Abstract
Enzymes purified from psychrophilic microorganisms prove to be efficient catalysts at low temperatures and possess a great potential for biotechnological applications. The low-temperature catalytic activity has to come from specific structural fluctuations involving the active site region, however, the relationship between protein conformational stability and enzymatic activity is subtle. We provide a survey of the thermodynamic stability of globular proteins and their rationalization grounded in a theoretical approach devised by one of us. Furthermore, we provide a link between marginal conformational stability and protein flexibility grounded in the harmonic approximation of the vibrational degrees of freedom, emphasizing the occurrence of long-wavelength and excited vibrations in all globular proteins. Finally, we offer a close view of three enzymes: chloride-dependent α-amylase, citrate synthase, and β-galactosidase.
1. Introduction
Despite our conception, Earth is a rather cold place. Temperatures below 10 °C characterize about 80% of Earth’s biosphere [1], of which 70% is made up of oceans (see Figure 1). The remaining percentage includes the polar regions, the mountainous regions, the mesosphere, and the stratosphere. A small contribution is also given by man and therefore by artificial niches attributable to the use of refrigerators and freezers [2,3]. Microorganisms have been discovered capable of living in conditions of extremely low temperatures compared to what we define “ambient temperature” [4]. Organisms living in these environments are known as psychrophiles; some of them reach maximum growth even at temperatures below 0 °C. Psychrophiles are mostly localized in marine ecosystems, particularly in the abysses, where the water temperature is permanently cold. Both cell lines and bacterial cultures can be stored in liquid nitrogen, where the temperature is −196 °C. Despite this, the temperature recordings carried out in monitoring the activities of these psychrophilic microorganisms did not report values below −20 °C [5]. There is an increasing interest in studying psychrophilic microorganisms to discover their adaptation mechanisms. The extreme environments are increasingly seen as a source of new microbes and biomolecules that could have potential biotechnological and biomedical applications [6,7,8]. For example: (a) enzymes active in the cold have been identified in microbial cultures from marine environments of polar regions [9,10]; (b) antibiotic-producing bacteria have been isolated from permafrost soils in the upper Arctic regions [11]. It is evident that psychrophiles have developed evolutionary defense mechanisms that support their survival in such inhospitable environments. The adaptation mechanisms allow these organisms to effectively cope with cold. Furthermore, physiological and genomic adaptations, such as the biosynthesis of cryoprotectants, changes in membrane composition and the functioning of enzymes, provide mechanisms to compensate for the kinetic and thermodynamic effects of low temperatures [12,13]. To maintain optimal membrane fluidity, for example, the cell must regulate membrane composition as well as the quantity and type of lipids (saturated or unsaturated). Since the membrane fluidity decreases as the temperature decreases, microorganisms show an increase in the ratio of unsaturated to saturated fatty acids. Furthermore, both the structure and functions of proteins are known to be influenced by temperature [14]. Thermophilic proteins are characterized by an increase in the content of ion-pairs, by the formation of higher-order oligomers and by a decrease in structural flexibility [15]. The latter seems to be the consequence of a decrease in surface loop length, optimization of electrostatic and van der Waals interactions, and amino acid substitutions aimed at increasing internal hydrophobicity [16,17,18].
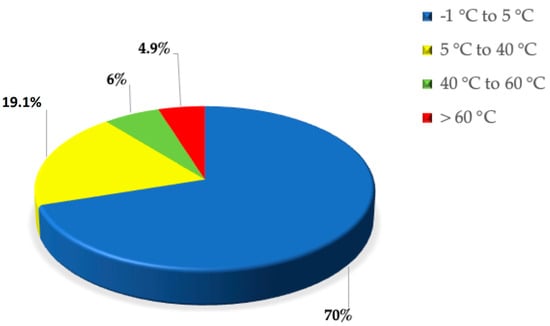
Figure 1.
Pie–plot of Earth temperature distribution.
On the other hand, the ability to withstand sub-zero temperatures relies on two strategies. The first one is to avoid the formation of ice in order to protect cells from freezing, synthetizing specific proteins called ice binding proteins, IBP [19,20]; the second one is to develop protective mechanisms to avoid damage during the thawing phase in case of still formed ice [5,21]. The proteins used in both processes are called “antifreeze” molecules and allow the freezing point of water to be lowered by more than 10 °C. The adaptation mechanisms of psychrophilic enzymes are different from those preferred by thermophilic ones [22]. The random thermal energy conferring structural flexibility and mobility to a protein chain is small at low temperature, nevertheless, the enzyme activity has to be as high as necessary to sustain the microorganism’s life. Thus, it is expected that psychrophilic enzymes have a greater structural flexibility to increase the catalytic rate constant, and a higher degree of conformational complementarity with substrates to allow a decrease in activation energy, when compared to mesophilic and thermophilic homologues [23]. In order to shed light on these fundamental issues and to be sure that significant differences emerge, several structural comparative studies have been performed on enzyme families belonging to psychrophiles, mesophiles and thermophiles [24,25,26,27,28,29,30,31,32,33,34,35]. Even though adaptation strategies are family-specific, the following common rules hold for psychrophilic proteins: (a) the structural differences between the folded states are small and subtle, notwithstanding large differences in thermal stability; (b) there are several charged residues on the surface of psychrophilic enzymes to increase solubility and flexibility, not the conformational stability; (c) the interior packing, on the average, does not change; (d) there is a higher percentage of tiny residues (i.e., Ala, Gly, Ser, and Thr), whose small side chains make psychrophilic enzymes less compact and more flexible than the mesophilic and thermophilic counterparts. In the present study we provide: (a) a view of metabolism adaptation to cope with cold environments; (b) a study of the thermodynamic characteristics of globular proteins and their conformational stability in general, with a specific application of the so-called geometric model to psychrophilic proteins; (c) an analysis of enzymatic activity with a specific focus on psychrophilic enzymes and closer inspection of the adaptation mechanisms of α-amylase, citrate synthase and β-galactosidase. It is worth noting that enzymes from psychrophilic microorganisms are largely used in industrial applications due to their high catalytic activity at low temperature [36,37]. For example, psychrophilic lipases play an important role in allowing a reduced washing temperature in laundry industry and so a significant energy saving; psychrophilic xylanases are a key ingredient to improve the bread quality for their ability to degrade the beta-1,4-xylan; psychrophilic α-amylases are largely used in food industry, for example to avoid haze formation in juice, and in baking industry processes [38,39,40]. Moreover, a DNA polymerase I, isolated from the psychrophilic Psychromonas grahamii, has very good catalytic activity at low temperature and is of potential interest for sequencing companies [41]. Readers interested in industrial and biotechnological applications of psychrophilic enzymes are referred to reviews specifically devoted to this topic [33,42,43,44,45].
2. Metabolism Adaptation
A Biokinetic Spectrum for Temperature, recently determined [46], represents the distribution of microorganism growth rate as a function of the environmental temperature, and covers a temperature range greater than 120 °C, emphasizing that all the biosphere hosts living organisms. This Biokinetic Spectrum for Temperature shows the maximum growth rate, at each temperature, for any life form on Earth, and it vividly clarifies that the growth rate is very small at low temperature [47,48]. Therefore, psychrophiles, the organisms capable of adapting to the coldest ecosystems on Earth and able to survive in conditions of low temperature, deserve great attention. The evolution of these organisms has made it possible to develop mechanisms that allow an adjustment of metabolic activities in adverse situations as, for example, the slowdown of enzymatic activities or modification in the function of proteins involved in metabolic cycles [7,49]. Cold shock largely influences cellular processes including: protein synthesis, absorption of nutrients, cell growth and membrane fluidity [50,51]. In very cold environments it is necessary to develop adaptive characteristics at the level of functionality and cellular structure [52]. Among these, one of the most important aspects concerns the alterations in expressed genes and, consequently, in proteins involved in specific metabolic pathways [26,53,54]. Cold shock leads to an up-regulation of genes coding for cold shock proteins (CSP). These proteins are expressed in mesophilic organisms when exposed to cold, while, in psychrophilic microorganisms, they are constitutively expressed and are classified as cold acclimatization proteins (CAP) [7]. Several studies have revealed that some psychrophilic communities are located in small ice pockets with high saline concentrations, rich in organic material that supports the population growth [55]. In some cases, the microorganism metabolism has to be compatible with temperatures below the freezing point of water [25]. Recent studies have shown that the induction of thermal stress drastically reduces the metabolic activity and that only a small portion of proteins is able to adapt to low temperatures [56]. Usually, in very cold environments, reactive oxygen species (ROS) accumulation is more pronounced. It has been found that genes coding for antioxidant enzymes, such as catalase and superoxide dismutase (SOD), are more expressed in psychrophiles [39,57]. Linked to an evident slowing down of the biosynthetic pathways, this is one of the regulatory mechanisms through which these organisms are able to avoid damage due to ROS accumulation [3,58]. In psychrophiles, the genes coding for proteins involved in flagellar motility and energy metabolism are down-regulated [59,60], while it has been evidenced an up-regulation in pathways involved in arginine degradation, ROS neutralization, and biosynthesis of proline, polyamines, and unsaturated fatty acids [61]; this is highlighted in Figure 2. Furthermore, a marked reduction in the expression levels of oxidative stress-related proteins was recorded in Pseudoalteromonas haloplanktis under cold stress growth conditions [9]. In addition to the repression of ROS-generating oxidative metabolic pathways, it was reported that the production level of several osmo-protectants [62], such as trehalose, increased [61]. The production of trehalose represents an adaptation mechanism in protecting microorganisms against a wide variety of lethal conditions [63,64]. It has been shown that, in cold stress, some cytoprotective polyamines are produced to stabilize nucleic acids and neutralize ROS [65]. Some of these polyamines come from arginine degradation [66], accomplished by the arginine decarboxylase enzyme in psychrophilic Pseudomonas helmanticensis [61].
