Enhancing Biocide Efficacy: Targeting Extracellular DNA for Marine Biofilm Disruption
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microorganisms
2.2. Sample Preparation and Surface Finish
2.3. Experimental Setup
2.4. Control Assessment and Inhibitor Dosing
2.5. Confocal Laser Scanning Microscopy (CLSM) and Post-Image Analysis
2.6. Scanning Electron Microscopy (SEM)
2.7. Standard Biofilm Colony Forming Unit (CFU) Enumeration
2.8. Biofilm Adenosine Triphosphate (ATP) Assays
3. Results
3.1. Biofilm Controls
3.2. Inhibitor Treatments
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Little, B.J.; Lee, J.S. Microbiologically Influenced Corrosion; Wiley-Interscience: Hoboken, NJ, USA, 2007. [Google Scholar]
- Skovhus, T.L.; Enning, D.; Lee, J.S. Microbiologically Influenced Corrosion in the Upstream Oil and Gas Industry, 1st ed.; Routledge: Milton, ON, Canada, 2017; Volume 1. [Google Scholar]
- Li, Y.; Xu, D.; Chen, C.; Li, X.; Jia, R.; Zhang, D.; Sand, W.; Wang, F.; Gu, T. Anaerobic microbiologically influenced corrosion mechanisms interpreted using bioenergetics and bioelectrochemistry: A review. J. Mater. Sci. Technol. 2018, 34, 1713–1718. [Google Scholar] [CrossRef]
- Santos, A.L.S.D.; Galdino, A.C.M.; Mello, T.P.d.; Ramos, L.d.S.; Branquinha, M.H.; Bolognese, A.M.; Neto, J.C.; Roudbary, M. What are the advantages of living in a community? A microbial biofilm perspective! Mem. Inst. Oswaldo. Cruz. 2018, 113, e180212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 76. [Google Scholar] [CrossRef]
- Nielsen, P.H.; Jahn, A.; Palmgren, R. Conceptual model for production and composition of exopolymers in biofilms. Water Sci. Technol. 1997, 36, 11–19. [Google Scholar] [CrossRef]
- Desmond, P.; Best, J.P.; Morgenroth, E.; Derlon, N. Linking composition of extracellular polymeric substances (EPS) to the physical structure and hydraulic resistance of membrane biofilms. Water Res. 2018, 132, 211–221. [Google Scholar] [CrossRef]
- Sutherland, I.W. Biofilm exopolysaccharides: A strong and sticky framework. Microbiology 2001, 147, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Whitchurch, C.B.; Tolker-Nielsen, T.; Ragas, P.C.; Mattick, J.S. Extracellular DNA Required for Bacterial Biofilm Formation. Science 2002, 295, 1487. [Google Scholar] [CrossRef]
- Mann, E.E.; Rice, K.C.; Boles, B.R.; Endres, J.L.; Ranjit, D.; Chandramohan, L.; Tsang, L.H.; Smeltzer, M.S.; Horswill, A.R.; Bayles, K.W. Modulation of eDNA Release and Degradation Affects Staphylococcus aureus Biofilm Maturation. PLoS ONE 2009, 4, e5822. [Google Scholar] [CrossRef] [Green Version]
- Thomas, V.C.; Hiromasa, Y.; Harms, N.; Thurlow, L.; Tomich, J.; Hancock, L.E. A fratricidal mechanism is responsible for eDNA release and contributes to biofilm development of Enterococcus faecalis. Mol. Microbiol. 2009, 72, 1022–1036. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Liu, X.; Liu, H.; Zhang, L.; Guo, Y.; Yu, S.; Wozniak, D.J.; Ma, L.Z. The exopolysaccharide Psl–eDNA interaction enables the formation of a biofilm skeleton in Pseudomonas aeruginosa. Environ. Microbiol. Rep. 2015, 7, 330–340. [Google Scholar] [CrossRef] [Green Version]
