The Presence of Mycobacterium leprae in Wild Rodents
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sampling
2.2. ELISA Method
2.3. Molecular Methods
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Imbiriba, E.N.B.; da Silva Neto, A.L.; de Souza, W.V.; Pedrosa, V.; da Graça Cunha, M.; Garnelo, L. Desigualdade social, crescimento urbano e hanseníase em Manaus: Abordagem espacial. Rev. Saúde Pública 2009, 43, 656–665. [Google Scholar] [CrossRef] [Green Version]
- Bratschi, M.W.; Steinmann, P.; Wickenden, A.; Gillis, T.P. Current Knowledge on Mycobacterium leprae Transmission: A Systematic Literature Review. Lepr. Rev. 2015, 86, 142–155. [Google Scholar] [CrossRef]
- Pine, R.H. Neotropical Rainforest Mammals: A Field Guide; JSTOR: New York, NY, USA, 1999; ISBN 0022-2372. [Google Scholar]
- Reis, N.R.; Peracchi, A.L.; Pedro, W.A.; Lima, I.P. Ordem Rodentia. In Mamíferos Do Brasil, 2nd ed.; UEL: Londrina, Brazil, 2011; pp. 347–439. [Google Scholar]
- Brewer, S.W.; Rejmánek, M. Small Rodents as Significant Dispersers of Tree Seeds in a Neotropical Forest. J. Veg. Sci. 1999, 10, 165–174. [Google Scholar] [CrossRef] [Green Version]
- Cáceres, N.C. Food Habits and Seed Dispersal by the White-Eared Opossum, Didelphis albiventris, in Southern Brazil. Stud. Neotrop. Fauna Environ. 2002, 37, 97–104. [Google Scholar] [CrossRef]
- Caceres, N.C.; Araujo Monteiro-Filho, E.L. The Common Opossum, Didelphis aurita, as a Seed Disperser of Several Plants in Southern Brazil. Ciênc. Cult. 2000, 52, 41–44. [Google Scholar]
- McLester, E. Golden-Bellied Mangabeys (Cercocebus chrysogaster) Consume and Share Mammalian Prey at LuiKotale, Democratic Republic of the Congo. J. Trop. Ecol. 2022, 1–5. [Google Scholar] [CrossRef]
- Bugir, C.K.; Butynski, T.M.; Hayward, M.W. Prey Preferences of the Chimpanzee (Pan troglodytes). Ecol. Evol. 2021, 11, 7138–7146. [Google Scholar] [CrossRef]
- Watts, D.P. Meat Eating by Nonhuman Primates: A Review and Synthesis. J. Hum. Evol. 2020, 149, 102882. [Google Scholar] [CrossRef]
- Cruz, A.C.R.; dos Prazeres, A.d.S.C.; Gama, E.C.; de Lima, M.F.; Azevedo, R.d.S.S.; Casseb, L.M.N.; Nunes Neto, J.P.; Martins, L.C.; Chiang, J.O.; Rodrigues, S.G.; et al. Vigilância sorológica para arbovírus em Juruti, Pará, Brasil. Cad. Saúde Pública 2009, 25, 2517–2523. [Google Scholar] [CrossRef] [Green Version]
- Rojas-Espinosa, O.; Løvik, M. Mycobacterium Leprae and Mycobacterium lepraemurium Infections in Domestic and Wild Animals. Rev. Sci. Tech. (Int. Off. Epizoot.) 2001, 20, 219–251. [Google Scholar] [CrossRef]
- Redford, K.H. Mammals of the Neotropics: The Central Neotropics: Ecuador, Peru, Bolivia, Brazil. The Central Neotropics: Ecuador, Peru, Bolivia, Brazil; University of Chicago Press: Chicago, IL, USA, 1999; ISBN 0-226-19541-4. [Google Scholar]
- Bonvicino, C.R.; de Oliveira, J.; D’Andrea, P.S. Guia Dos Roedores Do Brasil, Com Chaves Para Gêneros Baseadas em Caracteres Externos; Série de Manuais Técnicos 11; IRIS PAHO: Washington, DC, USA, 2008. [Google Scholar]
- Bührer, S.S.; Smits, H.L.; Gussenhoven, G.C.; Van Ingen, C.W.; Klatser, P.R. A Simple Dipstick Assay for the Detection of Antibodies to Phenolic Glycolipid-I of Mycobacterium leprae. Am. J. Trop. Med. Hyg. 1998, 58, 133–136. [Google Scholar] [CrossRef]
