Epidemiology and Characteristics of Elizabethkingia spp. Infections in Southeast Asia
Abstract
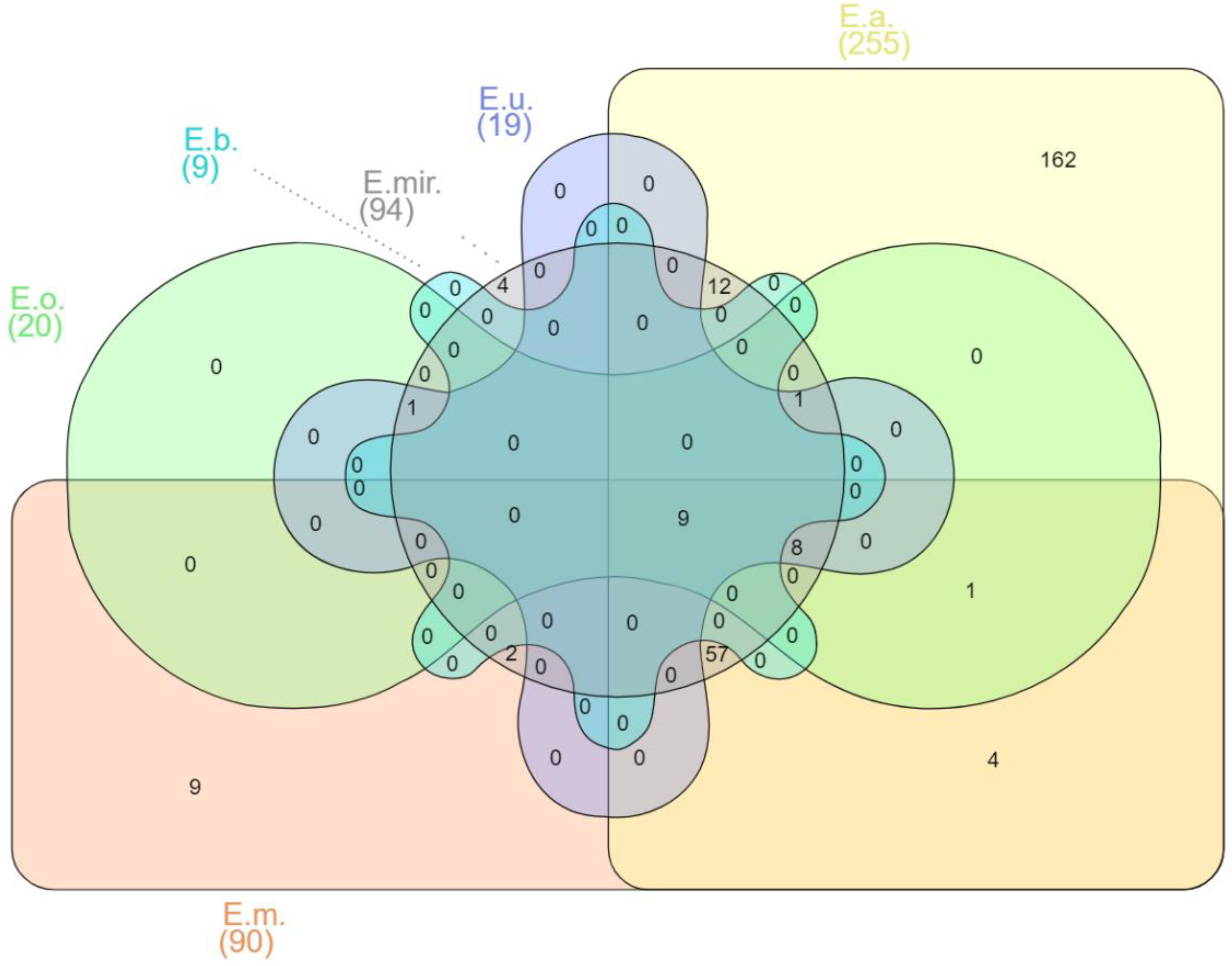
1. Introduction
2. Identification
3. Antibiotic Resistance
4. Virulence Factors
5. Sources of Isolation and Transmission
6. Malaysia Reports
7. Singapore Reports
8. Thailand Reports
9. Indonesia Reports
10. Cambodia Reports
11. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lau, S.K.; Chow, W.-N.; Foo, C.-H.; Curreem, S.O.; Lo, G.C.-S.; Teng, J.L.; Chen, J.H.; Ng, R.H.; Wu, A.K.; Cheung, I.Y.; et al. Elizabethkingia anophelis bacteremia is associated with clinically significant infections and high mortality. Sci. Rep. 2016, 6, 26045. [Google Scholar] [CrossRef] [PubMed]
- Dziuban, E.J.; Franks, J.L.; So, M.; Peacock, G.; Blaney, D.D. Elizabethkingia in children: A comprehensive review of symptomatic cases reported from 1944 to 2017. Clin. Infect. Dis. 2018, 67, 144–149. [Google Scholar] [CrossRef] [PubMed]
- Opota, O.; Diene, S.M.; Bertelli, C.; Prod’hom, G.; Eckert, P.; Greub, G. Genome of the carbapenemase-producing clinical isolate Elizabethkingia miricola EM_CHUV and comparative genomics with Elizabethkingia meningoseptica and Elizabethkingia anophelis: Evidence for intrinsic multidrug resistance trait of emerging pathogens. Int. J. Antimicrob. Agents 2017, 49, 93–97. [Google Scholar] [CrossRef] [PubMed]
- Swain, B.; Rout, S.; Otta, S.; Rakshit, A. Elizabethkingia meningoseptica: An unusual cause for septicaemia. JMM Case Rep. 2015, 2, e000005. [Google Scholar] [CrossRef]
- Lee, C.-H.; Lin, W.-C.; Chia, J.-H.; Su, L.-H.; Chien, C.-C.; Mao, A.-H.; Liu, J.-W. Community-acquired osteomyelitis caused by Chryseobacterium meningosepticum: Case report and literature review. Diagn. Microbiol. Infect. Dis. 2008, 60, 89–93. [Google Scholar] [CrossRef]
- Gupta, P.; Zaman, K.; Mohan, B.; Taneja, N. Elizabethkingia miricola: A rare non-fermenter causing urinary tract infection. World J. Clin. Cases 2017, 5, 187. [Google Scholar] [CrossRef]
- Raghavan, S.; Thomas, B.; Shastry, B. Elizabethkingia meningoseptica: Emerging multidrug resistance in a nosocomial pathogen. Case Rep. 2017, 2017, bcr-2017-221076. [Google Scholar] [CrossRef]
- Young, S.M.; Lingam, G.; Tambyah, P.A. Elizabethkingia meningoseptica Engodenous Endophthalmitis—A Case Report. Antimicrob. Resist. Infect. Control 2014, 3, 35. [Google Scholar] [CrossRef]
- Yang, J.; Xue, W.; Yu, X. Elizabethkingia meningosepticum endocarditis: A rare case and special therapy. Anatol. J. Cardiol. 2015, 15, 427. [Google Scholar] [CrossRef][Green Version]
- Chi, S.; Fekete, T. Epididymo-orchitis. In Clinical Infectious Disease, 2nd ed.; Schlossberg, D., Ed.; Cambridge University Press: Cambridge, UK, 2015; pp. 401–405. [Google Scholar]
- Gonzalez, C.; Coolen-Allou, N.; Allyn, J.; Esteve, J.; Belmonte, O.; Allou, N. Severe sepsis and pulmonary abscess with bacteremia due to Elizabethkingia miricola. Med. Mal. Infect. 2015, 46, 49–51. [Google Scholar] [CrossRef]