- Novotny, L.A.; Amer, A.O.; Brockson, M.E.; Goodman, S.D.; Bakaletz, L.O. Structural Stability of Burkholderia cenocepacia Biofilms Is Reliant on eDNA Structure and Presence of a Bacterial Nucleic Acid Binding Protein. PLoS ONE 2013, 8, e67629. [Google Scholar] [CrossRef] [PubMed]
- Jakubovics, N.S.; Shields, R.C.; Rajarajan, N.; Burgess, J.G. Life after death: The critical role of extracellular DNA in microbial biofilms. Lett. Appl. Microbiol. 2013, 57, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Płaza, G.; Achal, V. Biosurfactants: Eco-Friendly and Innovative Biocides against Biocorrosion. Int. J. Mol. Sci. 2020, 21, 2152. [Google Scholar] [CrossRef] [Green Version]
- Tuck, B.; Watkin, E.; Forsyth, M.; Somers, A.; Ghorbani, M.; Machuca, L.L. Evaluation of a novel, multi-functional inhibitor compound for prevention of biofilm formation on carbon steel in marine environments. Sci. Rep. 2021, 11, 15697. [Google Scholar] [CrossRef]
- Pakiet, M.; Kowalczyk, I.; Leiva Garcia, R.; Moorcroft, R.; Nichol, T.; Smith, T.; Akid, R.; Brycki, B. Gemini surfactant as multifunctional corrosion and biocorrosion inhibitors for mild steel. Bioelectrochemistry 2019, 128, 252–262. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Catubig, R.A.; Neil, W.C.; McAdam, G.; Yunis, R.; Forsyth, M.; Somers, A.E. Multifunctional Inhibitor Mixtures for Abating Corrosion on HY80 Steel under Marine Environments. J. Electrochem. Soc. 2020, 167, 21503. [Google Scholar] [CrossRef]
- Ghorbani, M.; Puelles, J.S.; Forsyth, M.; Catubig, R.A.; Ackland, L.; Machuca, L.; Terryn, H.; Somers, A.E. Corrosion Inhibition of Mild Steel by Cetrimonium trans-4-Hydroxy Cinnamate: Entrapment and Delivery of the Anion Inhibitor through Speciation and Micellar Formation. J. Phys. Chem. Lett. 2020, 11, 9886–9892. [Google Scholar] [CrossRef]
- Wang, H.; Chen, M.; Jin, C.; Niu, B.; Jiang, S.; Li, X.; Jiang, S. Antibacterial [2-(Methacryloyloxy) ethyl] Trimethylammonium Chloride Functionalized Reduced Graphene Oxide/Poly(ethylene-co-vinyl alcohol) Multilayer Barrier Film for Food Packaging. J. Agric. Food Chem. 2018, 66, 732–739. [Google Scholar] [CrossRef]
- Liu, G.; Wu, G.; Jin, C.; Kong, Z. Preparation and antimicrobial activity of terpene-based polyurethane coatings with carbamate group-containing quaternary ammonium salts. Prog. Org. Coat. 2015, 80, 150–155. [Google Scholar] [CrossRef]
- Seter, M.; Thomson, M.J.; Chong, A.; MacFarlane, D.R.; Forsyth, M. Cetrimonium Nalidixate as a Multifunctional Inhibitor to Combat Biofilm Formation and Microbiologically Influenced Corrosion. Aust. J. Chem. 2013, 66, 921–929. [Google Scholar] [CrossRef]
