Recent Advances in the Research on the Anticyanobacterial Effects and Biodegradation Mechanisms of Microcystis aeruginosa with Microorganisms
Abstract
:1. Introduction
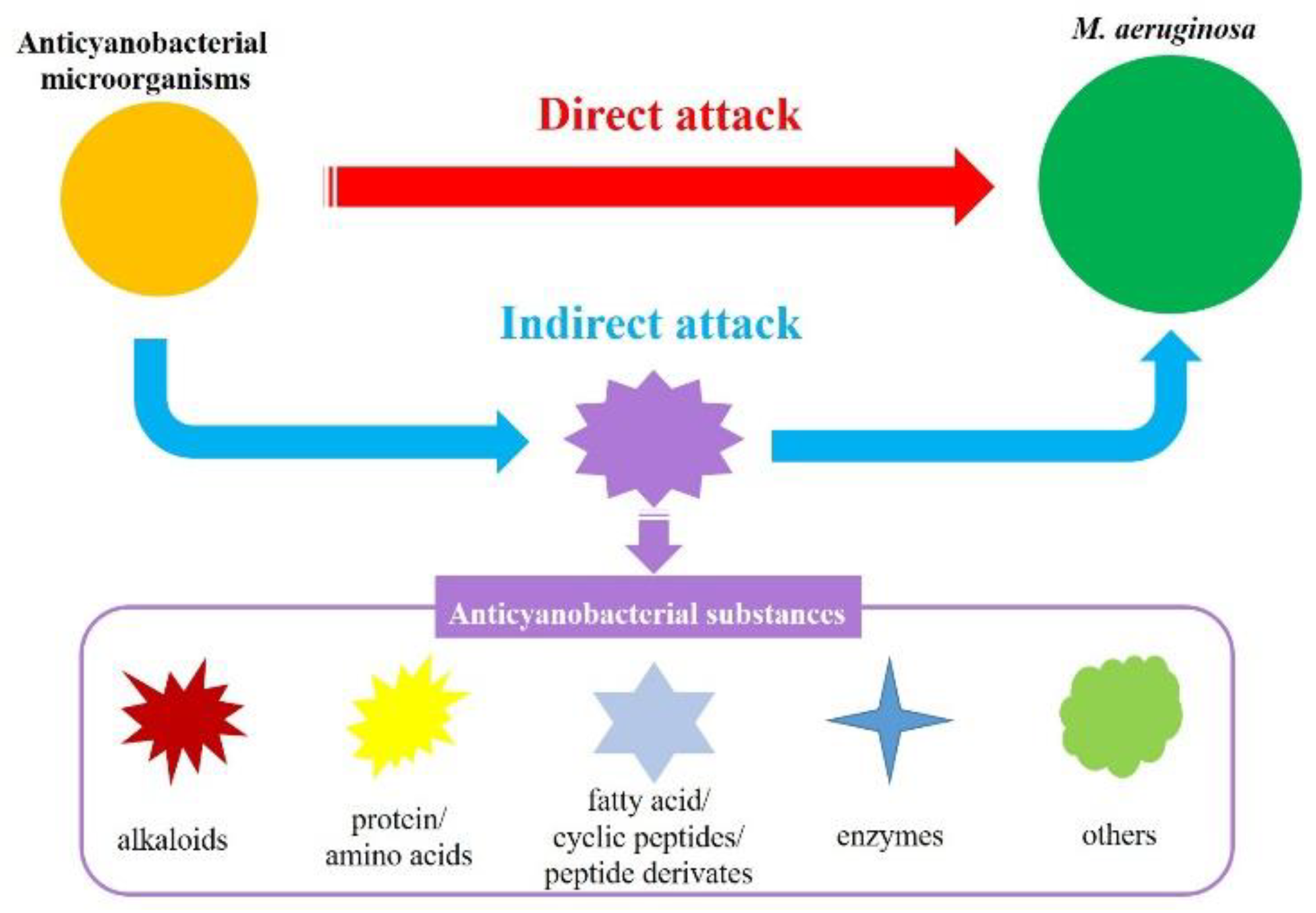
2. Anticyanobacterial Effects for M. aeruginosa
2.1. Anticyanobacterial Microorganisms
2.1.1. Anticyanobacteria
2.1.2. Anticyanobacterial Fungi
2.2. Anticyanobacterial Substances
3. Anticyanobacterial Modes and Mechanisms
3.1. Anticyanobacterial Modes
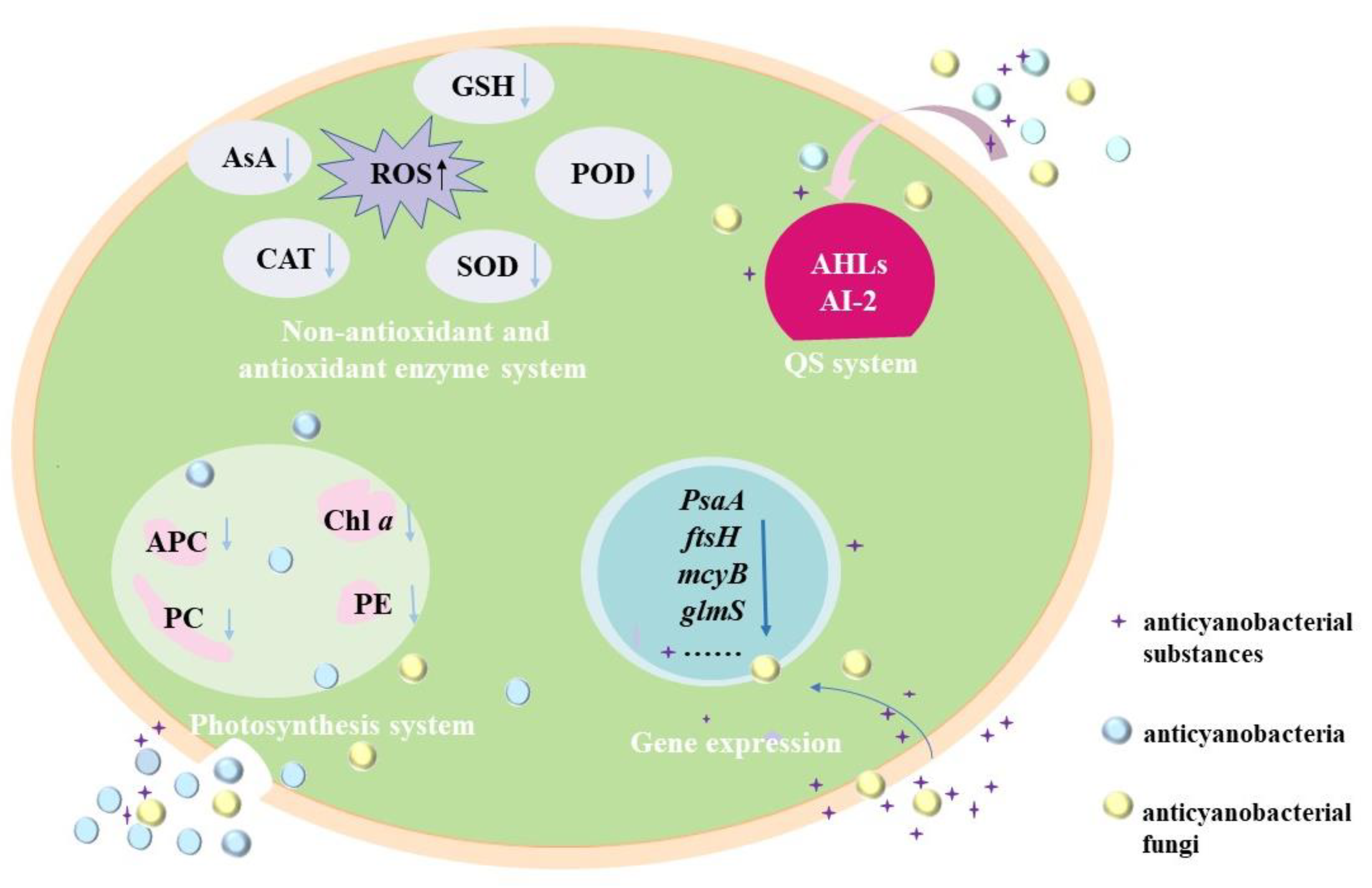
3.2. Anticyanobacterial Mechanisms
3.2.1. Effects of Anticyanobacterial Microorganisms on Photosynthesis
3.2.2. Effects of Anticyanobacterial Microorganisms on Antioxidant Enzymes System
3.2.3. Effects of Anticyanobacterial Microorganisms on Gene Expression
3.2.4. Regulating the Anticyanobacterial Activity by QS System
4. Application and Prospective
4.1. Application of Anticyanobacterial Microorganisms
4.2. Summary and Prospective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Harke, M.J.; Steffen, M.M.; Gobler, C.J.; Otten, T.G.; Wilhelm, S.; Wood, S.A.; Paerl, H.W. A review of the global ecology, genomics, and biogeography of the toxic cyanobacterium, Microcystis spp. Harmful Algae 2016, 54, 4–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Hou, X.; Wu, D.; Chang, W.; Zhang, X.; Dai, X.; Du, H.; Zhang, X.; Igarashi, Y.; Luo, F. The characteristics and algicidal mechanisms of cyanobactericidal bacteria, a review. World J. Microbiol. Biotechnol. 2020, 36, 188. [Google Scholar] [CrossRef]
- Ko, S.-R.; Lee, Y.-K.; Srivastava, A.; Park, S.-H.; Ahn, C.-Y.; Oh, H.-M. The Selective Inhibitory Activity of a Fusaricidin Derivative on a Bloom-Forming Cyanobacterium, Microcystis sp. J. Microbiol. Biotechnol. 2019, 29, 59–65. [Google Scholar] [CrossRef] [Green Version]
- Han, S.-I.; Kim, S.; Choi, K.Y.; Lee, C.; Park, Y.; Choi, Y.-E. Control of a toxic cyanobacterial bloom species, Microcystis aeruginosa, using the peptide HPA3NT3-A2. Environ. Sci. Pollut. Res. 2019, 26, 32255–32265. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Zeng, Y.; Li, J.; Yang, C.; Zhang, X.; Luo, F.; Dai, X. An algicidal Streptomyces amritsarensis strain against Microcystis aeruginosa strongly inhibits microcystin synthesis simultaneously. Sci. Total Environ. 2018, 650, 34–43. [Google Scholar] [CrossRef]
- Mohamed, Z.A.; Hashem, M.; Alamri, S.A. Growth inhibition of the cyanobacterium Microcystis aeruginosa and degradation of its microcystin toxins by the fungus Trichoderma citrinoviride. Toxicon 2014, 86, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Goslan, E.H.; Seigle, C.; Purcell, D.; Henderson, R.; Parsons, S.A.; Jefferson, B.; Judd, S.J. Carbonaceous and nitrogenous disinfection by-product formation from algal organic matter. Chemosphere 2016, 170, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Xin, H.; Yang, S.; Tang, Y.; Wu, M.; Deng, Y.; Xu, B.; Gao, N. Mechanisms and performance of calcium peroxide-enhanced Fe(ii) coagulation for treatment of Microcystis aeruginosa-laden water. Environ. Sci. Water Res. Technol. 2020, 6, 1272–1285. [Google Scholar] [CrossRef]
- Chen, Z.; Li, J.; Chen, M.; Koh, K.Y.; Du, Z.; Gin, K.Y.-H.; He, Y.; Ong, C.N.; Chen, J.P. Microcystis aeruginosa removal by peroxides of hydrogen peroxide, peroxymonosulfate and peroxydisulfate without additional activators. Water Res. 2021, 201, 117263. [Google Scholar] [CrossRef]
- Wang, M.; Chen, S.; Zhou, W.; Yuan, W.; Wang, D. Algal cell lysis by bacteria: A review and comparison to conventional methods. Algal Res. 2020, 46, 101794. [Google Scholar] [CrossRef]
- Matthijs, H.C.P.; Jančula, D.; Visser, P.M.; Maršálek, B. Existing and emerging cyanocidal compounds: New perspectives for cyanobacterial bloom mitigation. Aquat. Ecol. 2016, 50, 443–460. [Google Scholar] [CrossRef] [Green Version]
- Demuez, M.; González-Fernández, C.; Ballesteros, M. Algicidal microorganisms and secreted algicides: New tools to induce microalgal cell disruption. Biotechnol. Adv. 2015, 33, 1615–1625. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Sun, P.; Zhang, J.; Esquivel-Elizondo, S.; Wu, Y. Microorganisms-based methods for harmful algal blooms control: A review. Bioresour. Technol. 2018, 248, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Benegas, G.R.S.; Bernal, S.P.F.; de Oliveira, V.M.; Passarini, M.R.Z. Antimicrobial activity against Microcystis aeruginosa and degradation of microcystin-LR by bacteria isolated from Antarctica. Environ. Sci. Pollut. Res. 2021, 28, 52381–52391. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wu, X.; Jiang, X.; Liu, L.; Wang, H. Algicidal activity of Aspergillus niger induced by calcium ion as signal molecule on Microcystis aeruginosa. Algal Res. 2021, 60, 102536. [Google Scholar] [CrossRef]
- Meyer, N.; Bigalke, A.; Kaulfuß, A.; Pohnert, G. Strategies and ecological roles of algicidal bacteria. FEMS Microbiol. Rev. 2017, 41, 880–899. [Google Scholar] [CrossRef] [Green Version]
- Mohamed, Z.A.; Hashem, M.; Alamri, S.; Campos, A.; Vasconcelos, V. Fungal biodegradation and removal of cyanobacteria and microcystins: Potential applications and research needs. Environ. Sci. Pollut. Res. 2021, 28, 37041–37050. [Google Scholar] [CrossRef]
- Zhou, S.; Yin, H.; Tang, S.; Peng, H.; Yin, D.; Yang, Y.; Liu, Z.; Dang, Z. Physiological responses of Microcystis aeruginosa against the algicidal bacterium Pseudomonas aeruginosa. Ecotoxicol. Environ. Saf. 2016, 127, 214–221. [Google Scholar] [CrossRef]
- Zhang, H.; Yu, Z.; Huang, Q.; Xiao, X.; Wang, X.; Zhang, F.; Wang, X.; Liu, Y.; Hu, C. Isolation, identification and characterization of phytoplankton-lytic bacterium CH-22 against Microcystis aeruginosa. Limnologica 2011, 41, 70–77. [Google Scholar] [CrossRef] [Green Version]
- Park, B.S.; Park, C.-S.; Shin, Y.; Yoon, S.; Han, M.-S.; Kang, Y.-H. Different Algicidal Modes of the Two Bacteria Aeromonas bestiarum HYD0802-MK36 and Pseudomonas syringae KACC10292T against Harmful Cyanobacteria Microcystis aeruginosa. Toxins 2022, 14, 128. [Google Scholar] [CrossRef]
