M.G.; Lidstrom, M.E. QscR-mediated transcriptional activation of serine cycle genes in Methylobacterium extorquens AM1. J. Bacteriol. 2005, 187, 7511–7517. [Google Scholar] [CrossRef] [PubMed]
- Davagnino, J.; Springer, A.L.; Lidstrom, M.E. An RNA polymerase preparation from Methylobacterium extorquens AM1 capable of transcribing from a methylotrophic promoter. Microbiology 1998, 144, 177–182. [Google Scholar] [CrossRef][Green Version]
- Machlin, S.M.; Hanson, R.S. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J. Bacteriol. 1988, 170, 4739–4747. [Google Scholar] [CrossRef]
- Xu, H.H.; Viebahn, M.; Hanson, R.S. Identification of methanol-regulated promoter sequences from the facultative methylotrophic bacterium Methylobacterium organophilum XX. J. Gen. Microbiol. 1993, 139, 743–752. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bader, R.; Leisinger, T. Isolation and characterization of the Methylophilus sp. strain DM11 gene encoding dichloromethane dehalogenase/glutathione S-transferase. J. Bacteriol. 1994, 176, 3466–3473. [Google Scholar] [CrossRef]
- Irastortza-Olaziregi, M.; Amster-Choder, O. Coupled transcription-translation in Prokaryotes: An old couple with new surprises. Front. Microbiol. 2020, 11, 624830. [Google Scholar] [CrossRef] [PubMed]
- Sharma, C.M.; Vogel, J. Differential RNA-seq: The approach behind and the biological insight gained. Curr. Opin. Microbiol. 2014, 19, 97–105. [Google Scholar] [CrossRef]
- Irla, M.; Neshat, A.; Brautaset, T.; Rückert, C.; Kalinowski, J.; Wendisch, V.F. Transcriptome analysis of thermophilic methylotrophic Bacillus methanolicus MGA3 using RNA-sequencing provides detailed insights into its previously uncharted transcriptional landscape. BMC Genom. 2015, 16, 73. [Google Scholar] [CrossRef]
- Liao, Y.; Huang, L.; Wang, B.; Zhou, F.; Pan, L. The global transcriptional landscape of Bacillus amyloliquefaciens XH7 and high-throughput screening of strong promoters based on RNA-seq data. Gene 2015, 571, 252–262. [Google Scholar] [CrossRef]