- Lee, C.-C.; Chen, P.-L.; Wang, L.-R.; Lee, H.-C.; Chang, C.-M.; Lee, N.-Y.; Wu, C.-J.; Shih, H.-I.; Ko, W.-C. Fatal case of community-acquired bacteremia and necrotizing fasciitis caused by Chryseobacterium meningosepticum: Case report and review of the literature. J. Clin. Microbiol. 2006, 44, 1181–1183. [Google Scholar] [CrossRef] [PubMed]
- Taufiq Kadafi, K.; Yuliarto, S.; Aji Cahyono, H.; Ratridewi, I.; Khalasha, T. Cerebral Salt Wasting Due to Bacteremia Caused by Elizabethkingia meningoseptica: A Case Report. Arch. Pediatr. Infect. Dis. 2020, 8, e44832. [Google Scholar] [CrossRef]
- Kenna, D.T.; Fuller, A.; Martin, K.; Perry, C.; Pike, R.; Burns, P.J.; Narayan, O.; Wilkinson, S.; Hill, R.; Woodford, N.; et al. rpoB gene sequencing highlights the prevalence of an E. miricola cluster over other Elizabethkingia species among UK cystic fibrosis patients. Diagn. Microbiol. Infect. Dis. 2018, 90, 109–114. [Google Scholar] [CrossRef] [PubMed]
- Amir, A.; IC Sam, J.; Nawi, S. Elizabethkingia meningoseptica neonatal meningitis in a premature infant. Asian J. Med. Biomed. 2018, 2 (Suppl. 1), 22. [Google Scholar]
- Seong, H.; Kim, J.H.; Kim, J.H.; Lee, W.J.; Ahn, J.Y.; Ku, N.S.; Choi, J.Y.; Yeom, J.S.; Song, Y.G.; Jeong, S.J. Risk factors for mortality in patients with elizabethkingia infection and the clinical impact of the antimicrobial susceptibility patterns of elizabethkingia species. J. Clin. Med. 2020, 9, 1431. [Google Scholar] [CrossRef]
- Nori, P.; Cowman, K.; Chen, V.; Bartash, R.; Szymczak, W.; Madaline, T.; Katiyar, C.P.; Jain, R.; Aldrich, M.; Weston, G.; et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect. Control Hosp. Epidemiol. 2021, 42, 84–88. [Google Scholar] [CrossRef]
- Hayek, S.S.; Abd, T.T.; Cribbs, S.K.; Anderson, A.M.; Melendez, A.; Kobayashi, M.; Polito, C.; Wang, Y.F.W. Rare Elizabethkingia meningosepticum meningitis case in an immunocompetent adult. Emerg. Microbes Infect. 2013, 2, 1–4. [Google Scholar] [CrossRef]
- Sebastiampillai, B.S.; Luke, N.V.; Silva, S.; De Silva, S.T.; Premaratna, R. Septicaemia caused by Elizabethkingia-sp in a ‘healthy’Sri Lankan man. Trop. Dr. 2018, 48, 62–63. [Google Scholar] [CrossRef]
- Yang, C.; Liu, Z.; Yu, S.; Ye, K.; Li, X.; Shen, D. Comparison of three species of Elizabethkingia genus by whole-genome sequence analysis. FEMS Microbiol. Lett. 2021, 368, fnab018. [Google Scholar] [CrossRef]
- King, E.O. Studies on a group of previously unclassified bacteria associated with meningitis in infants. Am. J. Clin. Pathol. 1959, 31, 241–247. [Google Scholar] [CrossRef]
- Buttiaux, R.; Vandepitte, J. Flavobacterium in Epidemic Meningitis of New-Born Infants. Ann. Inst. Pasteur 1960, 98, 398–404. [Google Scholar]