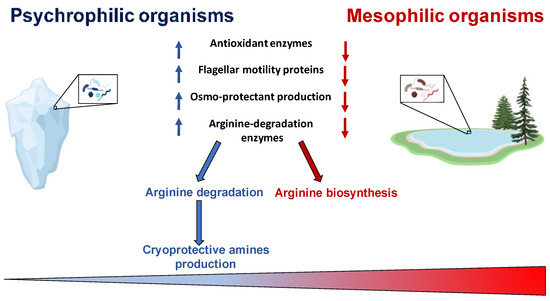
Figure 2.
Metabolic adaptation. Image created with a template from BioRender.com.
Following thermal shock, organisms trigger adaptive cryo-protection changes to reduce cell permeability and mitochondrial swelling [67]. The latter mitochondrial swelling occurs in mesophiles, but is not observed in psychrophiles. Another cryo-protection mechanism is the accumulation of proline. Proline is a membrane stabilizing agent, making direct interactions with phospholipids and rendering them less sensitive to low temperatures [68].
3. Thermodynamic Features
To gain the right perspective on enzymes extracted from psychrophilic microorganisms, it is necessary and useful to account for the main results obtained about the conformational stability of globular proteins in general. Circular dichroism (CD) and differential scanning calorimetry (DSC) measurements of globular protein thermal stability allowed the determination of the denaturation temperature, Td, the denaturation enthalpy change, ΔHd(Td), and the denaturation heat capacity change, ΔCp,d, for a large number of globular proteins and their mutant forms [69,70]. Normally, the temperature-induced denaturation of small globular proteins (i.e., single domain globular proteins; a structural domain should consist of less than 200 residues [71]) is a reversible and cooperative process, corresponding to a first-order phase transition between the native and denatured macro-states (N-state and D-state, respectively); it is an all-or-none, cooperative process [72]. The knowledge of the three thermodynamic parameters listed above, assuming ΔCp,d temperature-independent, makes it possible to calculate the denaturation Gibbs free energy change, ΔGd = G(D-state) − G(N-state), as a function of temperature, using the Gibbs-Helmholtz equation. The trend of ΔGd as a function of temperature is called the protein stability curve [73]. In the temperature range where ΔGd is positive, the N-state is stable; in the temperature range where ΔGd is negative, the D-state is stable; the temperature at which ΔGd = 0 corresponds to the denaturation temperature of the protein, where the two states have the same thermodynamic stability (i.e., the equilibrium constant Kd = [D-state]/[N-state] = exp[−ΔGd(Td)/RTd] = 1, and so the two macro-states are equally populated). In general, since ΔCp,d is a large positive quantity, the protein stability curve has a parabolic shape with two temperatures at which ΔGd = 0, corresponding to the unusual cold denaturation and the common hot denaturation, respectively. The former is unusual because: (a) it occurs on lowering the temperature from ambient conditions, and is characterized by a negative enthalpy change and a negative entropy change; (b) in the case of stable globular proteins, it occurs well below 0 °C, and so is not easy to study. Nevertheless, cold denaturation has directly been detected using DSC, CD and NMR measurements on destabilized mutant forms, or on wild-type proteins in destabilizing conditions, such as aqueous urea solutions [74,75]. The common hot denaturation occurs on increasing the temperature, is characterized by large positive enthalpy and entropy changes, and is driven by the large gain in conformational entropy of the polypeptide chain associated with unfolding. The Td values of mesophilic proteins are spread over a wide temperature range with an average at 60 °C, well above the living temperature of mesophiles [76]. Similarly, for the small number of psychrophilic proteins whose temperature-induced denaturation has carefully been investigated, the Td values are clustered around 40°C [26], well above the living temperature of psychrophiles. It seems that the N-state of both psychrophilic and mesophilic globular proteins is more stable than it would be strictly necessary. A survey over experimental data of a large set of globular proteins [77] has emphasized, in line with previous analyses [76,78], that the maximum of the stability curve is located at Tmax = 284 ± 19 K, and ΔGd(Tmax) ≈ 20–40 kJ mol−1 for a protein of about 100 residues. These two heuristic findings indicate that: (a) the maximal thermodynamic stability occurs around room temperature, regardless of the Td value, in all probability because the amino acid content of all proteins is similar, on the average; (b) the N-state is only marginally more stable than the D-state because a delicate balance between large stabilizing and large destabilizing interactions holds, and depends on temperature [69,76,78]. It is possible to calculate the stability curve for a mesophilic model protein and a psychrophilic model protein, both consisting of 100 residues, exploiting the relationships for mesophilic proteins reported by Robertson & Murphy [76]. For the psychrophilic model protein, the Td and ΔHd(Td) numbers have suitably been lowered with respect to those of Robertson & Murphy, whereas ΔCp,d has not been modified in view of the close structural similarity between a mesophilic enzyme and its psychrophilic homologue [27]. Specifically: (i) Td = 333 K, ΔHd(Td) = 292 kJ mol−1 and ΔCp,d = 5.8 kJ K−1 mol−1 for the mesophilic model; (ii) Td = 313 K, ΔHd(Td) = 176 kJ mol−1 and ΔCp,d = 5.8 kJ K−1 mol−1 for the psychrophilic model. The two calculated stability curves are shown in Figure 3; they practically possess the same Tmax ≈ 285 K, and ΔGd(Tmax) is around 8 kJ mol−1 for the psychrophilic model, and around 21 kJ mol−1 for the mesophilic model (note that in both cases cold denaturation occurs at very low temperatures). The two stability curves shown in Figure 3 closely resemble those obtained by Feller and colleagues for a psychrophilic α-amylase and its mesophilic counterpart [79]. It is clear that the knowledge of the macroscopic thermodynamic quantities associated with denaturation does not provide molecular-level information about the interactions stabilizing the N-state and those stabilizing the D-state. Since the magnitude of ΔCp,d, ΔHd, and ΔSd linearly depends on the number of protein residues [76,77], a mean-field theoretical approach, neglecting the subtleties of amino acid sequence and the differences in secondary structure content, can be successful in clarifying some important features of the conformational stability of globular proteins. A geometric model is able to explain the existence of cold denaturation [80,81], the extra-thermal stability of thermophilic proteins on entropic grounds [82], and the high thermal stability of very small globular proteins [83]. We would like to apply such an approach to the conformational stability of psychrophilic proteins. The starting point of the approach is the recognition that the solvent-excluded volume effect plays a pivotal role in processes occurring in the liquid phase, that is a condensed state of matter. The presence of a solute molecule reduces the configurational space accessible to solvent molecules because no other molecule can occupy the space occupied by the solute molecule. Cavity creation at a fixed position in the liquid is the theoretical means to account for this basic fact [84,85]. Cavity creation at a fixed position, keeping constant temperature and pressure, leads to an increase in liquid volume by the partial molar volume of the cavity; this, however, does not remove the solvent-excluded volume effect. The solvent molecule centers cannot penetrate the solvent-accessible-surface of the cavity if the latter has to exist [80]. The solvent-excluded volume is given by the shell between the van der Waals surface of the cavity and its solvent-accessible-surface, and its measure can be provided by the water-accessible-surface-area, WASA, of the cavity [86]. This means that the size of the statistical ensemble of liquid configurations is markedly reduced by cavity creation: relevant configurations are solely the ones in which it does exist a cavity suitable to host the solute molecule. The latter are a tiny fraction of the total in the case of molecular-sized cavities [87]. A loss in translational entropy of water molecules in associated with a decrease in accessible configurational space [84]. It is interesting that the solvent-excluded volume effect can be quantified by a geometric measure such as the WASA of the cavity hosting the solute. In this respect, proteins are special, because, being polymers, can populate conformations characterized by largely different WASA values, causing different solvent-excluded volume effects.
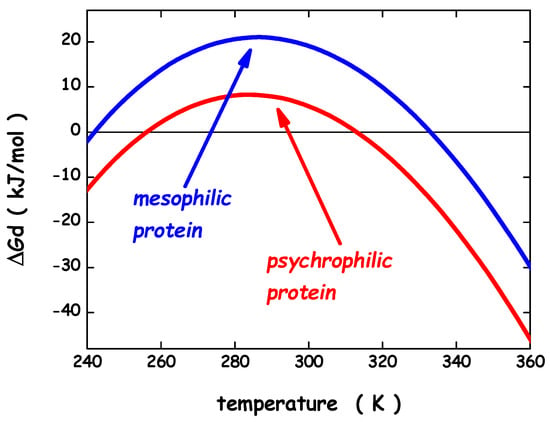
Figure 3.
Thermodynamic stability curves for a model psychrophilic protein and a model mesophilic one. See text for further details.