- Fan, B.; Li, L.; Chao, Y.; Förstner, K.; Vogel, J.; Borriss, R.; Wu, X.-Q. dRNA-Seq reveals genomewide TSSs and noncoding RNAs of plant beneficial Rhizobacterium Bacillus amyloliquefaciens FZB42. PLoS ONE 2015, 10, e0142002. [Google Scholar] [CrossRef]
- Čuklina, J.; Hahn, J.; Imakaev, M.; Omasits, U.; Förstner, K.U.; Ljubimov, N.; Goebel, M.; Pessi, G.; Fischer, H.-M.; Ahrens, C.H.; et al. Genome-wide transcription start site mapping of Bradyrhizobium japonicum grown free-living or in symbiosis—A rich resource to identify new transcripts, proteins and to study gene regulation. BMC Genom. 2016, 17, 302. [Google Scholar] [CrossRef]
- Martini, M.C.; Zhou, Y.; Sun, H.; Shell, S.S. Defining the transcriptional and post-transcriptional landscapes of Mycobacterium smegmatis in aerobic growth and hypoxia. Front. Microbiol. 2019, 10, 591. [Google Scholar] [CrossRef] [PubMed]
- Chaignaud, P.; Morawe, M.; Besaury, L.; Kröber, E.; Vuilleumier, S.; Bringel, F.; Kolb, S. Methanol consumption drives the bacterial chloromethane sink in a forest soil. ISME J. 2018, 12, 2681–2693. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-F.; Romero, A.D.; Guan, S.; Mamanova, L.; McDowall, K.J. A combination of improved differential and global RNA-seq reveals pervasive transcription initiation and events in all stages of the life-cycle of functional RNAs in Propionibacterium acnes, a major contributor to wide-spread human disease. BMC Genom. 2013, 14, 620. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Amman, F.; Wolfinger, M.T.; Lorenz, R.; Hofacker, I.L.; Stadler, P.F.; Findeiß, S. TSSAR: TSS annotation regime for dRNA-seq data. BMC Bioinform. 2014, 15, 89. [Google Scholar] [CrossRef]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.; Huber, W.; Pagès, H.; Aboyoun, P.; Carlson, M.; Gentleman, R.; Morgan, M.; Carey, V.J. Software for computing and annotating genomic ranges. PLoS Comput. Biol. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Charif, D.; Thioulouse, J.; Lobry, J.R.; Perrière, G. Online synonymous codon usage analyses with the ade4 and seqinR packages. Bioinformatics 2005, 21, 545–547. [Google Scholar] [CrossRef]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef]