- Das Nishu, S.; Kang, Y.; Han, I.; Jung, T.Y.; Lee, T.K. Nutritional status regulates algicidal activity of Aeromonas sp. L23 against cyanobacteria and green algae. PLoS ONE 2019, 14, e0213370. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Ai, H.; Kang, L.; Sun, X.; He, Q. Simultaneous Microcystis Algicidal and Microcystin Degrading Capability by a Single Acinetobacter Bacterial Strain. Environ. Sci. Technol. 2016, 50, 11903–11911. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Kang, X.; Chu, L.; Wang, Y.; Song, X.; Zhao, X.; Cao, X. Algicidal mechanism of Raoultella ornithinolytica against Microcystis aeruginosa: Antioxidant response, photosynthetic system damage and microcystin degradation. Environ. Pollut. 2021, 287, 117644. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wang, Y.; Huang, J.; Fan, Q.; Wei, J.; Wang, F.; Jia, Z.; Xiang, W.; Liang, W. Inhibition of Microcystis aeruginosa using Brevundimonas sp. AA06 immobilized in polyvinyl alcohol-sodium alginate beads. Desalination Water Treat. 2018, 111, 192–200. [Google Scholar] [CrossRef]
- Mu, R.; He, Y.; Liu, S.; Wang, X.; Fan, Z. The Algicidal Characteristics of One Algae-Lysing FDT5 Bacterium on Microcystis aeruginosa. Geomicrobiol. J. 2009, 26, 516–521. [Google Scholar] [CrossRef]
- Zhang, D.; Ye, Q.; Zhang, F.; Shao, X.; Fan, Y.; Zhu, X.; Li, Y.; Yao, L.; Tian, Y.; Zheng, T.; et al. Flocculating properties and potential of Halobacillus sp. strain H9 for the mitigation of Microcystis aeruginosa blooms. Chemosphere 2018, 218, 138–146. [Google Scholar] [CrossRef]
- Li, Z.; Lin, S.; Liu, X.; Tan, J.; Pan, J.; Yang, H. A freshwater bacterial strain, Shewanella sp. Lzh-2, isolated from Lake Taihu and its two algicidal active substances, hexahydropyrrolo[1,2-a]pyrazine-1,4-dione and 2, 3-indolinedione. Appl. Microbiol. Biotechnol. 2014, 98, 4737–4748. [Google Scholar] [CrossRef]
- Sun, P.; Esquivel-Elizondo, S.; Zhao, Y.; Wu, Y. Glucose triggers the cytotoxicity of Citrobacter sp. R1 against Microcystis aeruginosa. Sci. Total Environ. 2017, 603-604, 18–25. [Google Scholar] [CrossRef]
- Lin, S.; Geng, M.; Liu, X.; Tan, J.; Yang, H. On the control of Microcystis aeruginosa and Synechococccus species using an algicidal bacterium, Stenotrophomonas F6, and its algicidal compounds cyclo-(Gly-Pro) and hydroquinone. J. Appl. Phycol. 2015, 28, 345–355. [Google Scholar] [CrossRef]
- Liu, W.; Yang, J.; Tian, Y.; Zhou, X.; Wang, S.; Zhu, J.; Sun, D.; Liu, C. An in situ extractive fermentation strategy for enhancing prodigiosin production from Serratia marcescens BWL1001 and its application to inhibiting the growth of Microcystis aeruginosa. Biochem. Eng. J. 2020, 166, 107836. [Google Scholar] [CrossRef]
- Yang, K.; Chen, Q.; Zhang, D.; Zhang, H.; Lei, X.; Chen, Z.; Li, Y.; Hong, Y.; Ma, X.; Zheng, W.; et al. The algicidal mechanism of prodigiosin from Hahella sp. KA22 against Microcystis aeruginosa. Sci. Rep. 2017, 7, 7750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Y.; Wang, Q.; Chen, Y.; Xu, X.; Zhu, L.; Yao, H.; Pan, H. Anticyanobacterial process and action mechanism of Streptomyces sp. HJC-D1 on Microcystis aeruginosa. Environ. Prog. Sustain. Energy 2020, 39, e13392. [Google Scholar] [CrossRef]
- Luo, J.; Wang, Y.; Tang, S.; Liang, J.; Lin, W.; Luo, L. Isolation and Identification of Algicidal Compound from Streptomyces and Algicidal Mechanism to Microcystis aeruginosa. PLoS ONE 2013, 8, e76444. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-K.; Ahn, C.-Y.; Kim, H.-S.; Oh, H.-M. Cyanobactericidal effect of Rhodococcus sp. isolated from eutrophic lake on Microcystis sp. Biotechnol. Lett. 2010, 32, 1673–1678. [Google Scholar] [CrossRef]
- Chen, H.; Fu, L.; Luo, L.; Lu, J.; White, W.L.; Hu, Z. Induction and Resuscitation of the Viable but Nonculturable State in a Cyanobacteria-Lysing Bacterium Isolated from Cyanobacterial Bloom. Microb. Ecol. 2011, 63, 64–73. [Google Scholar] [CrossRef]
- Hua, X.-H.; Li, J.-H.; Li, J.-J.; Zhang, L.-H.; Cui, Y. Selective inhibition of the cyanobacterium, Microcystis, by a Streptomyces sp. Biotechnol. Lett. 2009, 31, 1531–1535. [Google Scholar] [CrossRef]
- Zhang, B.-H.; Chen, W.; Li, H.-Q.; Yang, J.-Y.; Zha, D.-M.; Duan, Y.-Q.; Hozzein, N.W.; Xiao, M.; Gao, R.; Li, W.-J. L-valine, an antialgal amino acid from Streptomyces jiujiangensis JXJ 0074T. Appl. Microbiol. Biotechnol. 2016, 100, 4627–4636. [Google Scholar] [CrossRef]
- Zhang, B.-H.; Ding, Z.-G.; Li, H.-Q.; Mou, X.-Z.; Zhang, Y.-Q.; Yang, J.-Y.; Zhou, E.-M.; Li, W.-J. Algicidal Activity of Streptomyces eurocidicus JXJ-0089 Metabolites and Their Effects on Microcystis Physiology. Appl. Environ. Microbiol. 2016, 82, 5132–5143. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-M.; Sheu, F.-S.; Sheu, S.-Y. Aquimarina salinaria sp. nov., a novel algicidal bacterium isolated from a saltpan. Arch. Microbiol. 2011, 194, 103–112. [Google Scholar] [CrossRef]
- Zhang, C.; Massey, I.Y.; Liu, Y.; Huang, F.; Gao, R.; Ding, M.; Xiang, L.; He, C.; Wei, J.; Li, Y.; et al. Identification and characterization of a novel indigenous algicidal bacterium Chryseobacterium species against Microcystis aeruginosa. J. Toxicol. Environ. Heal. Part A 2019, 82, 845–853. [Google Scholar] [CrossRef]
- Guo, X.; Liu, X.; Pan, J.; Yang, H. Synergistic algicidal effect and mechanism of two diketopiperazines produced by Chryseobacterium sp. strain GLY-1106 on the harmful bloom-forming Microcystis aeruginosa. Sci. Rep. 2015, 5, 14720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furusawa, G.; Iwamoto, K. Removal of Microcystis aeruginosa cells using the dead cells of a marine filamentous bacterium, Aureispira sp. CCB-QB1. PeerJ 2022, 10, e12867. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hongyi, W.; Komatsu, M.; Ishibashi, K.; Jinsan, L.; Ito, T.; Yoshikawa, T.; Maeda, H. Isolation and characterization of bacterial isolates algicidal against a harmful bloom-forming cyanobacterium Microcystis aeruginosa. Biocontrol Sci. 2012, 17, 107–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Liu, L.; Xu, Y.; Li, P.; Zhang, K.; Jiang, X.; Zheng, T.; Wang, H. Stress of algicidal substances from a bacterium Exiguobacterium sp. h10 on Microcystis aeruginosa. Lett. Appl. Microbiol. 2016, 64, 57–65. [Google Scholar] [CrossRef]
- Zhang, S.; Fan, C.; Xia, Y.; Li, M.; Wang, Y.; Cui, X.; Xiao, W. Characterization of a novel bacteriophage specific to Exiguobacterium indicum isolated from a plateau eutrophic lake. J. Basic Microbiol. 2018, 59, 206–214. [Google Scholar] [CrossRef]
- Li, Z.; Geng, M.; Yang, H. Algicidal activity of Bacillus sp. Lzh-5 and its algicidal compounds against Microcystis aeruginosa. Appl. Microbiol. Biotechnol. 2014, 99, 981–990. [Google Scholar] [CrossRef]
- Liu, J.; Yang, C.; Chi, Y.; Wu, D.; Dai, X.; Zhang, X.; Igarashi, Y.; Luo, F. Algicidal characterization and mechanism of Bacillus licheniformis Sp34 against Microcystis aeruginosa in Dianchi Lake. J. Basic Microbiol. 2019, 59, 1112–1124. [Google Scholar] [CrossRef]