- Chan, J.; Chong, C.; Thoon, K.; Tee, N.; Maiwald, M.; Lam, J.; Bhattacharya, R.; Chandran, S.; Yung, C.; Tan, N. Invasive paediatric Elizabethkingia meningoseptica infections are best treated with a combination of piperacillin/tazobactam and trimethoprim/sulfamethoxazole or fluoroquinolone. J. Med. Microbiol. 2019, 68, 1167–1172. [Google Scholar] [CrossRef] [PubMed]
- Saetiew, N.; Nilkate, S.; Suankratay, C. Elizabethkingia meningoseptica Infection: The First Case Series in Thailand. Presented at the 26th European Congress of Clinical Microbiology and Infectious Diseases, Bangkok, Thailand, 9–12 April 2016. [Google Scholar] [CrossRef]
- Agustini, N.M.A.; Wati, D.K.; Suparyatha, I.; Hartawan, I.N.B.; Utama, I.M.G.D.L.; Budayanti, N.N.S.; Tunas, I.K. The relationship between bacterial types and antibiotic resistance with the clinical outcomes of sepsis patients in Pediatric Intensive Care Unit at Sanglah Hospital Denpasar, Bali-Indonesia. Indones. J. Biomed. Sci. 2018, 12, 13–18. [Google Scholar] [CrossRef]
- Reed, T.A.; Watson, G.; Kheng, C.; Tan, P.; Roberts, T.; Ling, C.L.; Miliya, T.; Turner, P. Elizabethkingia anophelis Infection in Infants, Cambodia, 2012–2018. Emerg. Infect. Dis. 2020, 26, 320. [Google Scholar] [CrossRef] [PubMed]
- Trimpert, J.; Eichhorn, I.; Vladimirova, D.; Haake, A.; Schink, A.K.; Klopfleisch, R.; Lübke-Becker, A. Elizabethkingia miricola infection in multiple anuran species. Transbound. Emerg. Dis. 2020, 68, 931–940. [Google Scholar] [CrossRef] [PubMed]
- Karl, H.; Lehmann, I.; Rehbein, H.; Schubring, R. Composition and quality attributes of conventionally and organically farmed Pangasius fillets (Pangasius hypophthalmus) on the German market. Int. J. Food Sci. Technol. 2010, 45, 56–66. [Google Scholar] [CrossRef]
- Frederiksen, W.; Ursing, J. Proposed new bacterial taxa and proposed changes of bacterial names published during 1994 and considered to be of interest to medical or veterinary bacteriology. APMIS 1995, 103, 651–654. [Google Scholar] [CrossRef]
- Kim, K.; Kim, M.; Lim, J.; Park, H.; Lee, S. Transfer of Chryseobacterium meningosepticum and Chryseobacterium miricola to Elizabethkingia gen. nov. as Elizabethkingia meningoseptica comb. nov. and Elizabethkingia miricola comb. nov. Int. J. Syst. Evol. Microbiol. 2005, 55, 1287–1293. [Google Scholar] [CrossRef]
- Nicholson, A.C.; Gulvik, C.A.; Whitney, A.M.; Humrighouse, B.W.; Graziano, J.; Emery, B.; Bell, M.; Loparev, V.; Juieng, P.; Gartin, J.; et al. Revisiting the taxonomy of the genus Elizabethkingia using whole-genome sequencing, optical mapping, and MALDI-TOF, along with proposal of three novel Elizabethkingia species: Elizabethkingia bruuniana sp. nov., Elizabethkingia ursingii sp. nov., and Elizabethkingia occulta sp. nov. Antonie Van Leeuwenhoek 2018, 111, 55–72. [Google Scholar] [CrossRef] [PubMed]
- Hwang, J.-H.; Kim, J.; Kim, J.-H.; Mo, S. Elizabethkingia argenteiflava sp. nov., isolated from the pod of soybean, Glycine max. Int. J. Syst. Evol. Microbiol. 2021, 71, 004767. [Google Scholar] [CrossRef]