- Lima, L.N.G.C.; Frota, C.C.; Mota, R.M.S.; Almeida, R.L.F.; de Andrade Pontes, M.A.; de Sá Gonçalves, H.; Rodrigues, L.C.; Kendall, C.; Kerr, L. Widespread Nasal Carriage of Mycobacterium leprae among a Healthy Population in a Hyperendemic Region of Northeastern Brazil. Memórias Do Inst. Oswaldo Cruz 2015, 110, 898–905. [Google Scholar] [CrossRef] [Green Version]
- Junior, A.d.P.M.; Bergmann, J.A.G.; Heinemann, M.B.; da Silva, N. Cadernos Técnicos de Veterinária E Zootecnia; MVZ; Universidade Federal de Minas Gerais: Belo Horizonte, Brazil, 2014; Volume 72. [Google Scholar]
- Zanella, J.R.C. Zoonoses Emergentes e Reemergentes e Sua Importância Para Saúde e Produção Animal. Pesqui. Agropecuária Bras. 2016, 51, 510–519. [Google Scholar] [CrossRef] [Green Version]
- Howerth, E.W.; Stallknecht, D.E.; Davidson, W.R.; Wentworth, E.J. Survey for Leprosy in Nine-Banded Armadillos (Dasypus novemcinctus) from the Southeastern United States. J. Wildl. Dis. 1990, 26, 112–115. [Google Scholar] [CrossRef] [Green Version]
- Walsh, G.P.; Meyers, W.M.; Binford, C.H. Naturally Acquired Leprosy in the Nine-banded Armadillo: A Decade of Experience 1975–1985. J. Leukoc. Biol. 1986, 40, 645–656. [Google Scholar] [CrossRef]
- Balamayooran, G.; Pena, M.; Sharma, R.; Truman, R.W. The Armadillo as an Animal Model and Reservoir Host for Mycobacterium leprae. Clin. Dermatol. 2015, 33, 108–115. [Google Scholar] [CrossRef]
- Wahyuni, R.; Adriaty, D.; Prakoeswa, C.R.S.; Agusni, I. Mycobacterium Leprae in Daily Water Resources of Inhabitants Who Live in Leprosy Endemic Area of East Java. Indones. J. Trop. Infect. Dis. 2010, 1, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Kazda, J.; Irgens, L.M.; Kolk, A.H. Acid-Fast Bacilli Found in Sphagnum Vegetation of Coastal Norway Containing Mycobacterium leprae-Specific Phenolic Glycolipid-I. Int. J. Lepr. 1990, 58, 353–357. [Google Scholar]
- Sharma, R.; Singh, P.; Loughry, W.J.; Lockhart, J.M.; Inman, W.B.; Duthie, M.S.; Pena, M.T.; Marcos, L.A.; Scollard, D.M.; Cole, S.T. Zoonotic Leprosy in the Southeastern United States. Emerg. Infect. Dis. 2015, 21, 2127. [Google Scholar] [CrossRef]
- Truman, R.W.; Singh, P.; Sharma, R.; Busso, P.; Rougemont, J.; Paniz-Mondolfi, A.; Kapopoulou, A.; Brisse, S.; Scollard, D.M.; Gillis, T.P. Probable Zoonotic Leprosy in the Southern United States. N. Engl. J. Med. 2011, 364, 1626–1633. [Google Scholar] [CrossRef] [Green Version]
- Truman, R.W.; Krahenbuhl, J.L.; Viable, M. Leprae as a Research Reagent. Int. J. Lepr. Other Mycobact. Dis. 2001, 69, 1–12. [Google Scholar] [PubMed]
- Truman, R.; Fine, P.E. ‘Environmental’ sources of Mycobacterium leprae: Issues and Evidence. Lepr. Rev. 2010, 81, 89–95. [Google Scholar]
- Walsh, G.P.; Storrs, E.E.; Meyers, W.; Binford, C.H. Naturally Acquired Leprosy-like Disease in the Nine-Banded Armadillo (Dasypus novemcinctus): Recent Epizootiologic Findings. J. Reticuloendothel. Soc. 1977, 22, 363–367. [Google Scholar] [PubMed]