- Lee, C.; Jeon, M.S.; Vo, T.-T.; Park, C.; Choi, J.-S.; Kwon, J.; Roh, S.W.; Choi, Y.-E. Establishment of a new strategy against Microcystis bloom using newly isolated lytic and toxin-degrading bacteria. J. Appl. Phycol. 2018, 30, 1795–1806. [Google Scholar] [CrossRef]
- Yu, J.; Kong, Y.; Gao, S.; Miao, L.; Zou, P.; Xu, B.; Zeng, C.; Zhang, X. Bacillus amyloliquefaciens T1 as a potential control agent for cyanobacteria. J. Appl. Phycol. 2014, 27, 1213–1221. [Google Scholar] [CrossRef]
- Sun, P.; Hui, C.; Wang, S.; Khan, R.A.; Zhang, Q.; Zhao, Y.-H. Enhancement of algicidal properties of immobilized Bacillus methylotrophicus ZJU by coating with magnetic Fe3O4 nanoparticles and wheat bran. J. Hazard. Mater. 2015, 301, 65–73. [Google Scholar] [CrossRef]
- Xuan, H.; Dai, X.; Li, J.; Zhang, X.; Yang, C.; Luo, F. A Bacillus sp. strain with antagonistic activity against Fusarium graminearum kills Microcystis aeruginosa selectively. Sci. Total Environ. 2017, 583, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Kim, M.; Hong, M.; Park, W. Killing effect of deinoxanthins on cyanobloom-forming Microcystis aeruginosa: Eco-friendly production and specific activity of deinoxanthins. Environ. Res. 2021, 200, 111455. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhu, H.; Lei, X.; Zhang, H.; Cai, G.; Chen, Z.; Fu, L.; Xu, H.; Zheng, T. The death mechanism of the harmful algal bloom species Alexandrium tamarense induced by algicidal bacterium Deinococcus sp. Y35. Front. Microbiol. 2015, 6, 992. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, L.; Huo, M.; Sun, C.; Cui, X.; Zhou, D.; Crittenden, J.C.; Yang, W. Bioresources inner-recycling between bioflocculation of Microcystis aeruginosa and its reutilization as a substrate for bioflocculant production. Sci. Rep. 2017, 7, 43784. [Google Scholar] [CrossRef] [Green Version]
- Liu, H.; Guo, X.; Liu, L.; Yan, M.; Li, J.; Hou, S.; Wan, J.; Feng, L. Simultaneous Microcystin Degradation and Microcystis aeruginosa Inhibition with the Single Enzyme Microcystinase A. Environ. Sci. Technol. 2020, 54, 8811–8820. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Zou, P.; Yang, Q.; Xu, X.; Miao, L.; Zhu, L. Physiological responses of Microcystis aeruginosa under the stress of antialgal actinomycetes. J. Hazard. Mater. 2013, 262, 274–280. [Google Scholar] [CrossRef]
- Zhang, Y.; Chen, D.; Zhang, N.; Li, F.; Luo, X.; Li, Q.; Li, C.; Huang, X. Transcriptional Analysis of Microcystis aeruginosa Co-Cultured with Algicidal Bacteria Brevibacillus laterosporus. Int. J. Environ. Res. Public Health 2021, 18, 8615. [Google Scholar] [CrossRef] [PubMed]
- Pal, M.; Purohit, H.J.; Qureshi, A. Genomic insight for algicidal activity in Rhizobium strain AQ_MP. Arch. Microbiol. 2021, 203, 5193–5203. [Google Scholar] [CrossRef]
- Pathmalal, M.M.; Zenichiro, K.; Shin-ichi, N. Algicidal effect of the bacterium Alcaligenes denitrificans on Microcystis spp. Aquatic Microbial Ecology 2000, 22, 111–117. [Google Scholar]
- Xue, G.; Wang, X.; Xu, C.; Song, B.; Chen, H. Removal of harmful algae by Shigella sp. H3 and Alcaligenes sp. H5: Algicidal pathways and characteristics. Environ. Technol. 2021. [Google Scholar] [CrossRef]
- Van Le, V.; Ko, S.-R.; Kang, M.; Lee, S.-A.; Oh, H.-M.; Ahn, C.-Y. Algicide capacity of Paucibacter aquatile DH15 on Microcystis aeruginosa by attachment and non-attachment effects. Environ. Pollut. 2022, 302, 119079. [Google Scholar] [CrossRef]
- Crettaz-Minaglia, M.; Fallico, M.; Aranda, O.; Juarez, I.; Pezzoni, M.; Costa, C. Effect of temperature on microcystin-LR removal and lysis activity on Microcystis aeruginosa(cyanobacteria) by an indigenous bacterium belonging to the genus Achromobacter. Environ. Sci. Pollut. Res. 2020, 27, 44427–44439. [Google Scholar] [CrossRef]
- Wang, X.; Xie, M.; Wu, W.; Shi, L.; Luo, L.; Li, P. Differential sensitivity of colonial and unicellular Microcystis strains to an algicidal bacterium Pseudomonas aeruginosa. J. Plankton Res. 2013, 35, 1172–1176. [Google Scholar] [CrossRef]
- Kang, Y.-H.; Park, C.-S.; Han, M.-S. Pseudomonas aeruginosa UCBPP-PA14 a useful bacterium capable of lysing Microcystis aeruginosa cells and degrading microcystins. J. Appl. Phycol. 2012, 24, 1517–1525. [Google Scholar] [CrossRef]
- Chen, Q.; Wang, L.; Qi, Y.; Ma, C. Imaging mass spectrometry of interspecies metabolic exchange revealed the allelopathic interaction between Microcystis aeruginosa and its antagonist. Chemosphere 2020, 259, 127430. [Google Scholar] [CrossRef] [PubMed]
- Kodani, S.; Imoto, A.; Mitsutani, A.; Murakami, M. Isolation and identification of the antialgal compound, harmane (1-methyl-β-carboline), produced by the algicidal bacterium, Pseudomonas sp. K44-1. J. Appl. Phycol. 2002, 14, 109–114. [Google Scholar] [CrossRef]
- Zhang, X.; Song, T.; Ma, H.; Li, L. Physiological response of Microcystis aeruginosa to the extracellular substances from an Aeromonas sp. RSC Adv. 2016, 6, 103662–103667. [Google Scholar] [CrossRef]
- Liu, Y.-M.; Wang, M.-H.; Jia, R.-B.; Li, L. Removal of cyanobacteria by an Aeromonas sp. Desalination Water Treat. 2012, 47, 205–210. [Google Scholar] [CrossRef]
- Guo, X.; Liu, X.; Wu, L.; Pan, J.; Yang, H. The algicidal activity of Aeromonas sp. strain GLY-2107 against bloom-forming Microcystis aeruginosa is regulated by N-acyl homoserine lactone-mediated quorum sensing. Environ. Microbiol. 2016, 18, 3867–3883. [Google Scholar] [CrossRef]
- Yang, J.; Qiao, K.; Lv, J.; Liu, Q.; Nan, F.; Xie, S.; Feng, J. Isolation and Identification of Two Algae-Lysing Bacteria against Microcystis aeruginosa. Water 2020, 12, 2485. [Google Scholar] [CrossRef]
- Su, J.F.; Ma, M.; Wei, L.; Ma, F.; Lu, J.S.; Shao, S.C. Algicidal and denitrification characterization of Acinetobacter sp. J25 against Microcystis aeruginosa and microbial community in eutrophic landscape water. Mar. Pollut. Bull. 2016, 107, 233–239. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.-L.; Yu, X.-B.; Zhang, C.; Wang, G.-X. Growth inhibition and microcystin degradation effects of Acinetobacter guillouiae A2 on Microcystis aeruginosa. Res. Microbiol. 2015, 166, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Su, J.F.; Shao, S.C.; Ma, F.; Lu, J.S.; Zhang, K. Bacteriological control by Raoultella sp. R11 on growth and toxins production of Microcystis aeruginosa. Chem. Eng. J. 2016, 293, 139–150. [Google Scholar] [CrossRef]