- Catubig, R.; Michalczyk, A.; Neil, W.; McAdam, G.; Forsyth, J.; Ghorbani, M.; Yunis, R.; Ackland, L.; Forsyth, M.; Somers, A. Multifunctional Inhibitor Mixture for Reducing Bacteria Growth and Corrosion on Marine Grade Steel. ChemRxiv. Camb. Camb. Open Engage 2021. [Google Scholar] [CrossRef]
- Tuck, B.; Leinecker, N.; Watkin, E.; Somers, A.; Forsyth, M.; Machuca, L.L. Efficiency of a Novel Multifunctional Corrosion Inhibitor Against Biofilms Developed on Carbon Steel. Front. Bioeng. Biotechnol. 2022, 10, 803559. [Google Scholar] [CrossRef] [PubMed]
- Morente, E.O.; Fernández-Fuentes, M.A.; Burgos, M.J.G.; Abriouel, H.; Pulido, R.P.; Gálvez, A. Biocide tolerance in bacteria. Int. J. Food Microbiol. 2013, 162, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Salgar-Chaparro, S.J.; Darwin, A.; Kaksonen, A.H.; Machuca, L.L. Carbon steel corrosion by bacteria from failed seal rings at an offshore facility. Sci. Rep. 2020, 10, 12287. [Google Scholar] [CrossRef] [PubMed]
- Salgar-Chaparro, S.J.; Machuca, L.L.; Lepkova, K.; Pojtanabuntoeng, T.; Darwin, A. Investigating the Effect of Temperature in the Community Structure of an Oilfield Microbial Consortium, and Its Impact on Corrosion of Carbon Steel. In Proceedings of the Corrosion 2019, Nashville, TN, USA, 24–28 May 2019. [Google Scholar]
- Da Silva, N.; Taniwaki, M.H.; Junqueira, V.C.A.; de Arruda Silveira, N.F.; Okazaki, M.M.; Gomes, R.A.R. Microbiological Examination Methods of Food and Water: A Laboratory Manual; CRC Press: Boca Raton, FL, USA, 2018. [Google Scholar]
- Eguchi, M.; Nishikawa, T.; Macdonald, K.; Cavicchioli, R.; Gottschal, J.C.; Kjelleberg, S. Responses to Stress and Nutrient Availability by the Marine Ultramicrobacterium Sphingomonas sp. Strain RB2256. Appl. Environ. Microbiol. 1996, 62, 1287–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero, J.L.; Burgos, M.J.G.; Pérez-Pulido, R.; Gálvez, A.; Lucas, R. Resistance to Antibiotics, Biocides, Preservatives and Metals in Bacteria Isolated from Seafoods: Co-Selection of Strains Resistant or Tolerant to Different Classes of Compounds. Front. Microbiol. 2017, 8, 1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, K.; Pagedar Singh, A. Antibiofilm Effect of DNase against Single and Mixed Species Biofilm. Foods 2018, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Okshevsky, M.; Regina, V.R.; Meyer, R.L. Extracellular DNA as a target for biofilm control. Curr. Opin. Biotechnol. 2015, 33, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, R.; Banu, S.F.; Sowndarya, J.; Parveen, J.H.S.; Rubini, D.; Wilson, A.; Nithyanand, P. Biosurfactant synergized with marine bacterial DNase disrupts polymicrobial biofilms. Folia Microbiol. 2021, 66, 831–842. [Google Scholar] [CrossRef]
- Yan, Z.; Huang, M.; Melander, C.; Kjellerup, B.V. Dispersal and inhibition of biofilms associated with infections. J. Appl. Microbiol. 2020, 128, 1279–1288. [Google Scholar] [CrossRef] [Green Version]