It is the shape of the polypeptide chain to distinguish the conformations belonging to the N-state, from those belonging to the D-state. The simplest geometric choice is: a sphere can model the N-state, and a prolate spherocylinder can model the D-state. The two objects have equal VvdW, (i.e., the volume practically does not change on unfolding at P = 1 atm [88,89]), but different WASA (see Figure 4). The spherocylinder has larger WASA because it is well-known that, with a fixed volume, the sphere is the 3D object possessing the smallest surface area. As a consequence, the solvent-excluded volume experienced by water molecules will be different. The minimization of the loss in translational entropy is the reason why water molecules stabilize the N-state. Indeed, with a fixed cavity VvdW, the ΔGc magnitude rises with cavity WASA [90]. Thus, it is possible to calculate the difference between ΔGc to create in water a cavity suitable to host the prolate spherocylinder corresponding to the D-state, and ΔGc to create in water a cavity suitable to host the sphere corresponding to the N-state. The ΔΔGc = ΔGc(D-state) − ΔGc(N-state) contribution is always positive, stabilizing the N-state; it practically accounts for what is usually called the hydrophobic effect [91]. The large gain in conformational entropy upon denaturation destabilizes the N-state. It can be calculated by T⋅ΔSconf = T⋅Nres⋅ΔSconf(res), assuming equal and additive contributions by the chain residues. This rough approximation works well because it has been found that the denaturation entropy change ΔSd is a linear function of Nres [76,77]; note that ΔSconf is a large part of ΔSd.

Figure 4.
Simple geometric models for the two states of a globular protein: the sphere for the N-state and the prolate spherocylinder for the D-state.
Apart the two previous entropic contributions, it is necessary to account for an energetic term that differentiates the two macro-states; ΔEa = [Ea(D-state) − Ea(N-state) + ΔE(intra)] is the difference in energetic interactions among the D-state and the N-state and surrounding water molecules, and the difference in intra-chain energetic interactions between the D-state and the N-state [81]. This energetic contribution should not be large because: (a) the H-bonds that peptide groups make with water molecules in the D-state are largely reformed as intra-protein H-bonds in the secondary structure elements of the N-state; (b) side chains able to make H-bonds usually form them both with water molecules in the D-state, and intra-molecularly in the N-state; a H-bond satisfaction principle holds [92]; (c) van der Waals attractions between protein groups and water molecules are regained as intra-protein contacts in the close-packed interior of the N-state. In particular, a complete balance between protein-water energetic attractions in the D-state and N-state, respectively, and the intra-protein energetic attractions implies that ΔEa = 0. In this case, the ΔEa quantity should not contribute to the overall Gibbs free energy balance; this situation does appear realistic. Considering experimental data for 115 different globular proteins [77], the denaturation enthalpy change ΔHd proves to be zero at a temperature TH = (277.5 ± 25) K, corresponding to the characteristic maximum density temperature of H2O [93]; for detailed explanations the reader is referred to [82,83]. On this basis, it is suitable to assume that: (a) the ΔEa energetic term is temperature-independent; (b) ΔEa = 0 for a mesophilic model protein; (c) ΔEa < 0 for a psychrophilic model protein (i.e., energetic factors favor the D-state). This last point is reliable because structural comparative analyses have shown that the N-state interior of psychrophilic proteins possesses, on average, a smaller number of stabilizing energetic attractions (i.e., H-bonds and van der Waals contacts) with respect to mesophilic and thermophilic homologues [27,29]. Indeed, both Kanaya and co-workers on ribonuclease HI from the psychrotrophic bacterium Shewanella oneidensis MR-1 [94], and Feller and co-workers on the α-amylase from the Antarctic bacterium P. haloplanktis [95], have shown that, inserting in the cold-active protein some selected residues present in the mesophilic counterpart, thus allowing the formation of additional attractive interactions among side chains, the protein thermal stability increases significantly, and in an almost additive way. The latter feature is because each mutation affords a small contribution to the overall Gibbs free energy balance. The energetic destabilization mechanism should be the molecular basis of the higher structural flexibility attributed to psychrophilic enzymes. The approach provides the following formula for the denaturation Gibbs free energy change (note that the N-state is stable when ΔGd is positive):
ΔGd = ΔΔGc + ΔEa − T⋅ΔSconf
It is important to underscore that the ΔEa quantity does not correspond to ΔHd (i.e., the quantity carefully measured in a DSC experiment), because the latter takes into account also the enthalpy contribution coming from the reorganization of water-water H-bonds associated with protein denaturation. This reorganization process, according to different theoretical arguments and routes [96,97,98,99,100], produces compensating enthalpy and entropy changes. Since the structures of psychrophilic proteins are closely similar to those of mesophilic and thermophilic homologues [27], the same sphere can model their N-state. On similar grounds, the same prolate spherocylinder can model their D-state. The geometric sizes are: the sphere has radius a = 15 Å, VvdW = 14137 Å3 and WASA = 3380 Å2 (calculated using for the rolling sphere a radius of 1.4 Å [86]); the prolate spherocylinder has radius a = 6 Å, cylindrical length l = 117 Å, VvdW = 14137 Å3 and WASA = 6128 Å2. These values should represent a protein of 138 residues [101]. The analytical formulas of classic scaled particle theory [90,102] allow the calculation of the ΔΔGc function, using the experimental density of water [93]. The effective, temperature-independent, hard sphere diameter of water molecules has been fixed to σ (H2O) = 2.80 Å [81,84,103]. The conformational entropy contribution is calculated by fixing Nres = 138 and ΔSconf(res) = 19.0 J K−1 molres−1. The latter value is right for a mesophilic model protein [104], and should be suitable also for a psychrophilic model protein. For the latter, the energetic term has deliberately been fixed at ΔEa = −13 kJ mol−1 to account for the energetic destabilization emerged from structural comparative analyses [27,28]. It is important to underscore that the magnitude of the ΔEa energetic term is very small in comparison to that of the two entropic contributions; for instance, at 10 °C, ΔΔGc ≈ 768 kJ mol−1 and T⋅ΔSconf ≈ 743 kJ mol−1. This is because the energetic balance between the D-state and the N-state is always in action, even though it is not always complete. The calculated ΔΔGc function (the black curve in Figure 5) has a parabolic temperature dependence and its magnitude does not depend on the mesophilic or psychrophilic origin of the protein since their N-states and D-states share the same geometric sizes.
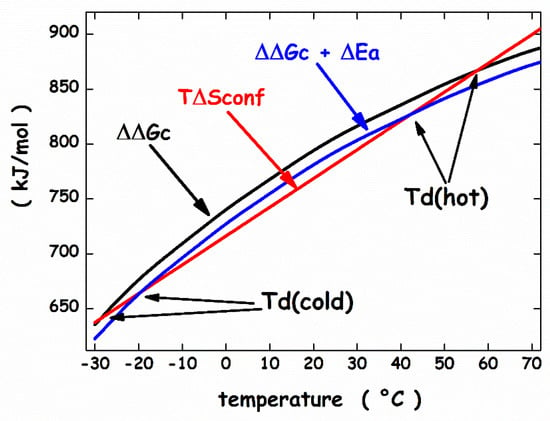
Figure 5.
Temperature dependence of the functions, ΔGc, ΔΔGc + ΔEa, and T⋅ΔSconf, calculated as described in the text.
The WASA increase associated with denaturation largely stabilizes the N-state. The ΔΔGc increase with temperature is because the water density is almost temperature-independent, and the cavity surface has to contrast the kinetic energy of bombarding water molecules [105,106]. The T⋅ΔSconf straight line (the red line in Figure 5) intersects the ΔΔGc parabola in two points that represent the temperature of cold denaturation and the temperature of hot denaturation, respectively. The subtraction of the T⋅ΔSconf straight line from the ΔΔGc function leads to a stability curve (the black line in Figure 6) reliable for a mesophilic model protein, with Td(hot) ≈ 57 °C.
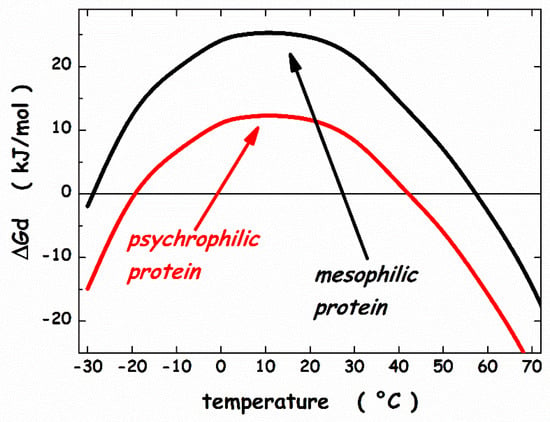
Figure 6.
Thermodynamic stability curves calculated by means of the geometric theoretical approach. See text for further details.
On the other hand, the same T⋅ΔSconf straight line intersects the ΔΔGc + ΔEa function (the blue curve in Figure 5) at two points that are shifted at higher temperature for cold denaturation and lower temperature for hot denaturation. The obtainaed stability curve (the red line in Figure 6) is reliable for a psychrophilic model protein, with Td(hot) ≈ 42 °C and a lower ΔGd at the maximum stability temperature. In particular, Tmax ≈ 10 °C and ΔGd(Tmax) ≈ 25 kJ mol−1 for the mesophilic model protein, while Tmax ≈ 10 °C and ΔGd(Tmax) ≈ 12 kJ mol−1 for the psychrophilic model protein. These numbers agree with experimental ones. It can be concluded that: (a) all globular proteins are characterized by a subtle balance between stabilizing and destabilizing terms [69,78]; (b) the inclusion of a destabilizing energetic term in the framework of the geometric theoretical approach leads to a model protein with psychrophilic features.