- Liu, Z.Z.; Zhu, J.P.; Li, M.; Xue, Q.Q.; Zeng, Y.; Wang, Z.P. Effects of freshwater bacterial siderophore on Microcystis and Anabaena. Biol. Control 2014, 78, 42–48. [Google Scholar] [CrossRef]
- Liao, C.; Liu, X. High-Cell-Density Cultivation and Algicidal Activity Assays of a Novel Algicidal Bacterium to Control Algal Bloom Caused by Water Eutrophication. Water Air Soil Pollut. 2014, 225, s11270–s12014. [Google Scholar] [CrossRef]
- Yang, F.; Wei, H.Y.; Li, Y.H.; Li, X.B.; Yin, L.H.; Pu, Y.P. Isolation and characterization of an algicidal bacterium indigenous to lake Taihu with a red pigment able to lyse Microcystis aeruginosa. Biomed. Environ. Sci. 2013, 26, 148–154. [Google Scholar] [CrossRef]
- Chen, W.M.; Sheu, F.S.; Sheu, S.Y. Novel l-amino acid oxidase with algicidal activity against toxic cyanobacterium Microcystis aeruginosa synthesized by a bacterium Aquimarina sp. Enzym. Microb. Technol. 2011, 49, 372–379. [Google Scholar] [CrossRef]
- Hong, G.Y.; Wang, J.; Zhang, J. Isolation and identification of an algicidal bacterium against Microcystis aeruginosa. Chem. Eng. Trans. 2015, 55, 139–144. [Google Scholar] [CrossRef]
- Wang, J.; Luo, L.; Chen, Y.; He, Q.; Zhan, L.; Zhao, X. Spectra characteristic and algicidal mechanism of Chryseobacterium sp. S7 on Microcystis aeruginosa. Spectrosc. Spectr. Anal. 2019, 39, 1817–1822. [Google Scholar]
- Choi, H.-J.; Kim, B.-H.; Kim, J.-D.; Han, M.-S. Streptomyces neyagawaensis as a control for the hazardous biomass of Microcystis aeruginosa (Cyanobacteria) in eutrophic freshwaters. Biol. Control 2005, 33, 335–343. [Google Scholar] [CrossRef]
- Phankhajon, K.; Somdee, A.; Somdee, T. Algicidal activity of an actinomycete strain, Streptomyces rameus, against Microcystis aeruginosa. Water Sci. Technol. 2016, 74, 1398–1408. [Google Scholar] [CrossRef] [PubMed]
- Somdee, T.; Sumalai, N.; Somdee, A. A novel actinomycete Streptomyces aurantiogriseus with algicidal activity against the toxic cyanobacterium Microcystis aeruginosa. J. Appl. Phycol. 2013, 25, 1587–1594. [Google Scholar] [CrossRef]
- Zeng, Y.; Wang, J.; Yang, C.; Ding, M.; Hamilton, P.B.; Zhang, X.; Yang, C.; Zhnag, L.; Dai, X. A Streptomyces globisporus strain kills Microcystis aeruginosa via cell-to-cell contact. Sci. Total Environ. 2021, 769, 144489. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.-H.; Chen, W.; Li, H.-Q.; Zhou, E.-M.; Hu, W.-Y.; Duan, Y.-Q.; Mohamad, O.A.; Gao, R.; Li, W.-J. An antialgal compound produced by Streptomyces jiujiangensis JXJ 0074T. Appl. Microbiol. Biotechnol. 2015, 99, 7673–7683. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Cai, G.; Wang, H.; Hu, Z.; Zheng, W.; Lei, X.; Zhu, X.; Chen, Y.; Chen, Q.; Din, H.; et al. Fast-growing algicidal Streptomyces sp. U3 and its potential in harmful algal bloom controls. J. Hazard. Mater. 2017, 341, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Ahn, C.-Y.; Joung, S.-H.; Jeon, J.-W.; Kim, H.-S.; Yoon, B.-D.; Oh, H.-M. Selective control of cyanobacteria by surfactin-containing culture broth of Bacillus subtilis C1. Biotechnol. Lett. 2003, 25, 1137–1142. [Google Scholar] [CrossRef]
- Mu, R.-M.; Fan, Z.-Q.; Pei, H.-Y.; Yuan, X.-L.; Liu, S.-X.; Wang, X.-R. Isolation and algae-lysing characteristics of the algicidal bacterium B5. J. Environ. Sci. 2007, 19, 1336–1340. [Google Scholar] [CrossRef]
- Wu, L.; Guo, X.; Liu, X.; Yang, H. NprR-NprX Quorum-Sensing System Regulates the Algicidal Activity of Bacillus sp. Strain S51107 against Bloom-Forming Cyanobacterium Microcystis aeruginosa. Front. Microbiol. 2017, 8, 1968. [Google Scholar] [CrossRef]
- Shunyu, S.; Yongding, L.; Yinwu, S.; Genbao, L.; Dunhai, L. Lysis of Aphanizomenon flos-aquae (Cyanobacterium) by a bacterium Bacillus cereus. Biol. Control 2006, 39, 345–351. [Google Scholar] [CrossRef]
- Gumbo, J.; Cloete, T.; van Zyl, G.; Sommerville, J. The viability assessment of Microcystis aeruginosa cells after co-culturing with Bacillus mycoides B16 using flow cytometry. Phys. Chem. Earth, Parts A/B/C 2014, 72–75, 24–33. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Wu, H.; Chen, L.; Xie, S.; Zang, H.; Borriss, R.; Gao, X. Bacilysin from Bacillus amyloliquefaciens FZB42 Has Specific Bactericidal Activity against Harmful Algal Bloom Species. Appl. Environ. Microbiol. 2014, 80, 7512–7520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, J.; He, Y.; Chen, A.; Peng, L.; Luo, S.; Wu, G.; Zou, H.; Li, R. Interactive effects of algicidal efficiency of Bacillus sp. B50 and bacterial community on susceptibility of Microcystis aeruginosa with different growth rates. Int. Biodeterior. Biodegradation 2015, 97, 1–6. [Google Scholar] [CrossRef]
- Shao, J.; Jiang, Y.; Wang, Z.; Peng, L.; Luo, S.; Gu, J.; Li, R. Interactions between algicidal bacteria and the cyanobacterium Microcystis aeruginosa: Lytic characteristics and physiological responses in the cyanobacteria. Int. J. Environ. Sci. Technol. 2013, 11, 469–476. [Google Scholar] [CrossRef] [Green Version]
- Xu, B.; Miao, L.; Yu, J.; Ji, L.; Lu, H.; Yang, J.; Gao, S.; Kong, Y. Isolation and identification of amino acids secreted by Bacillus amyloliquefaciens T1 with anti-cyanobacterial effect against cyanobacterium Microcystis aeruginosa. Desalination Water Treat. 2021, 231, 329–339. [Google Scholar] [CrossRef]
- Wang, S.; Yang, S.; Zuo, J.; Hu, C.; Song, L.; Gan, N.; Chen, P. Simultaneous Removal of the Freshwater Bloom-Forming Cyanobacterium Microcystis and Cyanotoxin Microcystins via Combined Use of Algicidal Bacterial Filtrate and the Microcystin-Degrading Enzymatic Agent, MlrA. Microorganisms 2021, 9, 1594. [Google Scholar] [CrossRef]
- Tian, C.; Liu, X.; Tan, J.; Lin, S.; Li, D.; Yang, H. Isolation, identification and characterization of an algicidal bacterium from Lake Taihu and preliminary studies on its algicidal compounds. J. Environ. Sci. 2012, 24, 1823–1831. [Google Scholar] [CrossRef]
- Han, S.; Zhou, Q.; Lilje, O.; Xu, W.; Zhu, Y.; van Ogtrop, F.F. Inhibition mechanism of Penicillium chrysogenum on Microcystis aeruginosa in aquaculture water. J. Clean. Prod. 2021, 299, 126829. [Google Scholar] [CrossRef]
- Mohamed, Z.A.; Alamri, S.; Hashem, M.; Mostafa, Y. Growth inhibition of Microcystis aeruginosa and adsorption of microcystin toxin by the yeast Aureobasidium pullulans, with no effect on microalgae. Environ. Sci. Pollut. Res. 2020, 27, 38038–38046. [Google Scholar] [CrossRef]
- Wang, Q.; Su, M.; Zhu, W.; Li, X.; Jia, Y.; Guo, P.; Chen, Z.; Jiang, W.; Tian, X. Growth inhibition of Microcystis aeruginosa by white-rot fungus Lopharia spadicea. Water Sci. Technol. 2010, 62, 317–323. [Google Scholar] [CrossRef]