- Waryah, C.B.; Wells, K.; Ulluwishewa, D.; Chen-Tan, N.; Gogoi-Tiwari, J.; Ravensdale, J.; Costantino, P.; Gökçen, A.; Vilcinskas, A.; Wiesner, J.; et al. In Vitro Antimicrobial Efficacy of Tobramycin Against Staphylococcus aureus Biofilms in Combination With or Without DNase I and/or Dispersin B: A Preliminary Investigation. Microb. Drug Resist. 2017, 23, 384–390. [Google Scholar] [CrossRef] [PubMed]
- Shukla, S.K.; Rao, T.S. Staphylococcus aureus biofilm removal by targeting biofilm-associated extracellular proteins. Indian J. Med. Res. 2017, 146, S1–S8. [Google Scholar] [CrossRef] [PubMed]
- Yarawsky, A.E.; Johns, S.L.; Schuck, P.; Herr, A.B. The biofilm adhesion protein Aap from Staphylococcus epidermidis forms zinc-dependent amyloid fibers. J. Biol. Chem. 2020, 295, 4411–4427. [Google Scholar] [CrossRef] [PubMed]
- Jennings, L.K.; Storek, K.M.; Ledvina, H.E.; Coulon, C.; Marmont, L.S.; Sadovskaya, I.; Secor, P.R.; Tseng, B.S.; Scian, M.; Filloux, A.; et al. Pel is a cationic exopolysaccharide that cross-links extracellular DNA in the Pseudomonas aeruginosa biofilm matrix. Proc. Natl. Acad. Sci. USA 2015, 112, 11353–11358. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Liu, H.; Mohamed, M.E.-S.; Saleh, M.A.; Gu, T. Mitigation of sulfate reducing Desulfovibrio ferrophilus microbiologically influenced corrosion of X80 using THPS biocide enhanced by Peptide A. J. Mater. Sci. Technol. 2022, 107, 43–51. [Google Scholar] [CrossRef]
- Wilson, C.; Lukowicz, R.; Merchant, S.; Valquier-Flynn, H.; Caballero, J.; Sandoval, J.; Okuom, M.; Huber, C.; Brooks, T.D.; Wilson, E.; et al. Quantitative and Qualitative Assessment Methods for Biofilm Growth: A Mini-review. Res. Rev. J. Eng. Technol. 2017, 6, 1–25. [Google Scholar]
- Pereira, B.M.P.; Salim, M.A.; Rai, N.; Tagkopoulos, I. Tolerance to Glutaraldehyde in Escherichia coli Mediated by Overexpression of the Aldehyde Reductase YqhD by YqhC. Front. Microbiol. 2021, 12, 680553. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Røder, H.L.; Madsen, J.S.; Bjarnsholt, T.; Sørensen, S.J.; Burmølle, M. Interspecific Bacterial Interactions are Reflected in Multispecies Biofilm Spatial Organization. Front. Microbiol. 2016, 7, 1366. [Google Scholar] [CrossRef] [Green Version]
- Elias, S.; Banin, E. Multi-species biofilms: Living with friendly neighbors. FEMS Microbiol. Rev. 2012, 36, 990–1004. [Google Scholar] [CrossRef]
- Nocker, A.; Cheung, C.-Y.; Camper, A.K. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J. Microbiol. Methods 2006, 67, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Nakata, K.; Tsuchido, T.; Matsumura, Y. Antimicrobial cationic surfactant, cetyltrimethylammonium bromide, induces superoxide stress in Escherichia coli cells. J. Appl. Microbiol. 2011, 110, 568–579. [Google Scholar] [CrossRef] [PubMed]