- Loughry, W.J.; Truman, R.W.; McDonough, C.M.; Tilak, M.-K.; Garnier, S.; Delsuc, F. Is Leprosy Spreading among Nine-Banded Armadillos in the Southeastern United States? J. Wildl. Dis. 2009, 45, 144–152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cardona-Castro, N.; Beltrán-Alzate, J.C.; Romero-Montoya, I.M.; Li, W.; Brennan, P.J.; Vissa, V. Mycobacterium leprae in Colombia Described by SNP7614 in GyrA, Two Minisatellites and Geography. Infect. Genet. Evol. 2013, 14, 375–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pedrini, S.C.B.; Rosa, P.S.; Medri, Í.M.; Mourão, G.; Bagagli, E.; de Magalhães Lopes, C.A. Search for Mycobacterium leprae in Wild Mammals. Braz. J. Infect. Dis. 2010, 14, 47–53. [Google Scholar] [CrossRef] [Green Version]
- Frota, C.C.; Lima, L.N.C.; Rocha, A.D.S.; Suffys, P.N.; Rolim, B.N.; Rodrigues, L.C.; Barreto, M.L.; Kendall, C.; Kerr, L.R.S. Mycobacterium Leprae in Six-Banded (Euphractus sexcinctus) and Nine-Banded Armadillos (Dasypus novemcinctus) in Northeast Brazil. Memórias Do Inst. Oswaldo Cruz 2012, 107, 209–213. [Google Scholar] [CrossRef] [Green Version]
- Gentile, R.; Cardoso, T.S.; Costa-Neto, S.F.; Teixeira, B.R.; D’Andrea, P.S. Community Structure and Population Dynamics of Small Mammals in an Urban-Sylvatic Interface Area in Rio de Janeiro, Brazil. Zoologia 2018, 35, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Cunha, A.A.; Vieira, M.V. Age, Season, and Arboreal Movements of the Opossum Didelphis aurita in an Atlantic Rain Forest of Brazil. Acta Theriol. 2005, 50, 551–560. [Google Scholar] [CrossRef]
- De Camargo, N.F. Uso Dos Estratos Verticais Por Pequenos Mamíferos Em Formações Florestais Do Cerrado Brasileiro: Padrões de Diversidade, Relação Com a Disponibilidade de Recursos, Seleção de Hábitat e Habilidade de Locomoção Arborícola Das Espécies. Ph.D. Thesis, University of Brasíli, Brasília, Brazil, 2015. [Google Scholar]
- Melo, G.L.; Miotto, B.; Peres, B.; Caceres, N.C. Microhabitat of Small Mammals at Ground and Understorey Levels in a Deciduous, Southern Atlantic Forest. An. Acad. Bras. Ciênc. 2013, 85, 727–736. [Google Scholar] [CrossRef] [Green Version]
- Rocha, M.F.; Passamani, M.; Louzada, J. A Small Mammal Community in a Forest Fragment, Vegetation Corridor and Coffee Matrix System in the Brazilian Atlantic Forest. PLoS ONE 2011, 6, e23312. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, S.H.R.; Vasconcellos, S.A.; Morais, Z.; Teixeira, A.d.A.; Dias, R.A.; Guimarães, M.A.d.B.V.; Ferreira, F.; Ferreira Neto, J.S. Epidemiologia Da Leptospirose em Animais Silvestres Na Fundação Parque Zoológico de São Paulo. Braz. J. Vet. Res. Anim. Sci. 2004, 41, 189–193. [Google Scholar] [CrossRef]
- Meerburg, B.G.; Singleton, G.R.; Kijlstra, A. Rodent-Borne Diseases and Their Risks for Public Health. Crit. Rev. Microbiol. 2009, 35, 221–270. [Google Scholar] [CrossRef]
- Hamdan, N.E.S.; Ng, Y.L.; Lee, W.B.; Tan, C.S.; Khan, F.A.A.; Chong, Y.L. Rodent Species Distribution and Hantavirus Seroprevalence in Residential and Forested Areas of Sarawak, Malaysia. Trop. Life Sci. Res. 2017, 28, 151. [Google Scholar] [CrossRef] [PubMed]
- Körting, K.S.; Flach, J.; Honscha, G.; de Martinez, A.M.B. Hantavirose: Patologia e Registro No Brasil. 2008. Available online: https://www.arca.fiocruz.br/bitstream/icict/46497/2/Hantavirose%20no%20Brasil.pdf (accessed on 31 March 2022).