- Zeng, G.; Gao, P.; Wang, J.; Zhang, J.; Zhang, M.; Sun, D. Algicidal Molecular Mechanism and Toxicological Degradation of Microcystis aeruginosa by White-Rot Fungi. Toxins 2020, 12, 406. [Google Scholar] [CrossRef]
- Zeng, G.; Zhang, M.; Gao, P.; Wang, J.; Sun, D. Algicidal Efficiency and Genotoxic Effects of Phanerochaete chrysosporium against Microcystis aeruginosa. Int. J. Environ. Res. Public Health 2020, 17, 4029. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Feng, X.; Jia, Y.; Wang, C.; He, X.; Zhou, Q.; Tian, X. Isolation and evaluation of terrestrial fungi with algicidal ability from Zijin Mountain, Nanjing, China. J. Microbiol. 2011, 49, 562–567. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Ma, H.; Ren, S.; Gao, X.; He, X.; Zhu, S.; Deng, R.; Zhang, S. Insights into the mechanism of cyanobacteria removal by the algicidal fungi Bjerkandera adusta and Trametes versicolor. Microbiol. Open 2020, 9, e1042. [Google Scholar] [CrossRef] [PubMed]
- Jin, P.; Wang, H.; Liu, W.; Zhang, S.; Lin, C.; Zheng, F.; Miao, W. Bactericidal metabolites from Phellinus noxius HN-1 against Microcystis aeruginosa. Sci. Rep. 2017, 7, 3132. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Wang, Q.; Chen, Z.; Jiang, W.; Zhang, P.; Tian, X. Inhibition of phytoplankton species by co-culture with a fungus. Ecol. Eng. 2010, 36, 1389–1391. [Google Scholar] [CrossRef]
- Jia, Y.; Han, G.; Wang, C.; Guo, P.; Jiang, W.; Li, X.; Tian, X. The efficacy and mechanisms of fungal suppression of freshwater harmful algal bloom species. J. Hazard. Mater. 2010, 183, 176–181. [Google Scholar] [CrossRef]
- Du, J.; Pu, G.; Shao, C.; Cheng, S.; Cai, J.; Zhou, L.; Jia, Y.; Tian, X. Potential of extracellular enzymes from Trametes versicolor F21a in Microcystis spp. degradation. Mater. Sci. Eng. C 2014, 48, 138–144. [Google Scholar] [CrossRef]
- Dai, W.; Chen, X.; Wang, X.; Xu, Z.; Gao, X.; Jiang, C.; Deng, R.; Han, G. The Algicidal Fungus Trametes versicolor F21a Eliminating Blue Algae via Genes Encoding Degradation Enzymes and Metabolic Pathways Revealed by Transcriptomic Analysis. Front. Microbiol. 2018, 9, 826. [Google Scholar] [CrossRef]
- Wei, J.; Xie, X.; Huang, F.; Xiang, L.; Wang, Y.; Han, T.; Massey, I.Y.; Liang, G.; Pu, Y.; Yang, F. Simultaneous Microcystis algicidal and microcystin synthesis inhibition by a red pigment prodigiosin. Environ. Pollut. 2019, 256, 113444. [Google Scholar] [CrossRef]
- Annett, H.; Kunimitsu, K.M.W.M. Selective control of Microcystis using an amino acid-a laboratory assay. J. Appl. Phycol. 2002, 14, 85–89. [Google Scholar]
- Kaya, K.; Liu, Y.-D.; Shen, Y.-W.; Xiao, B.-D.; Sano, T. Selective control of toxic Microcystis water blooms using lysine and malonic acid: An enclosure experiment. Environ. Toxicol. 2005, 20, 170–178. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Chen, M.; Ren, C.; Wang, Y.; Li, L. Anticyanobacterial effect of l-lysine on Microcystis aeruginosa. RSC Adv. 2018, 8, 21606–21612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, M.-H. Algicidal Activity of a Dibenzofuran-Degrader Rhodococcus sp. J. Microbiol. Biotechnol. 2013, 23, 260–266. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto, Y.; Kouchiwa, T.; Hodoki, Y.; Hotta, K.; Uchida, H.; Harada, K.-I. Distribution and identification of actinomycetes lysing cyanobacteria in a eutrophic lake. J. Appl. Phycol. 1998, 10, 391–397. [Google Scholar] [CrossRef]
- Liu, Y.-M. Inhibition of Microcystis aeruginosa by the Extracellular Substances from an Aeromonas sp. J. Microbiol. Biotechnol. 2013, 23, 1304–1307. [Google Scholar] [CrossRef] [Green Version]
- Weiss, G.; Kovalerchick, D.; Lieman-Hurwitz, J.; Murik, O.; De Philippis, R.; Carmeli, S.; Sukenik, A.; Kaplan, A. Increased algicidal activity of Aeromonas veronii in response to Microcystis aeruginosa: Interspecies crosstalk and secondary metabolites synergism. Environ. Microbiol. 2019, 21, 1140–1150. [Google Scholar] [CrossRef]
- Feng, Y.; Chang, X.; Zhao, L.; Li, X.; Li, W.; Jiang, Y. Nanaomycin A methyl ester, an actinomycete metabolite: Algicidal activity and the physiological response of Microcystis aeruginosa. Ecol. Eng. 2013, 53, 306–312. [Google Scholar] [CrossRef]
- Gerphagnon, M.; Macarthur, D.; Latour, D.; Gachon, C.; Van Ogtrop, F.; Gleason, F.H.; Sime-Ngando, T. Microbial players involved in the decline of filamentous and colonial cyanobacterial blooms with a focus on fungal parasitism. Environ. Microbiol. 2015, 17, 2573–2587. [Google Scholar] [CrossRef]
- Su, J.F.; Shao, S.C.; Huang, T.L.; Ma, F.; Lu, J.S.; Zhang, K. Algicidal effects and denitrification activities of Acinetobacter sp. J25 against Microcystis aeruginosa. J. Environ. Chem. Eng. 2016, 4, 1002–1007. [Google Scholar] [CrossRef]
- Chen, Y.-D.; Zhu, Y.; Xin, J.-P.; Zhao, C.; Tian, R.-N. Succinic acid inhibits photosynthesis of Microcystis aeruginosa via damaging PSII oxygen-evolving complex and reaction center. Environ. Sci. Pollut. Res. 2021, 28, 58470–58479. [Google Scholar] [CrossRef]
- Kong, Y.; Xu, X.; Zhu, L. Cyanobactericidal Effect of Streptomyces sp. HJC-D1 on Microcystis auruginosa. PLoS ONE 2013, 8, e57654. [Google Scholar] [CrossRef] [PubMed]
- Zhai, C.; Zhang, P.; Shen, F.; Zhou, C.; Liu, C. Does Microcystis aeruginosa have quorum sensing? FEMS Microbiol. Lett. 2012, 336, 38–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reading, N.C.; Sperandio, V. Quorum sensing: The many languages of bacteria. FEMS Microbiol. Lett. 2006, 254, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zheng, L.; Wang, S.; Zhao, Y.; Xu, X.; Han, B.; Hu, T. Quorum Sensing Bacteria in the Phycosphere of HAB Microalgae and Their Ecological Functions Related to Cross-Kingdom Interactions. Int. J. Environ. Res. Public Health 2021, 19, 163. [Google Scholar] [CrossRef] [PubMed]
- Dow, L. How Do Quorum-Sensing Signals Mediate Algae–Bacteria Interactions? Microorganisms 2021, 9, 1391. [Google Scholar] [CrossRef]
- Liu, J.; Liu, K.; Zhao, Z.; Wang, Z.; Wang, F.; Xin, Y.; Qu, J.; Song, F.; Li, Z. The LuxS/AI-2 Quorum-Sensing System Regulates the Algicidal Activity of Shewanella xiamenensis Lzh-2. Front. Microbiol. 2022, 12. [Google Scholar] [CrossRef]
- Zhang, S.-J.; Du, X.-P.; Zhu, J.-M.; Meng, C.-X.; Zhou, J.; Zuo, P. The complete genome sequence of the algicidal bacterium Bacillus subtilis strain JA and the use of quorum sensing to evaluate its antialgal ability. Biotechnol. Rep. 2020, 25, e00421. [Google Scholar] [CrossRef]