4. Enzymatic Adaptation
First Somero [107,108], and soon after Jaenicke [109] proposed that enzymes from microorganisms adapted to different environmental temperatures work in “corresponding states” conditions. Since the substrate-binding sites are strictly conserved from the structural point of view [26], and since to catalyze a given biochemical reaction the requested vibrations (i.e., structural movements) are always the same, a closely similar flexibility has to be associated with the psychrophilic enzyme at 5–20 °C, the mesophilic enzyme at 35–50 °C, and the thermophilic enzyme at 75–90 °C. The “corresponding states” hypothesis refers to the functioning of enzymes, not to their conformational stability. This hypothesis is reliable and has received some experimental confirmation [110]. It would be important to link the emerged marginal thermodynamic stability of the N-state with the significant structural fluctuations characterizing globular proteins. Such structural fluctuations can be described as vibrational normal modes: a molecule consisting of N atoms possesses 3N degrees of freedom, (3N − 6) of which are associated with vibrations, that can be treated as normal modes in the harmonic approximation. Clearly, the number of vibrational normal modes is very large also for a small globular protein. Their frequency distribution (i.e., the vibrational density of states, VDOS) should depend upon the molecular structure, and so should be different between the N-state and the D-state; more importantly, it is expected to be different among psychrophilic, mesophilic, and thermophilic enzymes. Actually, Song and colleagues [111] calculated the VDOS of 135 different globular proteins, starting from their crystallographic structures, and showed that it is universal, on average. The calculated VDOS is characterized by a large number of normal modes with frequency below 300 cm−1. Since RT = 2.5 kJ mol−1 at 300 K, and the latter energy corresponds to a frequency of 208 cm−1, a large number of vibrational normal modes are populated and excited at room temperature. This means that the N-state of globular proteins is highly dynamic and fluctuating, notwithstanding its ability to preserve a single average structure. Molecular vibrations, especially those involving the substrate-binding site, are necessary to perform catalytic activity [112]. Those useful for the catalytic function possess high frequency (i.e., greater than 1000 cm−1), and small wavelength, and should be localized in the substrate-binding region [113]. Note that a mechanism of energy transfer from the accessible normal modes (those with frequency smaller than 300 cm−1) to the un-accessible ones (those with a frequency greater than 1000 cm−1) has to be operative and should merit special attention. These functional vibrations should have the same degree of excitation at the “corresponding” working temperature of the psychrophilic, mesophilic and thermophilic enzyme (i.e., should possess a similar excess of energy to speed up the reaction). It should be clear that, if the enzymatic activity is used to interrogate and measure protein flexibility at room temperature, psychrophilic enzymes do appear flexible, whereas thermophilic ones do appear rigid [25,26,30]. However, this picture is not strictly correct or complete. As indicated by the universality of the calculated VDOS [111], the N-state of thermophilic enzymes is surely characterized by low frequency and long wavelength vibrational normal modes also at room temperature, whereas its functional high frequency and small wavelength vibrational normal modes are not populated at room temperature, but become sufficiently populated and excited only at higher temperature (note that RT ≈ 2.2 kJ mol−1 at 270 K, 2.5 kJ mol−1 at 300 K, and 3.1 kJ mol−1 at 373 K). In contrast, the N-state of psychrophilic enzymes has to be characterized by a sufficient population and excitation of functional high frequency and small wavelength vibrational normal modes also at very low temperatures. This fundamental dynamic property should be the consequence of subtle structural features, such as the absence of a strong coupling between the different parts of the folded structure for the presence of a higher fraction of residues with small side chains. The latter is emerged as a specific fingerprint of psychrophilic proteins in large-scale comparative investigations [27]. Clearly, relevant structural features can be more subtle and not so simple to unveil. For instance, cold adaptation driven by dynamic allostery (i.e., mutations in distant regions affect the functioning of the catalytic site) has been detected by means of both experimental and computational techniques in adenylate kinase [114], and malate dehydrogenase [115]. Now, we focus our attention on three specific enzymes: α-amylase (a monomer), citrate synthase (a dimer), and β-galactosidase (a tetramer).
4.1. α-Amylase
The family of cold-adapted enzymes more carefully investigated is surely that of chloride-dependent α-amylase. The latter enzyme is involved in the initial phase of starch digestion, specifically hydrolyzing starch into glucose. The research group of Feller and Gerday at the University of Liege has performed an in-depth characterization of the α-amylase from the psychrophilic microorganism P. haloplanktis, solving its 3D structure by means of X-ray diffraction [116,117], measuring its conformational stability by means of CD and DSC measurements, and characterizing its enzymatic properties [79,95,118,119]. A comparison between the kcat values at 20 °C of the protein from P. haloplanktis and of the mesophilic protein from Sus scrofa is shown in Figure 7, highlighting the marked difference in enzymatic activity, regardless of the substrate, at low temperature.
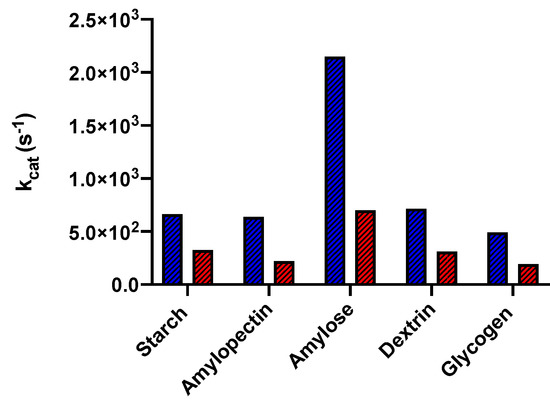
Figure 7.
Graphical representation of kcat values, at 20 °C, of α-amylase from the psychrophile P. haloplanktis (in blue) and from the mesophile S. scrofa (in red) on different substrates.
A careful comparison with mesophilic and thermophilic homologues has also been carried out, together with the production and characterization of several mutant forms [120,121]. The psychrophilic enzyme showed Td = 44 °C, both the mesophilic ones from D. melanogaster and S. scrofa showed Td = 58 °C, the thermophilic one from Thermobifida fusca showed Td = 77 °C [120]. Apart from the large difference in Td values, DSC measurements indicated that the temperature-induced denaturation of the psychrophilic enzyme is a cooperative, reversible two-state transition, whereas it is characterized by the presence of intermediate states in the case of mesophilic and thermophilic α-amylases [120]. The psychrophilic enzyme has maximal activity at 29 °C, and kcat = 179 s−1 at the physiological temperature of 5 °C; the mesophilic one from D. melanogaster has maximal activity at 54 °C, and kcat = 280 s−1 at the physiological temperature of 20 °C; the mesophilic one from S. scrofa has maximal activity at 54 °C, and kcat = 518 s−1 at the physiological temperature of 37 °C; the thermophilic one from T. fusca has maximal activity at 72 °C, and kcat = 1457 s−1 at the physiological temperature of 55 °C (in all cases the enzymatic activity has been measured using 1% soluble starch as substrate). These numbers underscore that the “corresponding states” hypothesis in the case of chloride-dependent α-amylases, even though qualitatively correct, is not quantitatively right. Since kcat increases exponentially with temperature (according to the Arrhenius equation), and the activation energy increases markedly on passing from the psychrophilic to the thermophilic enzyme [119], the latter possesses a kcat larger than that of the cold-adapted enzyme at the physiological temperature of the respective microorganisms. In all probability, the thermophilic chloride-dependent α-amylase is more efficient than the psychrophilic homologue because, at high temperatures, the availability of more thermal energy stored in the vibrational normal modes allows a marked increase in the reaction rate. Feller and colleagues have also engineered several point mutations in the psychrophilic α-amylase based on the structure of the mesophilic homologue from S. scrofa. The inserted additional interactions (i.e., H-bonds, salt-bridges, and van der Waals contacts) caused both an increase in the thermal stability of the mutant proteins, and a significant decrease of the catalytic activity at low temperature [121]. These findings confirm the strict relationships existing between conformational stability, chain dynamics, and enzymatic activity [122].
4.2. Citrate Synthase
The enzyme citrate synthase (CS) catalyzes the entry of carbon, as acetyl-CoA, into the citric acid cycle, and occurs in almost all organisms. CS has a characteristic dimeric structure [123,124]; each monomer consists of a small and a large domain, with the central part of the dimer formed by the association of the two large domains. In each monomer, the gap between the two domains forms an active site that binds oxaloacetate first, followed by acetyl-CoA, leading to citrate formation. A substantial movement of the small domain with respect to the large domain is involved in the catalytic function, closing and opening the flaps, as the substrate binds to the active site slot. The research group of Danson and Taylor at the University of Bath solved the 3D structure of five CSs from psychrophilic, mesophilic, and thermophilic sources [123,125,126,127]. Specifically, they solved the structures of: (a) the enzyme from the psychrophilic Arthrobacter DS2–3R, whose optimum growth temperature is 31 °C, ArCS; (b) the enzyme from pig, whose optimum growth temperature is 37 °C, PigCS; (c) the enzyme from Thermoplasma acidophilum, whose optimum growth temperature is 55 °C, TaCS; (d) the enzyme from Sulfolobus solfataricus, whose optimum growth temperature is 85 °C, SsCS; (e) the enzyme from Pyrococcus furiosus, whose optimum growth temperature is 100 °C, PfCS. The native structure of the five CSs proves to be very similar: the root mean square deviation between the Cα atoms for the complete dimeric structures solved via X-ray diffraction is at most 2.3 Å, even though the percentage of sequence identity is not large (i.e., 27% between ArCS and PigCS, 40% between ArCS and PfCS). The native structure of the five CSs possesses practically the same WASA, but there is a clear difference in the percentage of exposed hydrophobic WASA: the latter amounts to 29% in psychrophilic ArCS, 20% in mesophilic PigCS, and 18% in hyperthermophilic PfCS. There are a lot of ionic interactions (i.e., both isolated ion pairs and charge-charge networks) in both psychrophilic ArCS and thermophilic SsCS and PfCS. In the former, they occur mainly on the monomer surface, whereas, in the latter, they occur mainly at the interface between the two monomers. It appears that the stabilization of the dimer interface via ionic interactions is important to increase the thermal resistance in thermophilic CSs; in contrast, the significant presence of ionic interactions in the psychrophilic ArCS should not be important for the stability of the native state, but it should play an important role for protein solubility in aqueous solution at low temperature [127]. The non-trivial relationship between enzymatic activity and thermal stability was confirmed by performing point mutations in the active site region of ArCS, inspired by the structure of the hyperthermophilic PfCS. On reducing the active site accessibility, the enzymatic activity at low temperature does not decrease and the thermal resistance increases [124].