- Shine, J.; Dalgarno, L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: Complementarity to nonsense triplets and ribosome binding sites. Proc. Natl. Acad. Sci. USA 1974, 71, 1342–1346. [Google Scholar] [CrossRef]
- Su, W.; Liu, M.-L.; Yang, Y.-H.; Wang, J.-S.; Li, S.-H.; Lv, H.; Dao, F.-Y.; Yang, H.; Lin, H. PPD: A manually curated database for experimentally verified Prokaryotic promoters. J. Mol. Biol. 2021, 433, 166860. [Google Scholar] [CrossRef] [PubMed]
- D’Arrigo, I.; Bojanovič, K.; Yang, X.; Rau, M.H.; Long, K.S. Genome-wide mapping of transcription start sites yields novel insights into the primary transcriptome of Pseudomonas putida. Environ. Microbiol. 2016, 18, 3466–3481. [Google Scholar] [CrossRef] [PubMed]
- Thomason, M.K.; Bischler, T.; Eisenbart, S.K.; Förstner, K.U.; Zhang, A.; Herbig, A.; Nieselt, K.; Sharma, C.M.; Storz, G. Global transcriptional start site mapping using differential RNA sequencing reveals novel antisense RNAs in Escherichia coli. J. Bacteriol. 2015, 197, 18–28. [Google Scholar] [CrossRef]
- Georg, J.; Hess, W.R. Widespread antisense transcription in Prokaryotes. Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Kreitmeier, M.; Ardern, Z.; Abele, M.; Ludwig, C.; Scherer, S.; Neuhaus, K. Spotlight on alternative frame coding: Two long overlapping genes in Pseudomonas aeruginosa are translated and under purifying selection. iScience 2022, 25, 103844. [Google Scholar] [CrossRef] [PubMed]
- Schlüter, J.-P.; Reinkensmeier, J.; Barnett, M.J.; Lang, C.; Krol, E.; Giegerich, R.; Long, S.R.; Becker, A. Global mapping of transcription start sites and promoter motifs in the symbiotic α-proteobacterium Sinorhizobium meliloti 1021. BMC Genom. 2013, 14, 156. [Google Scholar] [CrossRef] [PubMed]