- Dziallas, C.; Grossart, H.-P. Temperature and biotic factors influence bacterial communities associated with the cyanobacterium Microcystis sp. Environ. Microbiol. 2011, 13, 1632–1641. [Google Scholar] [CrossRef]
- He, L.; Lin, Z.; Wang, Y.; He, X.; Zhou, J.; Guan, M.; Zhou, J. Facilitating harmful algae removal in fresh water via joint effects of multi-species algicidal bacteria. J. Hazard. Mater. 2020, 403, 123662. [Google Scholar] [CrossRef]
- Sun, P.; Lin, H.; Wang, G.; Lu, L.-L.; Zhao, Y.-H. Preparation of a new-style composite containing a key bioflocculant produced by Pseudomonas aeruginosa ZJU1 and its flocculating effect on harmful algal blooms. J. Hazard. Mater. 2014, 284, 215–221. [Google Scholar] [CrossRef]
- Kim, H.S.; Park, Y.H.; Kim, S.; Choi, Y.-E. Application of a polyethylenimine-modified polyacrylonitrile-biomass waste composite fiber sorbent for the removal of a harmful cyanobacterial species from an aqueous solution. Environ. Res. 2020, 190, 109997. [Google Scholar] [CrossRef] [PubMed]
- Paerl, H.W.; Gardner, W.S.; Havens, K.E.; Joyner, A.R.; McCarthy, M.J.; Newell, S.; Qin, B.; Scott, J.T. Mitigating cyanobacterial harmful algal blooms in aquatic ecosystems impacted by climate change and anthropogenic nutrients. Harmful Algae 2016, 54, 213–222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kong, Y.; Xu, X.; Zhu, L.; Miao, L. Control of the Harmful Alga Microcystis aeruginosa and Absorption of Nitrogen and Phosphorus by Candida utilis. Appl. Biochem. Biotechnol. 2012, 169, 88–99. [Google Scholar] [CrossRef] [PubMed]
| Strain Name | Target Cyanobacterium | Initial Cyanobacterial Cell Density (cells mL−1) | Dosage (v/v) | Duration Time | Inhibition Rate/Removal Efficiency | Anticyanobacterial Modes | References | |
|---|---|---|---|---|---|---|---|---|
| α-Proteobacteria | Brevibacillus laterosporus Bl-zj | M. aeruginosa FACHB- 905 | 1.0 × 107 | 1.0 × 107 ** | 3 d (4 d) | 72.36% (92.30%) | NA | [57] |
| Brevundimonas sp. AA06 | M. aeruginosa FACHB-905 | 2.0 × 109 | NA | 4 d | 70% | NA | [24] | |
| Ochrobactrum sp. FDT5 | M. aeruginosa | 2.0~6.0 × 106 | 4.0 × 107 ** | 5 d | 58.9% | indirect attack | [25] | |
| Stappia sp. F2 | M. aeruginosa FACHB-905 | 2.5 × 106 | 10% | 7 d | 94.9% | indirect attack | [35] | |
| Rhizobium sp. AQ_MP | M. aeruginosa | NA | 9% | 10 d | 100% | NA | [58] | |
| β-Proteobacteria | Alcaligenes denitrificans | M. aeruginosa NIES 298 | 2 × 105 | 0.7% | 4 d | 96.4% | direct contact | [59] |
| Alcaligenes sp. H3 | wild cyanobacterium | NA | 20% | 4 d | 93% | indirect attack | [60] | |
| Paucibacter aquatile DH15 | NA | 1.0 × 106 | NA | 36h | 94.9% | combination of direct and indirect attacks | [61] | |
| Achromobacter spp. LG1 | M. aeruginosa CAAT 2005-3 | 1.0 × 105 | 1.0 × 106 ** | 7 d | 29.0 ± 1.8~55.0 ± 3.8% | NA | [62] | |
| M. aeruginosa 24A | 25.3 ± 2.2~ 48.3 ± 5.5% | |||||||
| γ-Proteobacteria | Pseudomonas aeruginosa | M. aeruginosa FACHB-905 | 437 ± 21 * | 5% (10%) | 7 d | 81.21% (83.84%) | NA | [18] |
| P. aeruginosa ACB3 | M. aeruginosa FACHB-912 | 0.55~1.13 × 106 | 1.0 × 107 ** | 6 d | 96.5% | NA | [63] | |
| M. aeruginosa FACHB-924 | 82.6% | |||||||
| P. aeruginosa UCBPP-PA14 | M. aeruginosa NIES 298 | 1.0 × 105 | 1.0 × 105 ** | 10 d | 82.4 ± 2.4% | NA | [64] | |
| M. aeruginosa NIES 44 | 75.0 ± 2.7% | |||||||
| M. aeruginosa NIER 101 | 69.0 ± 3.7% | |||||||
| M. aeruginosa NIER 100001 | 67.0 ± 4.2% | |||||||
| P. grimontii A01 | M. aeruginosa FACHB-905 | 1.0 × 107 | 10% | 7 d | 91.81% | NA | [65] | |
| P. grimontii A14 | 78.25% | NA | ||||||
| P. putida CH-22 | M. aeruginosa FACHB-905 | 5.3 × 106 | 15% | 7 d | 98.8% | indirect attack | [19] | |
| Pseudomonas sp. K44-1 | M. aeruginosa NIES 299 | NA | NA | NA | NA | indirect attack | [66] | |
| P. syringae KACC10292T | M. aeruginosa NIES 298 | 1.1 × 105 | 10% | 10 d | 96% | indirect attack | [20] | |
| Aeromonas bestiarum HYD0802-MK36 | 91% | direct attack | ||||||
| Aeromonas sp. FM | M. aeruginosa | NA | 5% (10%) | 9 d | 70.7% (88.1%) | indirect attack | [67] | |
| Aeromonas sp. FM | M. aeruginosa FACHB 927 | 1.4 × 107 | 2.1 × 109 ** | 4 d | up to 85% | NA | [68] | |
| M. aeruginosa FACHB 975 | 5.88 × 106 | 7 d | 91.2% | NA | ||||
| Aeromonas sp. GLY-2107 | M. aeruginosa 9110 | 1.0 × 107 | 1% | 6 d | 96.5±1.1% | indirect attack | [69] | |
| M. aeruginosa PCC 7806 | 88.9±1.9% | |||||||
| Aeromonas sp. L23 | M. aeruginosa UTEX LB 2385 | 6.0 × 106 | 25% | 5 d | 88 ± 1.2% | indirect attack | [21] | |
| M. aeruginosa NHSB | 94 ± 2.6% | |||||||
| Aeromonas sp. | NA | NA | 8% | 5 d | 95% | indirect attack | [70] | |
| Acinetobacter sp. J25 | NA | NA | 10% | 24 d | 87.86% | NA | [71] | |
| Acinetobacter sp. CMDB-2 | M. aeruginosa FACHB-905 | 1.0 × 106 | 5% | 3 d | 87.5% | indirect attack | [22] | |
| A. guillouiae A2 | M. aeruginosa FACHB-905 | ~1.0 × 106 | 10% | 7 d | 91.6% | indirect attack | [72] | |
| Raoultella sp. R11 | M. aeruginosa FACHB-905 | NA | 15% (30%) | 6 d | 57.63% (93.58%) | NA | [73] | |
| R. planticola | M. aeruginosa FACHB-905 | NA | 4% (8%) | 9 d (3 d) | nearly 60% (83%) | indirect attack | [70] | |
| R. ornithinolytica S1 | M. aeruginosa FACHB-905 | NA | 5% | 3 d | 96.2% | indirect attack | [23] | |
| Halobacillus sp. H9 | M. aeruginosa PCC 7806 | 2.0 × 107 | 5% | 24h | 90% (93 ± 1%) | indirect attack | [26] | |
| M. aeruginosa TAIHU98 | 87 ± 2% | |||||||
| Shewanella sp. Lzh-2 | M. aeruginosa 9110 | 1.0 × 107 | 10% | 6 d | 92.3 ± 6.8% | indirect attack | [27] | |
| M. aeruginosa PCC 7806 | 84.9 ± 3.8% | |||||||
| Stenotrophomonas maltophilia 15 | M. aeruginosa FACHB-905 | 400 * | NA | 16 d | ~80% | indirect attack | [74] | |
| Hahella sp. KA22 | M. aeruginosa FACHB-1752 | NA | 0.01 *** | 3 d | 60% | indirect attack | [31] | |
| Citrobacter sp. R1 | M. aeruginosa FACHB-905 | 1.0 × 107 | 16.7% | 3 d | 81.6 ± 2.2% | NA | [28] | |
| Citrobacter sp. AzoR-1 | M. aeruginosa | 1.0 × 107 | NA | NA | ~95% | indirect attack | [54] | |
| Enterobacter sp. NP23 | M. aeruginosa | 1.0 × 108 | 1.0 × 108 ** | 20 d | ~70% | NA | [75] | |
| Shigella sp. H3 | wild cyanobacterium | NA | 20% | 10 d | 76% | direct attack | [60] | |
| Serratia marcescens LTH-2 | M. aeruginosa TH1 | 3.0 × 106 | 5% | 2 d (3 d) | 72.4% (79.0%) | indirect attack | [76] | |
| M. aeruginosa TH1 | 70.0% (74.6%) | |||||||
| M. aeruginosa FACHB-905 | 84.3% (87.7%) | |||||||
| S. marcescens BWL1001 | M. aeruginosa | NA | NA | 2 d | 91.1% | indirect attack | [30] | |