- Navea-Pérez, H.M.; Díaz-Sáez, V.; Corpas-López, V.; Merino-Espinosa, G.; Morillas-Márquez, F.; Martín-Sánchez, J. Leishmania Infantum in Wild Rodents: Reservoirs or Just Irrelevant Incidental Hosts? Parasitol. Res. 2015, 114, 2363–2370. [Google Scholar] [CrossRef] [PubMed]
- Echchakery, M.; Chicharro, C.; Boussaa, S.; Nieto, J.; Carrillo, E.; Sheila, O.; Moreno, J.; Boumezzough, A. Molecular Detection of Leishmania infantum and Leishmania tropica in Rodent Species from Endemic Cutaneous Leishmaniasis Areas in Morocco. Parasites Vectors 2017, 10, 454. [Google Scholar] [CrossRef] [Green Version]
- Caldart, E.T.; Freire, R.L.; Ferreira, F.P.; Ruffolo, B.B.; Sbeghen, M.R.; Mareze, M.; Garcia, J.L.; Mitsuka-Breganó, R.; Navarro, I.T. Leishmania in Synanthropic Rodents (Rattus rattus): New Evidence for the Urbanization of Leishmania (Leishmania) Amazonensis. Rev. Bras. Parasitol. Vet. 2017, 26, 17–27. [Google Scholar] [CrossRef]
- Durnez, L.; Eddyani, M.; Mgode, G.F.; Katakweba, A.; Katholi, C.R.; Machang’u, R.R.; Kazwala, R.R.; Portaels, F.; Leirs, H. First Detection of Mycobacteria in African Rodents and Insectivores, Using Stratified Pool Screening. Appl. Environ. Microbiol. 2008, 74, 768–773. [Google Scholar] [CrossRef] [Green Version]