- Beck, H.J.; Moll, I. Leaderless mRNAs in the spotlight: Ancient but not outdated! Microbiol. Spectr. 2018, 6. [Google Scholar] [CrossRef]
- Vera, J.M.; Ghosh, I.N.; Zhang, Y.; Hebert, A.S.; Coon, J.J.; Landick, R. Genome-scale transcription-translation mapping reveals features of Zymomonas mobilis transcription units and promoters. mSystems 2020, 5, e00250-20. [Google Scholar] [CrossRef]
- Zhou, B.; Schrader, J.; Kalogeraki, V.S.; Abeliuk, E.; Dinh, C.B.; Pham, J.Q.; Cui, Z.Z.; Dill, D.L.; McAdams, H.H.; Shapiro, L. The global regulatory architecture of transcription during the Caulobacter cell cycle. PLoS Genet. 2015, 11, e1004831. [Google Scholar] [CrossRef]
- Morris, C.J.; Lidstrom, M.E. Cloning of a methanol-inducible moxF promoter and its analysis in moxB mutants of Methylobacterium extorquens AM1rif. J. Bacteriol. 1992, 174, 4444–4449. [Google Scholar] [CrossRef]
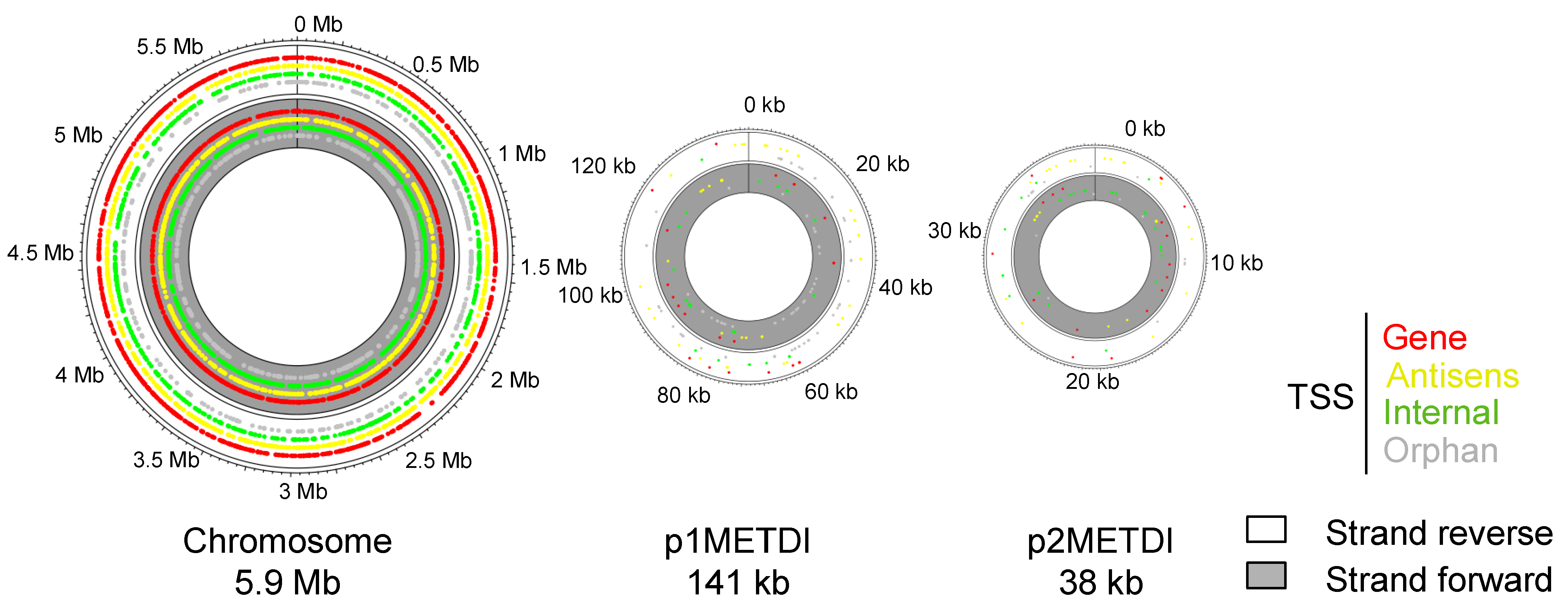
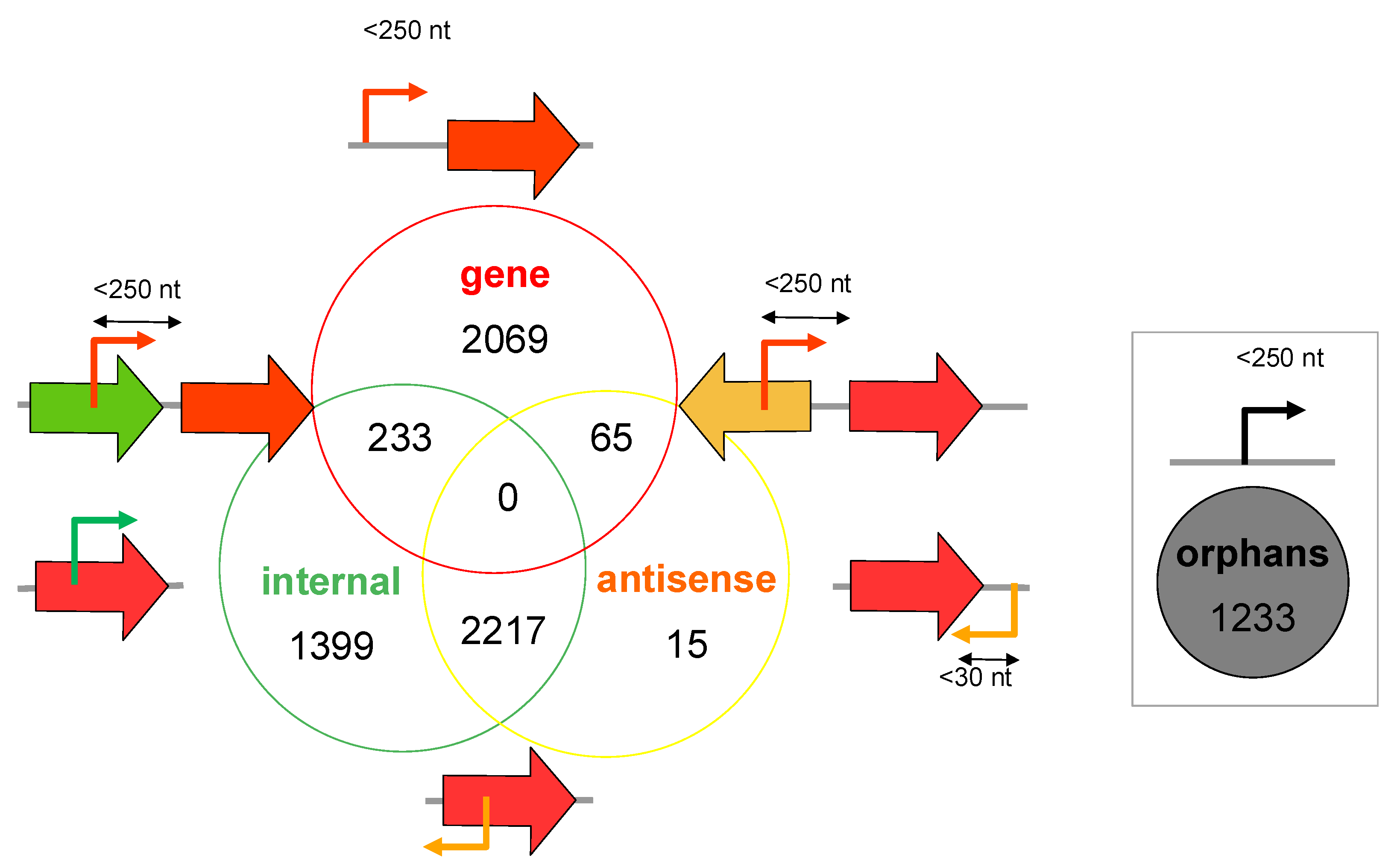

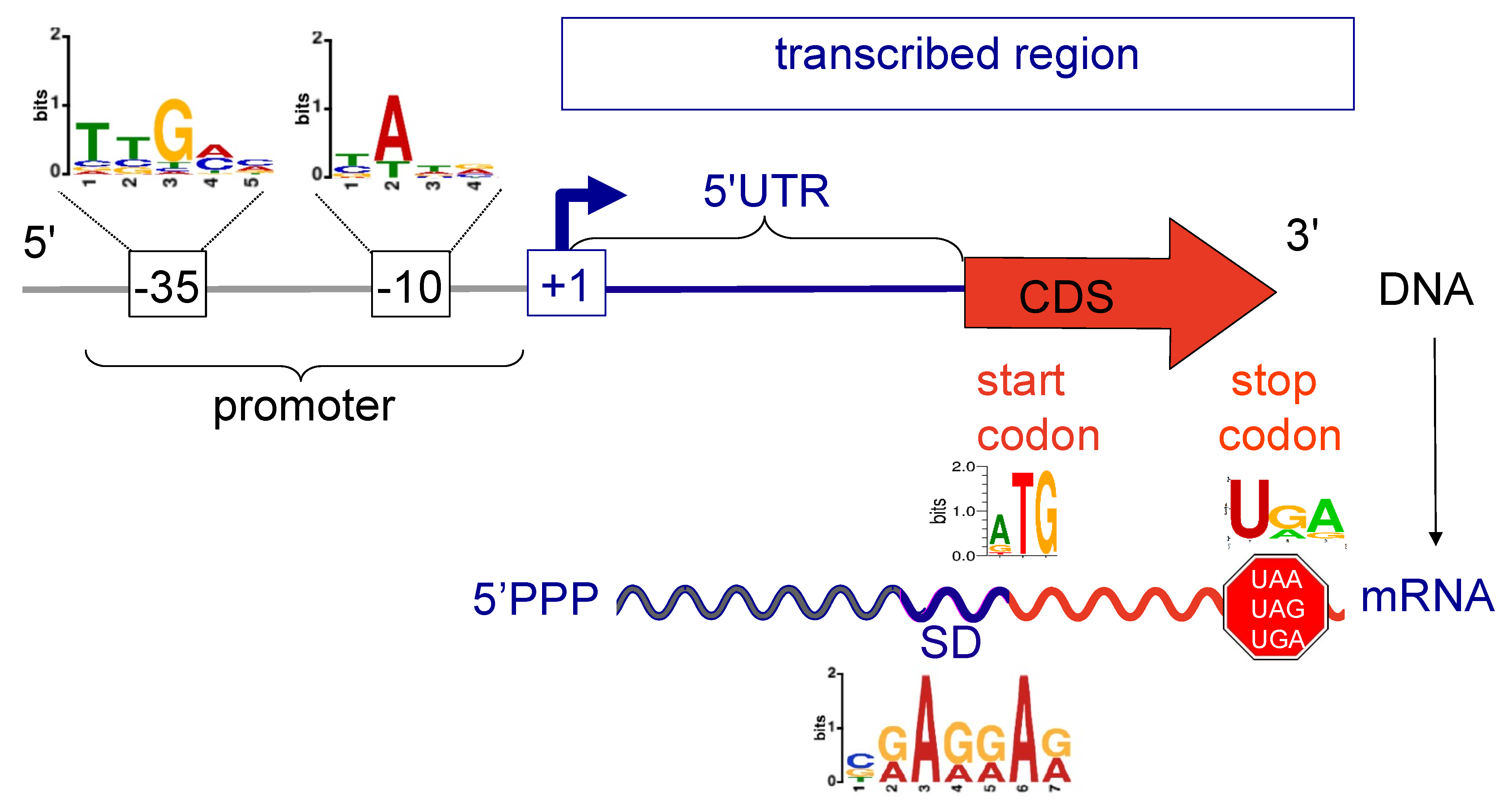
| Bank | Methanol | Dichloromethane | ||
|---|---|---|---|---|
| Replicate 1 | Replicate 2 | Replicate 1 | Replicate 2 | |
| “+” | 0.6 × 106 | 0.3 × 106 | 0.8 × 106 | 0.6 × 106 |
| (9%) | (5%) 1 | (14%) 1 | (8%) 1 | |
| “−” | 5.7 × 106 | 4.0 × 106 | 2.2 × 106 | 3.0 × 106 |
| (76%) | (74%) | (51%) | (64%) | |
| Gene | Product | Label in MaGe 1 | Promoter Region 2 | 5-URT (nt) | Approach 3 | Comments | ||
|---|---|---|---|---|---|---|---|---|
| −35 | −10 + 1 | /Name | ||||||
| dcmA | DCM dehalogenase | METDI2656 | TTGACA <16 nt> TATAGAactagccc G/PA TTGACA <16 nt> TATAGAactagc C/PA TTGACA <17 nt> TATAGTcaagtc A/PA | 173 175 122 | dRNA-seq nuclease S1 PE | M. extorquens DM4 (this study). TSS position 2562877 (plus) M. extorquens DM4 [16]. TSS identified from cloned DNA Methylophilus sp. DM11 [23] | ||