| Aquimarina salinaria | M. aeruginosa MTY01 | 1.0 × 105 | 10% | 3 d (6 d) | 80% (100%) | indirect attack | [39,77] | |
| Chryseobacterium sp. | M. aeruginosa FACHB-905 | 6.0 × 106 | 10% | 3 d | up to 80% | direct attack | [40] | |
| Bacteroidetes | Chryseobacterium sp. H2 | M. aeruginosa FACHB-905 | NA | 10% | 7 d | 85.3% | NA | [78] |
| Chryseobacterium sp. GLY-1106 | M. aeruginosa 9110 | 1.0 × 107 | NA | 6 d | 98.9% | indirect attack | [41] | |
| Chryseobacterium sp. S7 | M. aeruginosa FACHB-905 | 718 * | 28.5% | 7 d | 59.37% | indirect attack | [79] | |
| Aureispira sp. CCB-QB1 | M. aeruginosa NISE 102 | NA | NA | 3min | 75.39% | indirect attack | [42] | |
| Pedobacter sp. Mal 11-5 | M. aeruginosa NIES 843 | NA | 6.7% | 2 d (10 d) | exceeded 50% (75~85%) | NA | [43] | |
| Actinomycetes | Streptomyces sp. NT0401 | M. aeruginosa PCC 7806 | NA | 5% | 5 d | up to 85% | indirect attack | [36] |
| M. aeruginosa XW01 | ||||||||
| Streptomyces sp. L74 | M. aeruginosa FACHB-905 | 1.0 × 106 | 10% | 4 d | 71.48 ± 5.33% | indirect attack | [33] | |
| S. neyagawaensis | M. aeruginosa NIES 298 | NA | NA | 7 d | 84.5% | NA | [80] | |
| S. rameus KKU-A3 | M. aeruginosa KKU-13 | NA | 10% | 7 d | 81.56% | NA | [81] | |
| S. aurantiogriseus PK1 | M. aeruginosa KKU-13 | ~1.5 × 106 | 5% | 8 d | ~83.3% | indirect attack | [82] | |
| Streptomyces sp. KY-34 | M. aeruginosa FACHB-905 | 354.3 ± 13.8 * | 3% (10%) | 8 d | 81.2% (99.0%) | indirect attack | [56] | |
| Streptomyces sp. HJC-D1 | M. aeruginosa FACHB-905 | 637.5 ± 32.1 * | 5% (10%) | 5 d | 88.4 ± 2.8% (91.8 ± 1.2%) | indirect attack | [32] | |
| S. globisporus G9 | M. aeruginosa NIES 44 | 300 ± 60 * | 5% | 5 d | 95.1 ± 1.6% | direct attack | [83] | |
| M. aeruginosa NIES 90 | 88.8 ± 3.7% | |||||||
| M. aeruginosa NIES 843 | 94.6 ± 1.4% | |||||||
| M. aeruginosa FACHB-905 | 84.9 ± 0.3% | |||||||
| M. aeruginosa PCC 7806 | 86.5 ± 2.1% | |||||||
| S. amritsarensis | M. aeruginosa NIES 44 | 500 ± 100 * | 5% | 5 d (10 d) | 81.4 ± 0.57% (80.7 ± 0.87%) | NA | [5] | |
| M. aeruginosa NIES 90 | 51.3 ± 7.83% (80.9 ± 6.49%) | |||||||
| M. aeruginosa NIES 843 | 74.6 ± 0.00% (89.8 ± 2.89%) | |||||||
| M. aeruginosa FACHB-905 | 85.4 ± 2.21% (98.8 ± 1.05%) | |||||||
| M. aeruginosa DCM4 | 83.2 ± 0.00% (96.6 ± 4.79%) | |||||||
| S. jiujiangensis JXJ 0074 | M. aeruginosa FACHB-905 | 5.0 × 106 | 10% | 8 d | 90.50 ± 1.08% | indirect attack | [84] | |
| Streptomyces sp. U3 | M. aeruginosa PCC 1752 | NA | 5% | 3 d | 36.22% | indirect attack | [85] | |
| Rhodococcus sp. KWR2 | M. aeruginosa NIES 843 | 1.72 × 106 | 2% (filtrate) | 5 d | 97% | indirect attack | [34] | |
| M. aeruginosa UTEX 2388 | 94% | |||||||
| M. aeruginosa KW | 79% | |||||||
| M. aeruginosa Mi 0601 | 75% | |||||||
| Microbacterium sp. F3 | M. aeruginosa FACHB-905 | 2.5 × 106 | 10% | 7 d | 84.8% | indirect attack | [35] | |
| Arthrobacter sp. | M. aeruginosa | 2.0 × 106 | 9% | 10 d | 32.3 ±13.8% | NA | [14] | |
| Firmicutes | Bacillus subtilis C1 | M. aeruginosa | 1000 * | 1% | 2 d | 85% | NA | [86] |
| B. fusiformis B5 | M. aeruginosa | 412.3 * | 3.6 × 107 ** | 7 d | nearly 90% | indirect attack | [87] | |
| Bacillus sp. S51107 | M. aeruginosa 9110 | 1.0 × 106 | 10% | 6 d | 92.51 ± 2.79% | indirect attack | [88] | |
| M. aeruginosa PCC 7806 | 91.65 ± 1.00% | |||||||
| Bacillus sp. AF-1 | M. aeruginosa NIES 843 | 1.6 × 103 | 2% | 3 d (6 d) | 77% (93%) | indirect attack | [51] | |
| Bacillus sp. Lzh-5 | M. aeruginosa 9110 | 1.0 × 107 | 10% | 6 d | 91.2 ± 6.3% | indirect attack | [46] | |
| Bacillus sp. T4 | M. aeruginosa KW | 1.0 × 106 | 5% | 3 d | ~100% | indirect attack | [48] | |
| B. licheniformis Sp34 | M. aeruginosa DCM3 | 1.35 × 105 | 5% | 5 d (10 d) | 69.4 ± 0.67 (97.1 ± 0.86%) | indirect attack | [47] | |
| M. aeruginosa DCM4 | 5 d (10 d) | 60.8 ± 1.63 (82.4 ± 2.09) | ||||||
| M. aeruginosa NIES 843 | 5 d (10 d) | 78.7 ± 5.94% (97.1 ± 0.86%) | ||||||
| B. cereus DC22 | M. aeruginosa FACHB-905 | 1.0 × 108 | 10% | 4 d (7 d) | 74.89 ± 2.23% (78.45 ± 0.68%) | NA | [89] | |
| Bacillus mycoides B16 | M. aeruginosa PCC 7806 | ~1.0 × 106 | NA | 6 d | 97% | NA | [90] | |
| Bacillus methylotrophicus ZJU | M. aeruginosa | 1.0 × 107 | 16.7% | 3 d | 89 ± 0.5% | indirect attack | [50] | |
| Bacillus sp. Mal 11-2 | M. aeruginosa NIES 843 | NA | 6.7% | 10 d | up to 60% | NA | [43] | |
| Bacillus sp. Mal 11-10 | 10 d | 55~64% | ||||||
| B. amyloliquefaciens FZB42 | M. aeruginosa NIES 843 | 1.0 × 106 | NA | 7 d | 98.78% | NA | [91] | |
| B. amyloliquefaciens CH03 | 94.39% | NA | ||||||
| Bacillus sp. B50 | M. aeruginosa FACHB-905 | NA | 10% | 5 d | 100% | indirect attack | [92,93] | |
| M. aeruginosa FACHB-1023 | 62.52% | |||||||
| M. aeruginosa NIES 843 | 100% | |||||||
| M. aeruginosa PCC 7806 | 66.90% | |||||||
| M. aeruginosa CHAB-439 | 71.08% | |||||||
| M. aeruginosa CHAB-456 | 60.33% | |||||||
| B. amyloliquefaciens T1 | M. aeruginosa FACHB-905 | 1.0 × 106 | 5% | 6 d | 99.4% | indirect attack | [49,94] | |
| M. aeruginosa FACHB-907 | 2% | 4 d | 76.9 ± 3.1% | [49] | ||||
| M. aeruginosa FACHB-908 | 2% | 4 d | 78.2 ± 2.2% | |||||
| M. aeruginosa FACHB-912 | 2% | 4 d | 72.9 ± 3.0% | |||||
| M. aeruginosa PCC 7806 | 2% | 4 d | 85.1 ± 1.8% | |||||
| B. methylotrophicus ZJU | M. aeruginosa | 1.0 × 107 | 16.7% | 3 d | 89.0 ± 0.5% | NA | [50] | |
| Paenibacillus sp. SJ-73 | M. aeruginosa PCC 7806 | NA | 5% | 7 d | 83.97 ± 1.60% | indirect attack | [95] | |
| M. aeruginosa TH1701 | NA | 5% (10%) | 92.10% (94.38%) | |||||
| Exiguobacterium sp. h10 | M. aeruginosa PCC 7820 | NA | 5% | 2 d (6 d) | 43.4% (73.6%) | indirect attack | [44] | |
| Exiguobacterium sp. A27 | M. aeruginosa PCC 7806 | 1.0 × 107 | 10% | 2 d | 64.4 ± 10.3% | indirect attack | [96] | |
| M. aeruginosa 9110 | NA | 58.3 ± 8.2% | ||||||
| Exiguobacterium indicum EI9 | M. aeruginosa FACHB-905 | 4.4 × 107 | 1.1 × 108 ** | NA | NA | NA | [45] | |
| Staphylococcus sp. F1 | M. aeruginosa FACHB-905 | 2.5 × 106 | 10% | 7 d | 96.0% | indirect attack | [35] | |
| Thermus | Deinococcus metallilatus MA1002 | M. aeruginosa PCC 7806 | 6.0 × 106 | 10% | 3 d | up to 80% | indirect attack | [52] |
| Ascomycota | Trichoderma citrinoviride | M. aeruginosa | 3.2 × 104 | 10% | 2 d | 100% | NA | [6] |
| Aspergillus niger 7806F3 | M. aeruginosa PCC 7820 | 5.0 × 106 | 10% | 4 d | up to 80% | indirect attack | [15] | |
| Penicillium chrysogenum | M. aeruginosa | NA | 3.85% | 6 d | 69.56% | indirect attack | [97] | |