- Durnez, L.; Katakweba, A.; Sadiki, H.; Katholi, C.R.; Kazwala, R.R.; Machang’u, R.R.; Portaels, F.; Leirs, H. Mycobacteria in Terrestrial Small Mammals on Cattle Farms in Tanzania. Vet. Med. Int. 2011, 2011, 495074. [Google Scholar] [CrossRef] [Green Version]
- Cavanagh, R.; Begon, M.; Bennett, M.; Ergon, T.; Graham, I.M.; De Haas, P.E.W.; Hart, C.A.; Koedam, M.; Kremer, K.; Lambin, X.; et al. Mycobacterium microti Infection (Vole tuberculosis) in Wild Rodent Populations. J. Clin. Microbiol. 2002, 40, 3281–3285. [Google Scholar] [CrossRef] [Green Version]
- Galletti, G.; Cavicchi, G.; Ussia, G. Replication of Mycobacterium leprae in Hibernating Ground Squirrels (Citellus tridecemlineatus). Acta Leprol. 1982, 88, 23–31. [Google Scholar]
- Meredith, A.; Del Pozo, J.; Smith, S.; Milne, E.; Stevenson, K.; McLuckie, J. Leprosy in Red Squirrels in Scotland. Vet. Rec. 2014, 175, 285–286. [Google Scholar] [CrossRef] [PubMed]
- Avanzi, C.; Del-Pozo, J.; Benjak, A.; Stevenson, K.; Simpson, V.R.; Busso, P.; McLuckie, J.; Loiseau, C.; Lawton, C.; Schoening, J.; et al. Red Squirrels in the British Isles Are Infected with Leprosy Bacilli. Science 2016, 354, 744–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilling, A.-K.; Avanzi, C.; Ulrich, R.G.; Busso, P.; Pisanu, B.; Ferrari, N.; Romeo, C.; Mazzamuto, M.V.; McLuckie, J.; Shuttleworth, C.M.; et al. British Red Squirrels Remain the Only Known Wild Rodent Host for Leprosy Bacilli. Front. Vet. Sci. 2019, 6, 8. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, K.; Tanigawa, K.; Kawashima, A.; Miyamura, T.; Ishii, N. Chimpanzees Used for Medical Research Shed Light on the Pathoetiology of Leprosy. Future Microbiol. 2011, 6, 1151–1157. [Google Scholar] [CrossRef] [PubMed]
- Gorbunova, V.; Bozzella, M.J.; Seluanov, A. Rodents for Comparative Aging Studies: From Mice to Beavers. Age 2008, 30, 111–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kajin, M.; Cerqueira, R.; Vieira, M.V.; Gentile, R. Nine-Year Demography of the Black-Eared Opossum Didelphis aurita (Didelphimorphia: Didelphidae) Using Life Tables. Rev. Bras. Zool. 2008, 25, 206–213. [Google Scholar] [CrossRef]


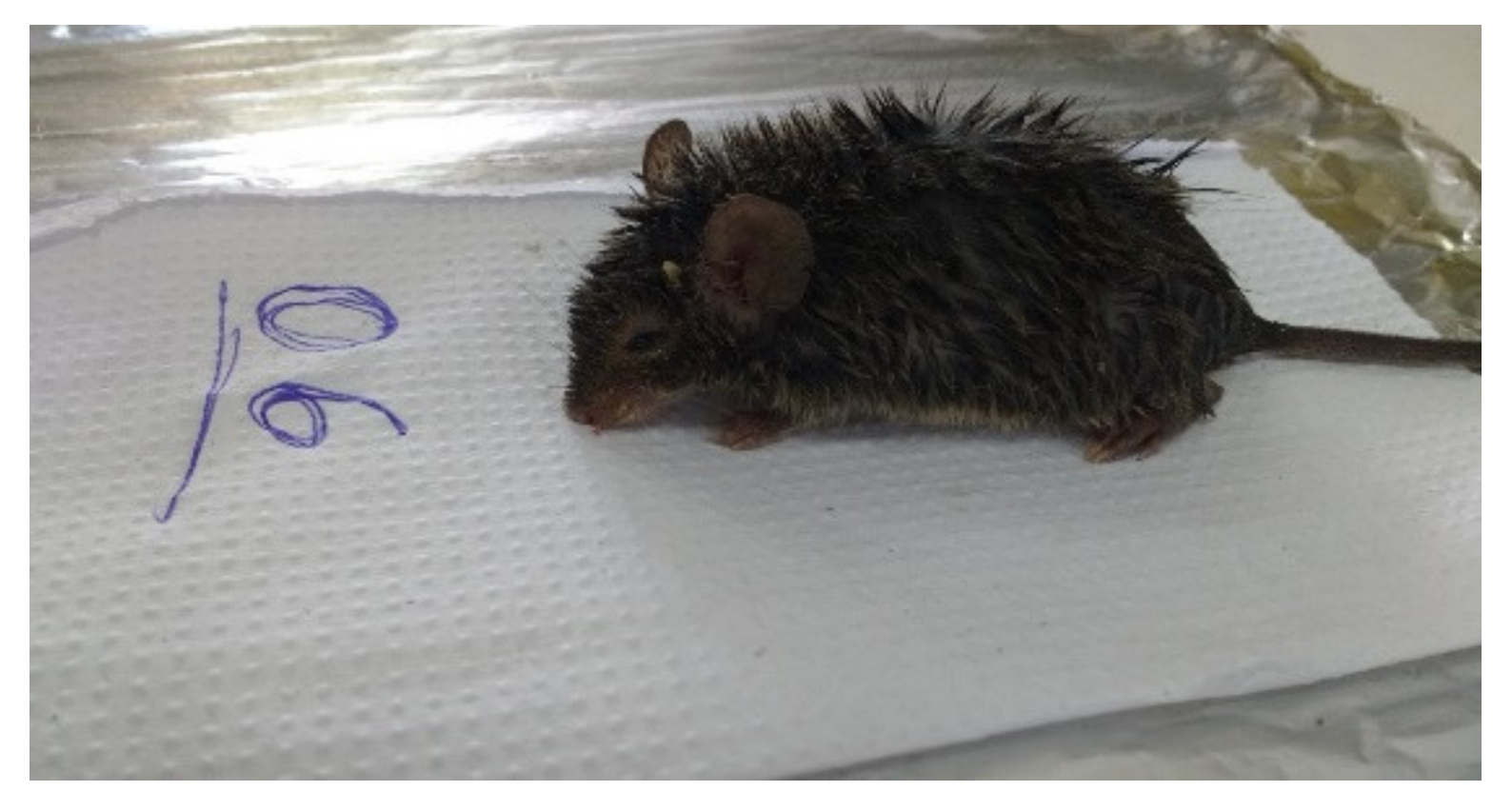

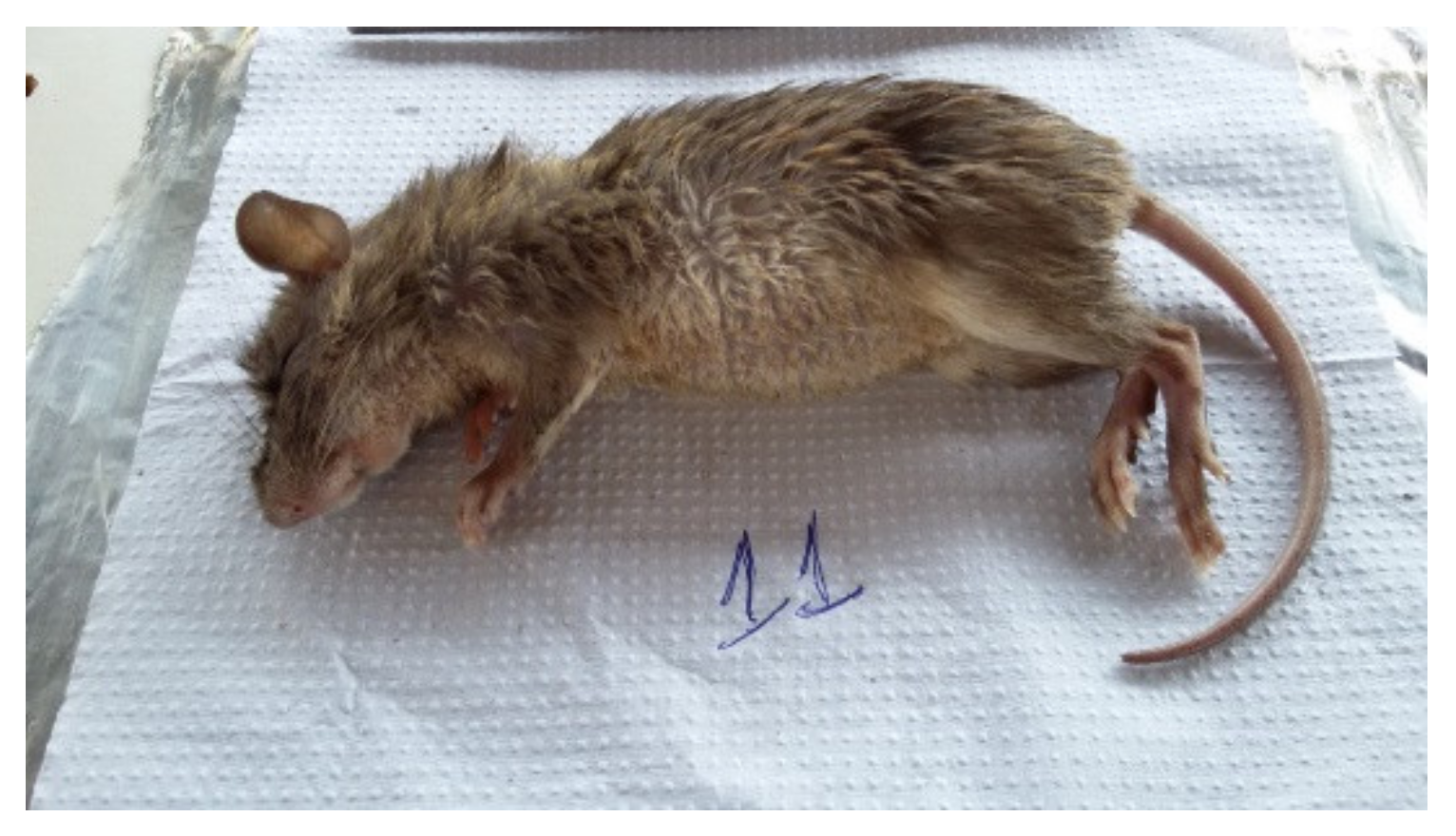
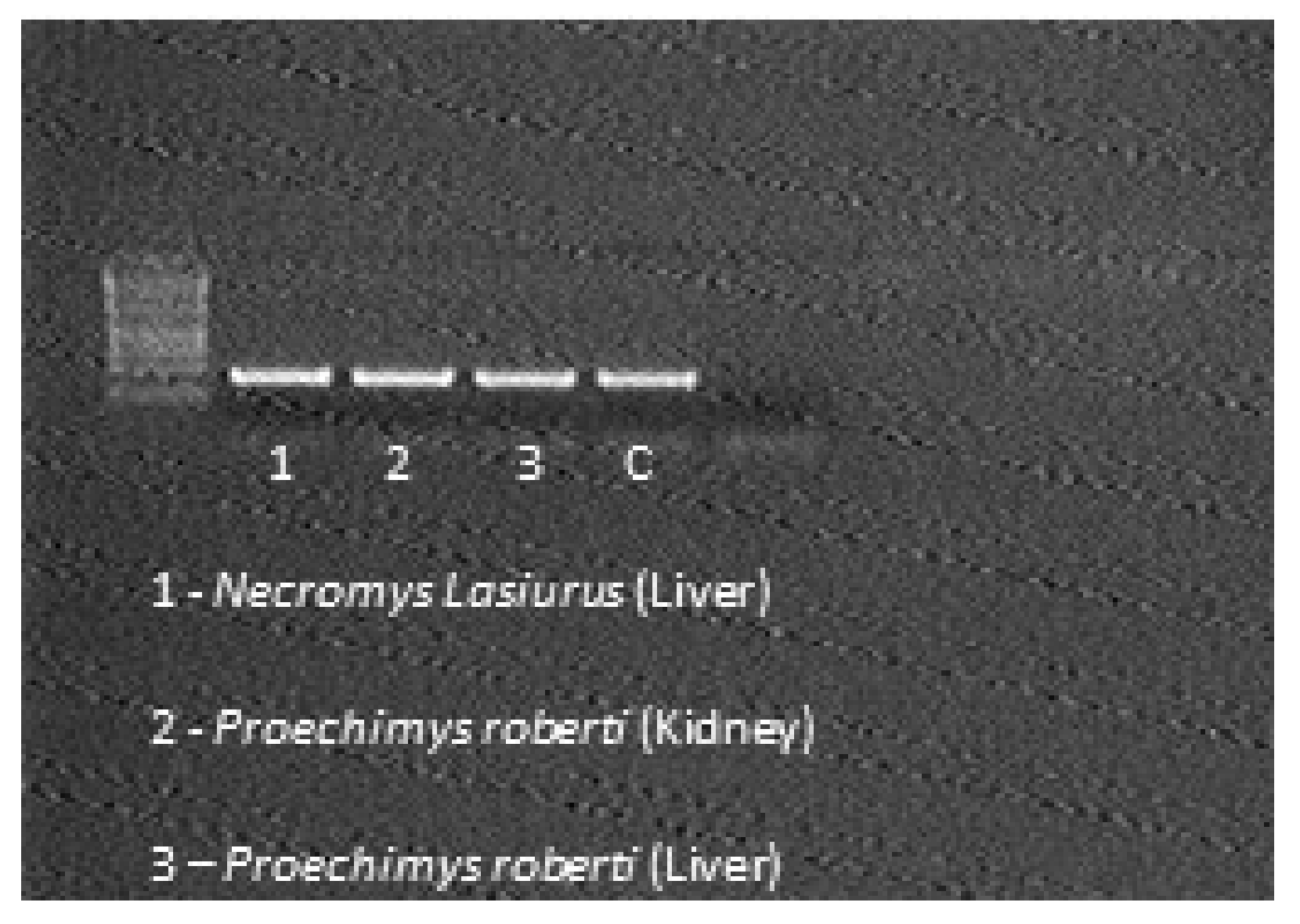
| N° | Municipality | Date | Specie | PCR | Trap/Number and Line |
|---|---|---|---|---|---|
| 1 | Curionópolis/Pará—Area Serra Leste—Location: Farm (Water Collection) | 2 November 2016 | Necromys lasiurus | Neg | Sherman—147/Line 4 |
| 2 | Curionópolis/Pará—Area Serra Leste—Location: Farm (Water Collection) | 3 November 2016 | Necromys lasiurus | Neg | Sherman—141/Line 4 |
| 3 | Curionópolis/Pará—Area Serra Leste—Location: Farm (Water Collection) | 3 November 2016 | Ameiva ameiva | Neg | Sherman—130/Line 4 |