4.3. β-Galactosidase
The enzyme β-galactosidase catalyzes the hydrolysis of lactose into its constituents monosaccharides, glucose, and galactose [128], and has attracted the interest of both researchers and the dairy industry due to nutritional, technological, and environmental problems associated with this important carbohydrate [129]. It is also used as a medical enzyme for producing Galalcto-Oligosaccharides, important for intestinal health [130]. The low-temperature catalytic activity of psychrophilic β-galactosidase is of relevant interest for potential industrial applications in diminishing lactose levels to avoid disorders in lactose-intolerant people [131]. Other applications are aimed at the preservation of heat-labile compounds [132,133]. A β-galactosidase was characterized by a strain of the Antarctic bacterium P. haloplanktis by the research group of Feller and Gerday at the University of Liege [134]. The psychrophilic microorganism shows the maximum production of β-galactosidase at around 4 °C. Such psychrophilic β-galactosidase proves to be very similar from the structural point of view with the mesophilic β-galactosidase from E. coli. The two enzymes have a tetrameric structure with a comparable subunit mass (each subunit consists of more than 1000 residues); they share 51% of sequence identity, the strict requirement for divalent metal ions (i.e., in line with the presence of two bound Mg2+ ions per monomer in the structure of the E. coli enzyme [135]), and a strong conservation of the amino acid residues constituting the active site. However, at 25 °C, the catalytic constants towards the natural substrate lactose are dramatically different: kcat = 33 s−1 for the enzyme from P. haloplanktis, and 2 s−1 for the enzyme from E. coli. In addition, upon 50 min of incubation at 4 °C, the psychrophilic enzyme is able to hydrolyze 33% of milk lactose, whereas the mesophilic one hydrolyzes only 12% of milk lactose. The surprise is that the thermal stability of the two proteins is not so different: the denaturation temperature is 50 °C for the enzyme from P. haloplanktis and 57 °C for the enzyme from E. coli. This confirms that the temperature dependence of the conformational stability and that of the enzymatic activity are governed by different rules. It is worth noting that recently: (a) a hexameric β-galactosidase has been isolated from the psychrophilic Marinomonas ef1, and has shown remarkable stability against temperature, notwithstanding its highest activity at 5 °C [136]; (b) a dimeric cold-active β-galactosidase has been isolated from Arthrobacter sp. 32cB, and is considered of relevant interest for food industry [137,138].
5. Conclusions
Psychrophilic enzymes represent a challenge to try to rationalize the relationship between their high catalytic activity at low temperature and their not-so-large conformational stability. The present analysis has offered some insight into the thermodynamics of the conformational stability of globular proteins, linking the marginal stability of the N-state to its large structural fluctuations. The latter have been discussed in the light of the harmonic approximation to describe the vibrational degrees of freedom of a molecule. In general, the enzymatic activity is associated with small wavelength vibrational normal modes localized in the active site region. Psychrophilic enzymes possess a sufficient excitation of such small wavelength vibrational normal modes to render operative the catalytic function at very low temperatures. This property should come from specific structural features that are difficult to decipher in view of the strong similarity existing between the native structure of homologous proteins from psychrophiles, mesophiles, and thermophiles.
Author Contributions
Conceptualization, G.G.; methodology, G.G. and R.R.; software, R.R.; validation, G.G. and R.R.; writing—original draft preparation, R.R. and G.G.; writing—review and editing, R.R. and G.G.; supervision, G.G.; project administration, G.G.; funding acquisition, G.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Università degli Studi del Sannio, FRA 2021.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Coker, J.A. Recent advances in understanding extremophiles. F1000Res 2019, 8, 1917. [Google Scholar] [CrossRef] [PubMed]
- Casanueva, A.; Tuffin, M.; Cary, C.; Cowan, D.A. Molecular adaptations to psychrophily: The impact of ‘omic’ technologies. Trends Microbiol. 2010, 18, 374–381. [Google Scholar] [CrossRef]
- Margesin, R.; Miteva, V. Diversity and ecology of psychrophilic microorganisms. Res. Microbiol. 2011, 162, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Murray, A.E.; Kenig, F.; Fritsen, C.H.; McKay, C.P.; Cawley, K.M.; Edwards, R.; Kuhn, E.; McKnight, D.M.; Ostrom, N.E.; Peng, V.; et al. Microbial life at −13 degrees C in the brine of an ice-sealed Antarctic lake. Proc. Natl. Acad. Sci. USA 2012, 109, 20626–20631. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.; Morris, G.J.; Fonseca, F.; Murray, B.J.; Acton, E.; Price, H.C. A Low Temperature Limit for Life on Earth. PLoS ONE 2013, 8, e66207. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, D.F.; Tiedje, J.M. Coping with our cold planet. Appl. Environ. Microbiol. 2008, 74, 1677–1686. [Google Scholar] [CrossRef]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef]
- Rizvi, A.; Ahmed, B.; Khan, M.S.; Umar, S.; Lee, J. Psychrophilic Bacterial Phosphate-Biofertilizers: A Novel Extremophile for Sustainable Crop Production under Cold Environment. Microorganisms 2021, 9, 2451. [Google Scholar] [CrossRef] [PubMed]
- Piette, F.; D’Amico, S.; Mazzucchelli, G.; Danchin, A.; Leprince, P.; Feller, G. Life in the cold: A proteomic study of cold-repressed proteins in the antarctic bacterium pseudoalteromonas haloplanktis TAC125. Appl. Environ. Microbiol. 2011, 77, 3881–3883. [Google Scholar] [CrossRef]
- Duarte, A.W.F.; Dos Santos, J.A.; Vianna, M.V.; Vieira, J.M.F.; Mallagutti, V.H.; Inforsato, F.J.; Wentzel, L.C.P.; Lario, L.D.; Rodrigues, A.; Pagnocca, F.C.; et al. Cold-adapted enzymes produced by fungi from terrestrial and marine Antarctic environments. Crit. Rev. Biotechnol. 2018, 38, 600–619. [Google Scholar] [CrossRef] [PubMed]
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167. [Google Scholar] [CrossRef]
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Tendulkar, S.; Hattiholi, A.; Chavadar, M.; Dodamani, S. Psychrophiles: A journey of hope. J. BioSci. 2021, 46, 64. [Google Scholar] [CrossRef] [PubMed]
- Jaenicke, R. Stability and folding of ultrastable proteins: Eye lens crystallins and enzymes from thermophiles. Faseb J. 1996, 10, 84–92. [Google Scholar] [CrossRef]
- Khan, S.; Farooq, U.; Kurnikova, M. Exploring Protein Stability by Comparative Molecular Dynamics Simulations of Homologous Hyperthermophilic, Mesophilic, and Psychrophilic Proteins. J. Chem. Inf. Model. 2016, 56, 2129–2139. [Google Scholar] [CrossRef]
- Peak, M.J.; Robb, F.T.; Peak, J.G. Extreme resistance to thermally induced DNA backbone breaks in the hyperthermophilic archaeon Pyrococcus furiosus. J. Bacteriol. 1995, 177, 6316–6318. [Google Scholar] [CrossRef][Green Version]
- Del Vecchio, P.; Graziano, G.; Granata, V.; Barone, G.; Mandrich, L.; Manco, G.; Rossi, M. Temperature- and denaturant-induced unfolding of two thermophilic esterases. Biochemistry 2002, 41, 1364–1371. [Google Scholar] [CrossRef] [PubMed]
- Granata, V.; Graziano, G.; Ruggiero, A.; Raimo, G.; Masullo, M.; Arcari, P.; Vitagliano, L.; Zagari, A. Stability against temperature of Sulfolobus solfataricus elongation factor 1 alpha, a multi-domain protein. Bioch. Biophys. Acta 2008, 1784, 573–581. [Google Scholar] [CrossRef]
- Bialkowska, A.; Majewska, E.; Olczak, A.; Twarda-Clapa, A. Ice Binding Proteins: Diverse Biological Roles and Applications in Different Types of Industry. Biomolecules 2020, 10, 274. [Google Scholar] [CrossRef]
- Munoz, P.A.; Marquez, S.L.; Gonzalez-Nilo, F.D.; Marquez-Miranda, V.; Blamey, J.M. Structure and application of antifreeze proteins from Antarctic bacteria. Microb. Cell Fact. 2017, 16, 138. [Google Scholar] [CrossRef]