| dcmR | transcriptional regulator of DCM dehalogenase | METDI2655 | TTGCGC <17 nt> TAACTAcaagg G/PR1 TTGCGC <17 nt> TAACTAcaagggtct C/PR1 TTTACT <16 nt> TTTACTcatcgg A/PR2 TTTACT <16 nt> TTTACTcatcgg A/PR2 | 66 62 157 157 | dRNA-seq nuclease S1 dRNA-seq nuclease S1 | M. extorquens DM4 (this study). TSS position 2562332 (minus) M. extorquens DM4 [16]. TSS identified from a cloned DNA M. extorquens DM4 (this study). TSS position 2562423 (minus) M. extorquens DM4 [16]. TSS identified from cloned DNA | ||
| glyA | serine hydroxyl-methyltransferase | METDI3959 META1_3384 | TTGGCC <18 nt> ACGAATagtgc C ATCACC <16 nt> TGCCGCggcgtgta C | 75 84 | dRNA-seq PE | M. extorquens DM4 (this study). TSS position 3888433 (minus) M. extorquens AM1 [19]. Other TSS with 63 and 38 nt 5′UTR | ||
| lspA4 | lipoprotein signal peptidase | METDI3108 META1_2328 (old name ORF181) | TTCCCC <17 nt> TAGAAGcgctcca A/PileS TCGACG <19 nt> GGTGCCcgagcggg C/Porf181 | 150 129 | dRNA-seq PE | M. extorquens DM4 (this study). TSS position 3023525 (plus), 150 nt upstream of ileS. No TSS found upstream (0.9 kb) lspA M. extorquens AM1. Porf181 deduced from a faint primer extension band, with low promoter activity (225 bp upstream of orf181pqqFG fused to xylE [18] | ||
| mxaF | methanol dehydrogenase alpha SU precursor | METDI5145 META1p4538 nd 5 | AAGACA <18 nt> TAGAAAatatag G AAGACA <18 nt> TAGAAAatata GG AAGACA <18 nt> TAGAAAcgat A | 168 167–168 170 | dRNA-seq nuclease S1, run-off PE | M. extorquens DM4 (this study). TSS position 5068109 (minus) M. extorquens AM1 [15]. Gene mxaF also named moxF M. organophilum XX [21] | ||
| mxaW | uncharacterized conserved exported protein | METDI5146 META1p4539 nd | TTGACC <18 nt> ACCGTTgtcgtcAacgggC TTGGCA <nd> ACCCAT <nd> G 6 TTGACC <18 nt> ACCACTaggcgg A | 41 52 54 | dRNA-seq PE PE | M. extorquens DM4 (this study). TSS position 5068271 (plus) M. extorquens AM1 [18]. Gene mxaW also named moxW M. organophilum XX [22] | ||
| mtkA | malate thiokinase large SU | METDI2482 META1_1730 | TTCCCG <17 nt> GAAGGTcggcccaa C TTGAGA <19 nt> AGTAATttttcc G/Pqsc2-1 AAGTCA < 7 > AAGAAAaattga G/Pqsc2-2 | 31 46 80 | dRNA-seq PE | M. extorquens DM4 (this study). TSS position 2393413 (plus) M. extorquens AM1 of operon qsc2 (mtkA-mtkB-ppc-mcl) [19]. Predominance of the two TSS varies upon growth conditions | ||