| 4 | Curionópolis/Pará—Area Serra Leste—Location: Farm (Water Collection) | 4 November 2016 | Ameiva ameiva | Neg | Sherman—36/Line 3 |
| 5 | Curionópolis/Pará—Area Serra Leste—Location: Farm (Water Collection) | 5 November 2016 | Necromys lasiurus | Neg | Sherman—141/Line 4 |
| 6 | Curionópolis/Pará—Area Serra Leste—Location: Farm (Water Collection) | 9 November 2016 | Necromys lasiurus | Pos | Sherman—143/Line 4 |
| 7 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 11 November 2016 | Proechimys roberti | Pos | Tomahawk—16/Line 1 |
| 8 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 12 November 2016 | Marmosops parvidens (Br-1311) | Neg | Sherman—04/Line 1 |
| 9 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 12 November 2016 | Proechimys cf. roberti | Neg | Sherman—16/Line 1 |
| 10 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 12 November 2016 | Oxymycterus amazonicus | Neg | Sherman—78/Line 1 |
| 11 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 12 November 2016 | Proechimys roberti | Pos | Sherman—131/Line 2 |
| 12 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 15 November 2016 | Marmosops parvidens (Br-1313) | Neg | Sherman—17/Line 1 |
| 13 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 15 November 2016 | Didelphis aurita (1314) | Neg | Tomahawk—34/Line 1 |