- Eskandari, A.; Leow, T.C.; Rahman, M.B.A.; Oslan, S.N. Antifreeze Proteins and Their Practical Utilization in Industry, Medicine, and Agriculture. Biomolecules 2020, 10, 1649. [Google Scholar] [CrossRef] [PubMed]
- Russell, N.J. Toward a molecular understanding of cold activity of enzymes from psychrophiles. Extremophiles 2000, 4, 83–90. [Google Scholar] [CrossRef]
- Arcus, V.L.; Mulholland, A.J. Temperature, Dynamics, and Enzyme-Catalyzed Reaction Rates. Annu. Rev. Biophys. 2020, 49, 163–180. [Google Scholar] [CrossRef] [PubMed]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Molecular basis of cold adaptation. Cell. Mol. Life Sci. 1997, 53, 830–841. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Claverie, P.; Collins, T.; Georlette, D.; Gratia, E.; Hoyoux, A.; Meuwis, M.A.; Feller, G.; Gerday, C. Molecular basis of cold adaptation. Philos. Trans. R Soc. Lond. B Biol. Sci. 2002, 357, 917–925. [Google Scholar] [CrossRef] [PubMed]
- Struvay, C.; Feller, G. Optimization to low temperature activity in psychrophilic enzymes. Int. J. Mol. Sci. 2012, 13, 11643–11665. [Google Scholar] [CrossRef]
- Gianese, G.; Bossa, F.; Pascarella, S. Comparative structural analysis of psychrophilic and meso- and thermophilic enzymes. Proteins 2002, 47, 236–249. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef]
- Jahandideh, S.; Abdolmaleki, P.; Jahandideh, M.; Barzegari Asadabadi, E. Sequence and structural parameters enhancing adaptation of proteins to low temperatures. J. Theor. Biol. 2007, 246, 159–166. [Google Scholar] [CrossRef]
- De Vendittis, E.; Castellano, I.; Cotugno, R.; Ruocco, M.R.; Raimo, G.; Masullo, M. Adaptation of model proteins from cold to hot environments involves continuous and small adjustments of average parameters related to amino acid composition. J. Theor. Biol. 2008, 250, 156–171. [Google Scholar] [CrossRef] [PubMed]
- Paredes, D.I.; Watters, K.; Pitman, D.J.; Bystroff, C.; Dordick, J.S. Comparative void-volume analysis of psychrophilic and mesophilic enzymes: Structural bioinformatics of psychrophilic enzymes reveals sources of core flexibility. BMC Struct. Biol. 2011, 11, 42. [Google Scholar] [CrossRef]
- Siddiqui, K.S. Defying the activity-stability trade-off in enzymes: Taking advantage of entropy to enhance activity and thermostability. Crit. Rev. Biotechnol. 2017, 37, 309–322. [Google Scholar] [CrossRef] [PubMed]
- Parvizpour, S.; Hussin, N.; Shamsir, M.S.; Razmara, J. Psychrophilic enzymes: Structural adaptation, pharmaceutical and industrial applications. Appl. Microbiol. Biotechnol. 2021, 105, 899–907. [Google Scholar] [CrossRef] [PubMed]
- Shokrollahzade, S.; Sharifi, F.; Vaseghi, A.; Faridounnia, M.; Jahandideh, S. Protein cold adaptation: Role of physico-chemical parameters in adaptation of proteins to low temperatures. J. Theor. Biol. 2015, 383, 130–137. [Google Scholar] [CrossRef] [PubMed]
- Metpally, R.P.; Reddy, B.V. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: Insights into the molecular basis of cold adaptation of proteins. BMC Genom. 2009, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mukhia, S.; Kumar, R. Industrial applications of cold-adapted enzymes: Challenges, innovations and future perspective. 3 Biotech. 2021, 11, 426. [Google Scholar] [CrossRef] [PubMed]
- Mhetras, N.; Mapare, V.; Gokhale, D. Cold Active Lipases: Biocatalytic Tools for Greener Technology. Appl. Biochem. Biotechnol. 2021, 193, 2245–2266. [Google Scholar] [CrossRef]
- Park, H.J.; Lee, C.W.; Kim, D.; Do, H.; Han, S.J.; Kim, J.E.; Koo, B.H.; Lee, J.H.; Yim, J.H. Crystal structure of a cold-active protease (Pro21717) from the psychrophilic bacterium, Pseudoalteromonas arctica PAMC 21717, at 1.4 A resolution: Structural adaptations to cold and functional analysis of a laundry detergent enzyme. PLoS ONE 2018, 13, e0191740. [Google Scholar] [CrossRef]
- Yang, G.; Mozzicafreddo, M.; Ballarini, P.; Pucciarelli, S.; Miceli, C. An In-Silico Comparative Study of Lipases from the Antarctic Psychrophilic Ciliate Euplotes focardii and the Mesophilic Congeneric Species Euplotes crassus: Insight into Molecular Cold-Adaptation. Mar. Drugs 2021, 19, 67. [Google Scholar] [CrossRef]
- Al-Ghanayem, A.A.; Joseph, B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 2020, 104, 2871–2882. [Google Scholar] [CrossRef]
- Xue, Y.; Braslavsky, I.; Quake, S.R. Temperature effect on polymerase fidelity. J. Biol. Chem. 2021, 297, 101270. [Google Scholar] [CrossRef] [PubMed]
- Feller, G. Psychrophilic enzymes: From folding to function and biotechnology. Scientifica 2013, 2013, 512840. [Google Scholar] [CrossRef] [PubMed]
- Dhaulaniya, A.S.; Balan, B.; Kumar, M.; Agrawal, P.K.; Singh, D.K. Cold survival strategies for bacteria, recent advancement and potential industrial applications. Arch. Microbiol. 2019, 201, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Furhan, J. Adaptation, production, and biotechnological potential of cold-adapted proteases from psychrophiles and psychrotrophs: Recent overview. J. Genet. Eng. Biotechnol. 2020, 18, 36. [Google Scholar] [CrossRef] [PubMed]
- Mesbah, N.M. Industrial Biotechnology Based on Enzymes From Extreme Environments. Front. Bioeng. Biotechnol. 2022, 10, 870083. [Google Scholar] [CrossRef]
- Corkrey, R.; Macdonald, C.; McMeekin, T. The Biokinetic Spectrum for Temperature and optimal Darwinian fitness. J. Theor. Biol. 2019, 462, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Corkrey, R.; McMeekin, T.A.; Bowman, J.P.; Ratkowsky, D.A.; Olley, J.; Ross, T. The Biokinetic Spectrum for Temperature. PLoS ONE 2016, 11, e0153343. [Google Scholar] [CrossRef]
- Corkrey, R.; McMeekin, T.A.; Bowman, J.P.; Olley, J.; Ratkowsky, D.; Ross, T. The maximum growth rate of life on Earth. Int. J. Astrobiol. 2018, 17, 17–33. [Google Scholar] [CrossRef]
- Tribelli, P.M.; Lopez, N.I. Reporting Key Features in Cold-Adapted Bacteria. Life 2018, 8, 8. [Google Scholar] [CrossRef]
- Phadtare, S. Recent developments in bacterial cold-shock response. Curr. Issues Mol. Biol. 2004, 6, 125–136. [Google Scholar]
- Collins, T.; Margesin, R. Psychrophilic lifestyles: Mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 2019, 103, 2857–2871. [Google Scholar] [CrossRef] [PubMed]
- Brininger, C.; Spradlin, S.; Cobani, L.; Evilia, C. The more adaptive to change, the more likely you are to survive: Protein adaptation in extremophiles. Semin. Cell Dev. Biol. 2018, 84, 158–169. [Google Scholar] [CrossRef] [PubMed]
- Piszkin, L.; Bowman, J. Extremophile enzyme optimization for low temperature and high salinity are fundamentally incompatible. Extremophiles 2021, 26, 5. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Lucas, J.; Acebron, I.; Wu, R.Y.; Alfaro, Y.; Acosta, J.; Kaminski, P.A.; Arroyo, M.; Joachimiak, A.; Nocek, B.P.; De la Mata, I.; et al. Biochemical and structural studies of two tetrameric nucleoside 2′-deoxyribosyltransferases from psychrophilic and mesophilic bacteria: Insights into cold-adaptation. Int. J. Biol. Macromol. 2021, 192, 138–150. [Google Scholar] [CrossRef]
- Mock, T.; Thomas, D.N. Recent advances in sea-ice microbiology. Environ. Microbiol. 2005, 7, 605–619. [Google Scholar] [CrossRef]
- Beckering, C.L.; Steil, L.; Weber, M.H.; Volker, U.; Marahiel, M.A. Genomewide transcriptional analysis of the cold shock response in Bacillus subtilis. J. Bacteriol. 2002, 184, 6395–6402. [Google Scholar] [CrossRef] [PubMed]
- Castellano, I.; Di Maro, A.; Ruocco, M.R.; Chambery, A.; Parente, A.; Di Martino, M.T.; Parlato, G.; Masullo, M.; De Vendittis, E. Psychrophilic superoxide dismutase from Pseudoalteromonas haloplanktis: Biochemical characterization and identification of a highly reactive cysteine residue. Biochimie 2006, 88, 1377–1389. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Yu, H.; Li, L.; Hu, S.; Dong, X. The genome and transcriptome of a newly described psychrophilic archaeon, Methanolobus psychrophilus R15, reveal its cold adaptive characteristics. Environ. Microbiol. Rep. 2012, 4, 633–641. [Google Scholar] [CrossRef]