| pqqA | coenzyme PQQ biosynthesis protein A | METDI2503 META1_1751 | TGGCGC <19 nt> TGATGGcgcc A/PmxbM TTGCAG <16 nt> CGATATacctccg G/PpqqD | 119 95 | dRNA-seq PE | M. extorquens DM4 (this study). TSS position 2416334 (minus) upstream of adjacent gene mxbM (METDI2504). No TSS detected upstream of METDI2503 M. extorquens AM1 [17]. Promoter checked using xylE fusion | ||
| sga | serine-glyoxylate aminotransferase | METDI2478 META1_1726 | TTGCGC <16 nt> CGGGATcgccccc G/Psga GTGCCC <18 nt> CCGGCAgaggtg C/Pqsc1 TTGAAT <17 nt> CATCGAgggtt C/Pqsc1 | 46 356 343 | dRNA-seq PE | M. extorquens DM4 (this study). TSS position 2388788 (plus, P class) and 2388435 (plus, O class) M. extorquens AM1 of operon qsc1 (sga-hpr-mtdA-fch) [19] | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maucourt, B.; Roche, D.; Chaignaud, P.; Vuilleumier, S.; Bringel, F. Genome-Wide Transcription Start Sites Mapping in Methylorubrum Grown with Dichloromethane and Methanol. Microorganisms 2022, 10, 1301. https://doi.org/10.3390/microorganisms10071301
Maucourt B, Roche D, Chaignaud P, Vuilleumier S, Bringel F. Genome-Wide Transcription Start Sites Mapping in Methylorubrum Grown with Dichloromethane and Methanol. Microorganisms. 2022; 10(7):1301. https://doi.org/10.3390/microorganisms10071301
Chicago/Turabian StyleMaucourt, Bruno, David Roche, Pauline Chaignaud, Stéphane Vuilleumier, and Françoise Bringel. 2022. "Genome-Wide Transcription Start Sites Mapping in Methylorubrum Grown with Dichloromethane and Methanol" Microorganisms 10, no. 7: 1301. https://doi.org/10.3390/microorganisms10071301
APA StyleMaucourt, B., Roche, D., Chaignaud, P., Vuilleumier, S., & Bringel, F. (2022). Genome-Wide Transcription Start Sites Mapping in Methylorubrum Grown with Dichloromethane and Methanol. Microorganisms, 10(7), 1301. https://doi.org/10.3390/microorganisms10071301