- Guéroult, M.; Picot, D.; Abi-Ghanem, J.; Hartmann, B.; Baaden, M. How Cations Can Assist DNase I in DNA Binding and Hydrolysis. PLoS Comput. Biol. 2010, 6, e1001000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Flint, S.; Palmer, J. Magnesium and calcium ions: Roles in bacterial cell attachment and biofilm structure maturation. Biofouling 2019, 35, 959–974. [Google Scholar] [CrossRef] [PubMed]


 ), are compared against CTA-4OHcinn treated biofilms (
), are compared against CTA-4OHcinn treated biofilms (  ) and dual CTA-4OHcinn and DNase treated biofilms (
) and dual CTA-4OHcinn and DNase treated biofilms (  ). Error bars represent the standard deviation of the data.
). Error bars represent the standard deviation of the data.
 ), are compared against CTA-4OHcinn treated biofilms (
), are compared against CTA-4OHcinn treated biofilms (  ) and dual CTA-4OHcinn and DNase treated biofilms (
) and dual CTA-4OHcinn and DNase treated biofilms (  ). Error bars represent the standard deviation of the data.
). Error bars represent the standard deviation of the data.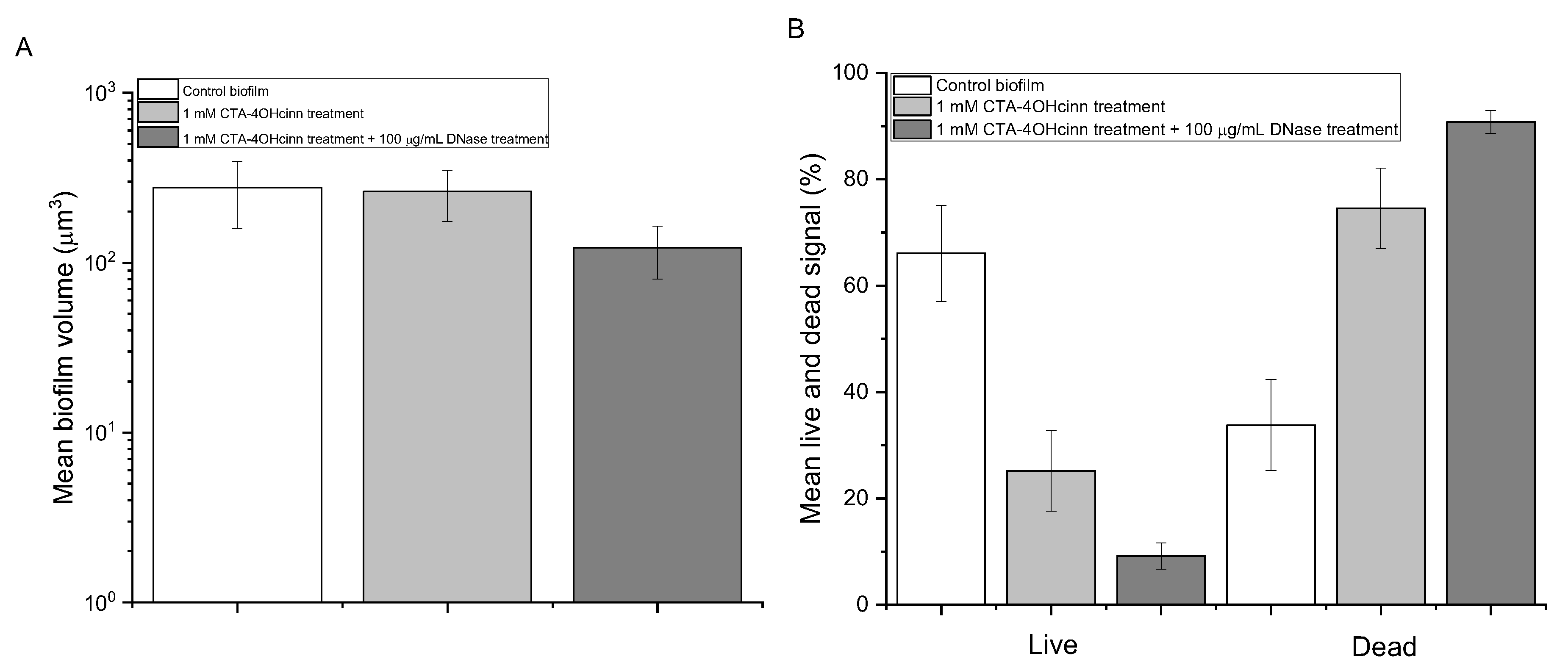

 ), 1 mM CTA-4OHcinn (
), 1 mM CTA-4OHcinn (  ) and 1 mM 1 mM CTA-4OHcinn + DNase 1 enzyme (
) and 1 mM 1 mM CTA-4OHcinn + DNase 1 enzyme (  ). Each bar represents measurements taken from triplicate biofilms with two technical replicates. Error bars represent the standard deviation of this data.
). Each bar represents measurements taken from triplicate biofilms with two technical replicates. Error bars represent the standard deviation of this data.
 ), 1 mM CTA-4OHcinn (
), 1 mM CTA-4OHcinn (  ) and 1 mM 1 mM CTA-4OHcinn + DNase 1 enzyme (
) and 1 mM 1 mM CTA-4OHcinn + DNase 1 enzyme (  ). Each bar represents measurements taken from triplicate biofilms with two technical replicates. Error bars represent the standard deviation of this data.
). Each bar represents measurements taken from triplicate biofilms with two technical replicates. Error bars represent the standard deviation of this data.
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tuck, B.; Watkin, E.; Somers, A.; Forsyth, M.; Machuca, L.L. Enhancing Biocide Efficacy: Targeting Extracellular DNA for Marine Biofilm Disruption. Microorganisms 2022, 10, 1227. https://doi.org/10.3390/microorganisms10061227
Tuck B, Watkin E, Somers A, Forsyth M, Machuca LL. Enhancing Biocide Efficacy: Targeting Extracellular DNA for Marine Biofilm Disruption. Microorganisms. 2022; 10(6):1227. https://doi.org/10.3390/microorganisms10061227
Chicago/Turabian StyleTuck, Benjamin, Elizabeth Watkin, Anthony Somers, Maria Forsyth, and Laura L. Machuca. 2022. "Enhancing Biocide Efficacy: Targeting Extracellular DNA for Marine Biofilm Disruption" Microorganisms 10, no. 6: 1227. https://doi.org/10.3390/microorganisms10061227
APA StyleTuck, B., Watkin, E., Somers, A., Forsyth, M., & Machuca, L. L. (2022). Enhancing Biocide Efficacy: Targeting Extracellular DNA for Marine Biofilm Disruption. Microorganisms, 10(6), 1227. https://doi.org/10.3390/microorganisms10061227