- Gao, H.; Yang, Z.K.; Wu, L.; Thompson, D.K.; Zhou, J. Global transcriptome analysis of the cold shock response of Shewanella oneidensis MR-1 and mutational analysis of its classical cold shock proteins. J. Bacteriol. 2006, 188, 4560–4569. [Google Scholar] [CrossRef] [PubMed]
- Campanaro, S.; Williams, T.J.; Burg, D.W.; De Francisci, D.; Treu, L.; Lauro, F.M.; Cavicchioli, R. Temperature-dependent global gene expression in the Antarctic archaeon Methanococcoides burtonii. Environ. Microbiol. 2011, 13, 2018–2038. [Google Scholar] [CrossRef]
- Kumar, S.; Suyal, D.C.; Yadav, A.; Shouche, Y.; Goel, R. Psychrophilic Pseudomonas helmanticensis proteome under simulated cold stress. Cell Stress Chaperones 2020, 25, 1025–1032. [Google Scholar] [CrossRef]
- Hou, Y.; Qiao, C.; Wang, Y.; Wang, Y.; Ren, X.; Wei, Q.; Wang, Q. Cold-Adapted Glutathione S-Transferases from Antarctic Psychrophilic Bacterium Halomonas sp. ANT108: Heterologous Expression, Characterization, and Oxidative Resistance. Mar. Drugs 2019, 17, 147. [Google Scholar] [CrossRef]
- Kandror, O.; DeLeon, A.; Goldberg, A.L. Trehalose synthesis is induced upon exposure of Escherichia coli to cold and is essential for viability at low temperatures. Proc. Natl. Acad. Sci. USA 2002, 99, 9727–9732. [Google Scholar] [CrossRef]
- Kaushik, J.K.; Bhat, R. Why is trehalose an exceptional protein stabilizer? An analysis of the thermal stability of proteins in the presence of the compatible osmolyte trehalose. J. Biol. Chem. 2003, 278, 26458–26465. [Google Scholar] [CrossRef] [PubMed]
- Limsuwun, K.; Jones, P.G. Spermidine acetyltransferase is required to prevent spermidine toxicity at low temperatures in Escherichia coli. J. Bacteriol 2000, 182, 5373–5380. [Google Scholar] [CrossRef]
- Koh, H.Y.; Park, H.; Lee, J.H.; Han, S.J.; Sohn, Y.C.; Lee, S.G. Proteomic and transcriptomic investigations on cold-responsive properties of the psychrophilic Antarctic bacterium Psychrobacter sp. PAMC 21119 at subzero temperatures. Environ. Microbiol. 2017, 19, 628–644. [Google Scholar] [CrossRef] [PubMed]
- Strubbe-Rivera, J.O.; Schrad, J.R.; Pavlov, E.V.; Conway, J.F.; Parent, K.N.; Bazil, J.N. The mitochondrial permeability transition phenomenon elucidated by cryo-EM reveals the genuine impact of calcium overload on mitochondrial structure and function. Sci. Rep. 2021, 11, 1037. [Google Scholar] [CrossRef]
- Anchordoguy, T.J.; Rudolph, A.S.; Carpenter, J.F.; Crowe, J.H. Modes of interaction of cryoprotectants with membrane phospholipids during freezing. Cryobiology 1987, 24, 324–331. [Google Scholar] [CrossRef]
- Privalov, P.L. Stability of proteins: Small globular proteins. Adv. Protein Chem. 1979, 33, 167–241. [Google Scholar] [CrossRef]
- Makhatadze, G.I.; Privalov, P.L. Energetics of protein structure. Adv. Protein Chem. 1995, 47, 307–425. [Google Scholar] [CrossRef]
- Spassov, V.Z.; Karshikoff, A.D.; Ladenstein, R. The optimization of protein-solvent interactions: Thermostability and the role of hydrophobic and electrostatic interactions. Protein Sci. 1995, 4, 1516–1527. [Google Scholar] [CrossRef] [PubMed]
- Privalov, P.L. Thermodynamic problems of protein structure. Annu. Rev. Biophys. Biophys. Chem. 1989, 18, 47–69. [Google Scholar] [CrossRef] [PubMed]
- Becktel, W.J.; Schellman, J.A. Protein stability curves. Biopolymers 1987, 26, 1859–1877. [Google Scholar] [CrossRef] [PubMed]
- Privalov, P.L.; Griko Yu, V.; Venyaminov, S.; Kutyshenko, V.P. Cold denaturation of myoglobin. J. Mol. Biol. 1986, 190, 487–498. [Google Scholar] [CrossRef]
- Pastore, A.; Martin, S.R.; Politou, A.; Kondapalli, K.C.; Stemmler, T.; Temussi, P.A. Unbiased cold denaturation: Low- and high-temperature unfolding of yeast frataxin under physiological conditions. J. Am. Chem. Soc. 2007, 129, 5374–5375. [Google Scholar] [CrossRef]
- Robertson, A.D.; Murphy, K.P. Protein Structure and the Energetics of Protein Stability. Chem. Rev. 1997, 97, 1251–1268. [Google Scholar] [CrossRef] [PubMed]
- Sawle, L.; Ghosh, K. How do thermophilic proteins and proteomes withstand high temperature? Biophys. J. 2011, 101, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Rees, D.C.; Robertson, A.D. Some thermodynamic implications for the thermostability of proteins. Protein Sci. 2001, 10, 1187–1194. [Google Scholar] [CrossRef]
- D’Amico, S.; Marx, J.C.; Gerday, C.; Feller, G. Activity-stability relationships in extremophilic enzymes. J. Biol. Chem. 2003, 278, 7891–7896. [Google Scholar] [CrossRef]
- Graziano, G. On the molecular origin of cold denaturation of globular proteins. Phys. Chem. Chem. Phys. 2010, 12, 14245–14252. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. On the mechanism of cold denaturation. Phys. Chem. Chem. Phys. 2014, 16, 21755–21767. [Google Scholar] [CrossRef] [PubMed]
- Pica, A.; Graziano, G. Shedding light on the extra thermal stability of thermophilic proteins. Biopolymers 2016, 105, 856–863. [Google Scholar] [CrossRef]
- Graziano, G. Why small proteins tend to have high denaturation temperatures. Phys. Chem. Chem. Phys. 2020, 22, 16258–16266. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. Contrasting the hydration thermodynamics of methane and methanol. Phys. Chem. Chem. Phys. 2019, 21, 21418–21430. [Google Scholar] [CrossRef]
- Graziano, G. A purely geometric derivation of the scaled particle theory formula for the work of cavity creation in a liquid. Chem. Phys. Lett. 2007, 440, 221–223. [Google Scholar] [CrossRef]
- Lee, B.; Richards, F.M. The interpretation of protein structures: Estimation of static accessibility. J. Mol. Biol. 1971, 55, 379–400. [Google Scholar] [CrossRef]
- Graziano, G. On the cavity size distribution in water and n-hexane. Biophys. Chem. 2003, 104, 393–405. [Google Scholar] [CrossRef]
- Royer, C.A. Revisiting volume changes in pressure-induced protein unfolding. Biochim. Biophys. Acta 2002, 1595, 201–209. [Google Scholar] [CrossRef]
- Chalikian, T.V. Volumetric properties of proteins. Annu. Rev. Biophys. Biomol. Struct. 2003, 32, 207–235. [Google Scholar] [CrossRef] [PubMed]
- Graziano, G. The Gibbs energy cost of cavity creation depends on geometry. J. Mol. Liq. 2015, 211, 1047–1051. [Google Scholar] [CrossRef]
- Graziano, G. Shedding light on the hydrophobicity puzzle. Pure Appl. Chem. 2016, 88, 177–188. [Google Scholar] [CrossRef]
- Rose, G.D. Protein folding—Seeing is deceiving. Protein Sci. Publ. Protein Soc. 2021, 30, 1606–1616. [Google Scholar] [CrossRef] [PubMed]
- Kell, G.S. Density, thermal expansivity, and compressibility of liquid water from 0.deg. to 150.deg. Correlations and tables for atmospheric pressure and saturation reviewed and expressed on 1968 temperature scale. J. Chem. Eng. Data 1975, 20, 97–105. [Google Scholar] [CrossRef]
- Tadokoro, T.; Matsushita, K.; Abe, Y.; Rohman, M.S.; Koga, Y.; Takano, K.; Kanaya, S. Remarkable stabilization of a psychrotrophic RNase HI by a combination of thermostabilizing mutations identified by the suppressor mutation method. Biochemistry 2008, 47, 8040–8047. [Google Scholar] [CrossRef] [PubMed]
- D’Amico, S.; Gerday, C.; Feller, G. Structural determinants of cold adaptation and stability in a large protein. J. Biol. Chem. 2001, 276, 25791–25796. [Google Scholar] [CrossRef]
- Ben-Naim, A. Hydrophobic interaction and structural changes in the solvent. Biopolym. Orig. Res. Biomol. 1975, 14, 1337–1355. [Google Scholar] [CrossRef]
- Yu, H.A.; Karplus, M. A thermodynamic analysis of solvation. J. Chem. Phys. 1988, 89, 2366–2379. [Google Scholar] [CrossRef]
- Lee, B. Enthalpy-entropy compensation in the thermodynamics of hydrophobicity. Biophys. Chem. 1994, 51, 271–278. [Google Scholar] [CrossRef]
- Dunitz, J.D. Win some, lose some: Enthalpy-entropy compensation in weak intermolecular interactions. Chem. Biol. 1995, 2, 709–712. [Google Scholar] [CrossRef]
- Graziano, G. Case study of enthalpy-entropy noncompensation. J. Chem. Phys. 2004, 120, 4467–4471. [Google Scholar] [CrossRef] [PubMed]
- Liquori, A.M.; Sadun, C. Close packing of amino acid residues in globular proteins: Specific volume and site binding of water molecules. Int. J. Biol. Macromol. 1981, 3, 56–59. [Google Scholar] [CrossRef]