| 14 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 15 November 2016 | Proechimys roberti | Neg | Tomahawk—64/Line 1 |
| 15 | Canaã dos Carajás/Pará—Area Serra Sul—Location: Plateau S11D | 17 November 2016 | Marmosops parvidens (Br-1315) | Neg | Sherman—12/Line 1 |
| Total | 15 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lima, M.F.d.; Silvestre, M.d.P.S.A.; Santos, E.C.d.; Martins, L.C.; Quaresma, J.A.S.; de Barros, B.d.C.V.; Silva, M.J.A.; Lima, L.N.G.C. The Presence of Mycobacterium leprae in Wild Rodents. Microorganisms 2022, 10, 1114. https://doi.org/10.3390/microorganisms10061114
Lima MFd, Silvestre MdPSA, Santos ECd, Martins LC, Quaresma JAS, de Barros BdCV, Silva MJA, Lima LNGC. The Presence of Mycobacterium leprae in Wild Rodents. Microorganisms. 2022; 10(6):1114. https://doi.org/10.3390/microorganisms10061114
Chicago/Turabian StyleLima, Maxwell Furtado de, Maria do Perpétuo Socorro Amador Silvestre, Everaldina Cordeiro dos Santos, Lívia Caricio Martins, Juarez Antônio Simões Quaresma, Bruno de Cássio Veloso de Barros, Marcos Jessé Abrahão Silva, and Luana Nepomuceno Gondim Costa Lima. 2022. "The Presence of Mycobacterium leprae in Wild Rodents" Microorganisms 10, no. 6: 1114. https://doi.org/10.3390/microorganisms10061114