| Aureobasidium pullulans KKUY070 | M. aeruginosa DRCK1 | 5.0 × 104 | 1.2 × 106 ** | 1 d (3 d) | 84% (100%) | NA | [98] | |
| Basidiomycetes | Lopharia spadicea | M. aeruginosa FACHB-912 | 798 ± 13 * | NA | 39h | 100% | NA | [99] |
| Phanerochaete chrysosporium | M. aeruginosa | about 1.57 × 107 | 500 *** | NA | 88.6 ± 0.52% | NA | [100,101] | |
| Irpex lacteus T2b | M. aeruginosa PCC 7806 | 646.25±19.11 * | 5% | 30h | 96.82% | direct attack | [102] | |
| Trametes hirsuta T24 | 705.19±15.45 * | 39h | 60.19% | |||||
| T. versicolor F21a | 701.33±13.50 * | 30h | 100% | [102,103] | ||||
| Bjerkandera adusta T1 | 656.28±26.78 * | 39h | 98.35% | |||||
| Phellinus noxius HN-1 | M. aeruginosa NIES 843 | 656.28 ± 26.78 * | NA | NA | NA | NA | [104] | |
| Trichaptum abietinum 1302BG | M. aeruginosa FACHB-918 | 750 * | NA | 2 d | 100% | direct attack | [105] | |
| M. aeruginosa PCC 7806 | 1300 * | NA | 36h | 100% |
| Anticyanobacterial Substances | Strain Name | Target Cyanobacterium | Initial Cyanobacterial Cell Density (cells mL−1) | EC50 (mg L−1) | References | |
|---|---|---|---|---|---|---|
| Alkaloids | Harmane (1-methyl-β-carboline) | Pseudomonas sp. K44-1 | M. aeruginosa NIES 299 | NA | NA | [66] |
| prodigiosin (C20H25N3O) | S. marcescens LTH-2 | M. aeruginosa TH1 | 3.0 × 106 | 0.048 ± 0.004 (24 h) | [76,109] | |
| M. aeruginosa TH2 | 0.089 ± 0.011 (24 h) | |||||
| M. aeruginosa FACHB-905 | 0.25 (24 h)/0.16 (72 h) | |||||
| Hahella sp. KA22 | M. aeruginosa FACHB-1752 | NA | 5.87 (72 h) | [31] | ||
| S. marcescens BWL1001 | M. aeruginosa | NA | NA | [30] | ||
| 2-(3, 4-dihydroxy2-methoxyphenyl)-1, 3-benzodioxole-5-carbaldehyde | Phellinus noxius HN-1 | M. aeruginosa NIES 843 | 656.28 ± 26.78 * | 20.6 (72 h) | [104] | |
| 3, 4-dihydroxybenzalacetone(C10H10O3) | 5.1 (72 h) | |||||
| Bacilysin (L-alanyl-[2,3-epoxycyclohexanone-4]-L-alanine) | Bacillus amyloliquefaciens FZB42 | M. aeruginosa NIES 843 | 1.0 × 106 | 4.13 (96h) | [91] | |
| tryptamine (C10H12N2) | Streptomyces eurocidicus JXJ-0089 | NA | NA | 3.00 ± 0.09 (72 h) | [38] | |
| Tryptoline (C11H12N2) | 2.54 ± 0.05 (72 h) | |||||
| 3-methylindole | Aeromonas sp. GLY-2107 | M. aeruginosa 9110 | 1.0 × 107 | 1.10 (24 h) | [69] | |
| indole-3-carboxaldehyde | Bacillus sp. S51107 | M. aeruginosa 9110 | 1.0 × 106 | 6.55 (24 h) | [88] | |
| 2′-deoxyadenosine (C10H13N5O3) | Streptomyces jiujiangensis JXJ 0074 | M. aeruginosa FACHB-905 | 5.0 × 106 | 6.42 (72 h) | [84] | |
| adenosine | 53.75 (72 h) | |||||
| 2, 3-indolinedione | Shewanella sp. Lzh-2 | M. aeruginosa 9110 | 1.0 × 107 | 12.5 | [27] | |
| 4-hydroxyphenethylamine (C8H11NO) | Acinetobacter guillouiae A2 | M. aeruginosa FACHB-905 | ~1.0 × 106 | 22.5 ± 1.9 (72 h) | [72] | |
| Fatty acid/Cyclic peptides/peptide derivates | cyclo(Gly-Pro) | Stenotrophomonas sp. F6 | M. aeruginosa 9110 | NA | 5.9 (24 h) | [29] |
| cyclo(Pro-Phe) | Bacillus sp. S51107 | M. aeruginosa 9110 | 1.0 × 106 | 1.85 (24 h) | [88] | |
| cyclo(4-OH-Pro-Leu) (C11H18N2O3) | Chryseobacterium sp. GLY-1106 | M. aeruginosa 9110 | 1.0 × 107 | 1.26 (24 h) | [41] | |
| cyclo(Pro-Leu) (C11H18N2O2) | 2.70 (24 h) | |||||
| Cyclo(Gly-Pro) | Bacillus sp. Lzh-5 | M. aeruginosa 9110 | 1.0 × 107 | 5.7 (24 h) | [46] | |
| Cyclo(Pro-Val) | 19.4 (24 h) | |||||
| cyclo(Gly-Pro) | Shewanella sp. Lzh-2 | M. aeruginosa 9110 | 1.0 × 107 | 5.7 (24 h) | [27] | |
| cyclo(Gly-Phe) | Aeromonas sp. GLY-2107 | M. aeruginosa 9110 | 1.0 × 107 | 4.72 (24 h) | [69] | |
| trans-3-indoleacrylic acid | Rhodococcus sp. p52 | M. aeruginosa | 7.3 × 106 | NA | [113] | |
| DL-pipecolic acid | NA | |||||
| L-pyroglutamic acid | NA | |||||
| fusaricidins | Paenibacillus polymyxa E681 | M. aeruginosa KW | 2.37 ± 0.15 ×107 | NA | [3] | |
| Protein/Amino acids | protein | Raoultella planticola | M. aeruginosa FACHB-905 | NA | NA | [70] |
| Aeromonas sp. | ||||||
| L-lysine and L-phenylalanine | Bacillus amyloliquefaciens T1 | M. aeruginosa FACHB-905 | 1.0 × 106 | NA | [94] | |
| L-valine | Streptomyces jiujiangensis JXJ 0074 | M. aeruginosa FACHB-905 | 5.0 × 106 | NA | [37] | |
| L-lysine | Streptomyces phaeofaciens S-9 | M. aeruginosa NIES 112 | NA | NA | [114] | |
| M. aeruginosa NIES 298 | ||||||
| lysine | Aeromonas sp. FM | M. aeruginosa FACHB-905 | NA | NA | [115] | |
| Enzymes | enzyme | Streptomyces neyagawaensis | M. aeruginosa NIES 298 | NA | NA | [80] |
| L-amino acid oxidase | Aquimarina spongiae | M. aeruginosa MTY01 | NA | NA | [77] | |
| microcystinase A | Sphingopyxis sp. C1 | M. aeruginosa FACHB-905 | 3.75 × 106 | NA | [55] | |
| Others | active flocculating substance | Halobacillus sp. H9 | M. aeruginosa PCC 7806 | 2.0 × 107 | NA | [26] |
| M. aeruginosa TAIHU98 | ||||||
| clavulanate | Aeromonas sp. FM | M. aeruginosa FACHB-905 | NA | NA | [115] | |
| biosurfactant | Bacillus subtilis C1 | M. aeruginosa | 1000 * | NA | [86] | |
| lumichrome | Aeromonas veronii A134 | M. aeruginosa MGK | NA | NA | [116] | |
| triterpenoid saponin (C42H70O13) | Streptomyces sp. L74 | M. aeruginosa FACHB-905 | 1×106 | NA | [33] | |
| hydroquinone | Stenotrophomonas sp. F6 | M. aeruginosa 9110 | NA | 0.96 (24 h) | [29] | |
| nanaomycin A methyl ester | Streptomyces hebeiensis YIM 001T | M. aeruginosa FACHB-905 | ~1.0 × 106 | 2.97 (72 h) | [117] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, Y.; Wang, Y.; Miao, L.; Mo, S.; Li, J.; Zheng, X. Recent Advances in the Research on the Anticyanobacterial Effects and Biodegradation Mechanisms of Microcystis aeruginosa with Microorganisms. Microorganisms 2022, 10, 1136. https://doi.org/10.3390/microorganisms10061136
Kong Y, Wang Y, Miao L, Mo S, Li J, Zheng X. Recent Advances in the Research on the Anticyanobacterial Effects and Biodegradation Mechanisms of Microcystis aeruginosa with Microorganisms. Microorganisms. 2022; 10(6):1136. https://doi.org/10.3390/microorganisms10061136
Chicago/Turabian StyleKong, Yun, Yue Wang, Lihong Miao, Shuhong Mo, Jiake Li, and Xing Zheng. 2022. "Recent Advances in the Research on the Anticyanobacterial Effects and Biodegradation Mechanisms of Microcystis aeruginosa with Microorganisms" Microorganisms 10, no. 6: 1136. https://doi.org/10.3390/microorganisms10061136
APA StyleKong, Y., Wang, Y., Miao, L., Mo, S., Li, J., & Zheng, X. (2022). Recent Advances in the Research on the Anticyanobacterial Effects and Biodegradation Mechanisms of Microcystis aeruginosa with Microorganisms. Microorganisms, 10(6), 1136. https://doi.org/10.3390/microorganisms10061136