- Cotter, M.A.; Martire, D.E. Statistical Mechanics of Rodlike Particles. II. A Scaled Particle Investigation of the Aligned→Isotropic Transition in a Fluid of Rigid Spherocylinders. J. Chem. Phys. 1970, 52, 1909–1919. [Google Scholar] [CrossRef]
- Head-Gordon, T.; Hura, G. Water structure from scattering experiments and simulation. Chem. Rev. 2002, 102, 2651–2670. [Google Scholar] [CrossRef]
- Baxa, M.C.; Haddadian, E.J.; Jumper, J.M.; Freed, K.F.; Sosnick, T.R. Loss of conformational entropy in protein folding calculated using realistic ensembles and its implications for NMR-based calculations. Proc. Natl. Acad. Sci. USA 2014, 111, 15396–15401. [Google Scholar] [CrossRef] [PubMed]
- Tomasi, J.; Persico, M. Molecular interactions in solution: An overview of methods based on continuous distributions of the solvent. Chem. Rev. 1994, 94, 2027–2094. [Google Scholar] [CrossRef]
- Hummer, G.; Garde, S.; Garcia, A.; Paulaitis, M.E.; Pratt, L.R. Hydrophobic effects on a molecular scale. J. Phys. Chem. B 1998, 102, 10469–10482. [Google Scholar] [CrossRef]
- Graves, J.E.; Somero, G.N. Electrophoretic and Functional Enzymic Evolution in Four Species of Eastern Pacific Barracudas from Different Thermal Environments. Evolution 1982, 36, 97–106. [Google Scholar] [CrossRef]
- Somero, G.N. Proteins and temperature. Annu. Rev. Physiol. 1995, 57, 43–68. [Google Scholar] [CrossRef] [PubMed]
- Jaenicke, R.; Zavodszky, P. Proteins under extreme physical conditions. FEBS Lett. 1990, 268, 344–349. [Google Scholar] [CrossRef]
- Svingor, A.; Kardos, J.; Hajdu, I.; Nemeth, A.; Zavodszky, P. A better enzyme to cope with cold. Comparative flexibility studies on psychrotrophic, mesophilic, and thermophilic IPMDHs. J. Biol. Chem. 2001, 276, 28121–28125. [Google Scholar] [CrossRef] [PubMed]
- Na, H.; Song, G.; ben-Avraham, D. Universality of vibrational spectra of globular proteins. Phys. Biol. 2016, 13, 016008. [Google Scholar] [CrossRef]
- Callender, R.; Dyer, R.B. The dynamical nature of enzymatic catalysis. Acc. Chem. Res. 2015, 48, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Chalopin, Y. The physical origin of rate promoting vibrations in enzymes revealed by structural rigidity. Sci. Rep. 2020, 10, 17465. [Google Scholar] [CrossRef] [PubMed]
- Saavedra, H.G.; Wrabl, J.O.; Anderson, J.A.; Li, J.; Hilser, V.J. Dynamic allostery can drive cold adaptation in enzymes. Nature 2018, 558, 324–328. [Google Scholar] [CrossRef]
- Dong, Y.W.; Liao, M.L.; Meng, X.L.; Somero, G.N. Structural flexibility and protein adaptation to temperature: Molecular dynamics analysis of malate dehydrogenases of marine molluscs. Proc. Natl. Acad Sci. USA 2018, 115, 1274–1279. [Google Scholar] [CrossRef] [PubMed]
- Feller, G.; Payan, F.; Theys, F.; Qian, M.; Haser, R.; Gerday, C. Stability and structural analysis of alpha-amylase from the antarctic psychrophile Alteromonas haloplanctis A23. Eur. J. Biochem. 1994, 222, 441–447. [Google Scholar] [CrossRef] [PubMed]
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level. Structure 1998, 6, 1503–1516. [Google Scholar] [CrossRef]
- Feller, G.; d’Amico, D.; Gerday, C. Thermodynamic stability of a cold-active alpha-amylase from the Antarctic bacterium Alteromonas haloplanctis. Biochemistry 1999, 38, 4613–4619. [Google Scholar] [CrossRef]
- Cipolla, A.; Delbrassine, F.; Da Lage, J.L.; Feller, G. Temperature adaptations in psychrophilic, mesophilic and thermophilic chloride-dependent alpha-amylases. Biochimie 2012, 94, 1943–1950. [Google Scholar] [CrossRef]
- D’Amico, S.; Gerday, C.; Feller, G. Temperature adaptation of proteins: Engineering mesophilic-like activity and stability in a cold-adapted alpha-amylase. J. Mol. Biol. 2003, 332, 981–988. [Google Scholar] [CrossRef] [PubMed]
- Cipolla, A.; D’Amico, S.; Barumandzadeh, R.; Matagne, A.; Feller, G. Stepwise adaptations to low temperature as revealed by multiple mutants of psychrophilic alpha-amylase from Antarctic Bacterium. J. Biol. Chem. 2011, 286, 38348–38355. [Google Scholar] [CrossRef]
- Li, Q.; Yan, Y.; Liu, X.; Zhang, Z.; Tian, J.; Wu, N. Enhancing thermostability of a psychrophilic alpha-amylase by the structural energy optimization in the trajectories of molecular dynamics simulations. Int. J. Biol. Macromol. 2020, 142, 624–633. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Hough, D.W.; Danson, M.J.; Taylor, G.L. The crystal structure of citrate synthase from the thermophilic archaeon, Thermoplasma acidophilum. Structure 1994, 2, 1157–1167. [Google Scholar] [CrossRef]
- Gerike, U.; Danson, M.J.; Hough, D.W. Cold-active citrate synthase: Mutagenesis of active-site residues. Protein Eng. 2001, 14, 655–661. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Ferguson, J.M.; Hough, D.W.; Danson, M.J.; Taylor, G.L. The crystal structure of citrate synthase from the hyperthermophilic archaeon pyrococcus furiosus at 1.9 A resolution. Biochemistry 1997, 36, 9983–9994. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.J.; Gerike, U.; Danson, M.J.; Hough, D.W.; Taylor, G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure 1998, 6, 351–361. [Google Scholar] [CrossRef]
- Bell, G.S.; Russell, R.J.; Connaris, H.; Hough, D.W.; Danson, M.J.; Taylor, G.L. Stepwise adaptations of citrate synthase to survival at life’s extremes. From psychrophile to hyperthermophile. Eur. J. Biochem. 2002, 269, 6250–6260. [Google Scholar] [CrossRef] [PubMed]
- Juers, D.H.; Matthews, B.W.; Huber, R.E. LacZ beta-galactosidase: Structure and function of an enzyme of historical and molecular biological importance. Protein Sci. 2012, 21, 1792–1807. [Google Scholar] [CrossRef] [PubMed]
- Triveni Piasad Shukla, L.E.W. Beta-galactosidase technology: A solution to the lactose problem. CRC Crit. Rev. Food Technol. 1975, 5, 325–356. [Google Scholar] [CrossRef]
- Li, S.; Zhu, X.; Xing, M. A New beta-Galactosidase from the Antarctic Bacterium Alteromonas sp. ANT48 and Its Potential in Formation of Prebiotic Galacto-Oligosaccharides. Mar. Drugs 2019, 17, 599. [Google Scholar] [CrossRef]
- Mangiagalli, M.; Lotti, M. Cold-Active beta-Galactosidases: Insight into Cold Adaption Mechanisms and Biotechnological Exploitation. Mar. Drugs 2021, 19, 43. [Google Scholar] [CrossRef] [PubMed]
- Bruno, S.; Coppola, D.; di Prisco, G.; Giordano, D.; Verde, C. Enzymes from Marine Polar Regions and Their Biotechnological Applications. Mar. Drugs 2019, 17, 544. [Google Scholar] [CrossRef] [PubMed]
- Mangiagalli, M.; Brocca, S.; Orlando, M.; Lotti, M. The “cold revolution”. Present and future applications of cold-active enzymes and ice-binding proteins. New Biotechnol. 2020, 55, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Hoyoux, A.; Jennes, I.; Dubois, P.; Genicot, S.; Dubail, F.; Francois, J.M.; Baise, E.; Feller, G.; Gerday, C. Cold-adapted beta-galactosidase from the Antarctic psychrophile Pseudoalteromonas haloplanktis. Appl. Environ. Microbiol. 2001, 67, 1529–1535. [Google Scholar] [CrossRef] [PubMed]
- Jacobson, R.H.; Zhang, X.J.; DuBose, R.F.; Matthews, B.W. Three-dimensional structure of beta-galactosidase from E. coli. Nature 1994, 369, 761–766. [Google Scholar] [CrossRef] [PubMed]
- Mangiagalli, M.; Lapi, M.; Maione, S.; Orlando, M.; Brocca, S.; Pesce, A.; Barbiroli, A.; Camilloni, C.; Pucciarelli, S.; Lotti, M.; et al. The co-existence of cold activity and thermal stability in an Antarctic GH42 beta-galactosidase relies on its hexameric quaternary arrangement. FEBS J. 2021, 288, 546–565. [Google Scholar] [CrossRef]
- Rutkiewicz, M.; Bujacz, A.; Wanarska, M.; Wierzbicka-Wos, A.; Cieslinski, H. Active Site Architecture and Reaction Mechanism Determination of Cold Adapted beta-d-galactosidase from Arthrobacter sp. 32cB. Int. J. Mol. Sci. 2019, 20, 4301. [Google Scholar] [CrossRef] [PubMed]
- Rutkiewicz, M.; Wanarska, M.; Bujacz, A. Mapping the Transglycosylation Relevant Sites of Cold-Adapted beta-d-Galactosidase from Arthrobacter sp. 32cB. Int. J. Mol. Sci. 2020, 21, 5354. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).