- Hem, S.; Jarocki, V.M.; Baker, D.J.; Charles, I.G.; Drigo, B.; Aucote, S.; Donner, E.; Burnard, D.; Bauer, M.J.; Harris, P.N.; et al. Genomic analysis of Elizabethkingia species from aquatic environments: Evidence for potential clinical transmission. Curr. Res. Microb. Sci. 2022, 3, 100083. [Google Scholar] [CrossRef] [PubMed]
- Kukutla, P.; Lindberg, B.G.; Pei, D.; Rayl, M.; Yu, W.; Steritz, M.; Faye, I.; Xu, J. Insights from the genome annotation of Elizabethkingia anophelis from the malaria vector Anopheles gambiae. PLoS ONE 2014, 9, e97715. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.-N.; Lai, C.-H.; Yang, C.-H.; Huang, Y.-H.; Lin, H.-F.; Lin, H.-H. Comparison of four automated microbiology systems with 16S rRNA gene sequencing for identification of Chryseobacterium and Elizabethkingia species. Sci. Rep. 2017, 7, 13824. [Google Scholar] [CrossRef] [PubMed]
- Ekcharoenkul, K.; Ngamskulrungroj, P.; Joyjamras, K.; Leelaporn, A.; Harun, A.; Kiratisin, P. Identification of Uncommon Pathogenic Bacteria by MALDI-TOF Mass Spectrometry Using a Custom Library of Siriraj Hospital. Siriraj Med. J. 2018, 70, 127–130. [Google Scholar]
- Han, M.-S.; Kim, H.; Lee, Y.; Kim, M.; Ku, N.S.; Choi, J.Y.; Yong, D.; Jeong, S.H.; Lee, K.; Chong, Y. Relative prevalence and antimicrobial susceptibility of clinical isolates of Elizabethkingia species based on 16S rRNA gene sequencing. J. Clin. Microbiol. 2017, 55, 274–280. [Google Scholar] [CrossRef]
- Burnard, D.; Gore, L.; Henderson, A.; Ranasinghe, A.; Bergh, H.; Cottrell, K.; Sarovich, D.S.; Price, E.P.; Paterson, D.L.; Harris, P.N. Comparative Genomics and Antimicrobial Resistance Profiling of Elizabethkingia Isolates Reveal Nosocomial Transmission and In Vitro Susceptibility to Fluoroquinolones, Tetracyclines, and Trimethoprim-Sulfamethoxazole. J. Clin. Microbiol. 2020, 58, e00730-20. [Google Scholar] [CrossRef]
- González, L.J.; Vila, A.J. Carbapenem resistance in Elizabethkingia meningoseptica is mediated by metallo-β-lactamase BlaB. Antimicrob. Agents Chemother. 2012, 56, 1686–1692. [Google Scholar] [CrossRef]
- Teo, J.; Tan, S.Y.-Y.; Liu, Y.; Tay, M.; Ding, Y.; Li, Y.; Kjelleberg, S.; Givskov, M.; Lin, R.T.P.; Yang, L. Comparative Genomic Analysis of Malaria Mosquito Vector-Associated Novel Pathogen Elizabethkingia anophelis. Genome Biol. Evol. 2014, 6, 1158–1165. [Google Scholar] [CrossRef]
- Breurec, S.; Criscuolo, A.; Diancourt, L.; Rendueles, O.; Vandenbogaert, M.; Passet, V.; Caro, V.; Rocha, E.P.; Touchon, M.; Brisse, S. Genomic epidemiology and global diversity of the emerging bacterial pathogen Elizabethkingia anophelis. Sci. Rep. 2016, 6, 30379. [Google Scholar] [CrossRef]
- Hu, R.; Zhang, Q.; Gu, Z. Molecular diversity of chromosomal metallo-β-lactamase genes in Elizabethkingia genus. Int. J. Antimicrob. Agents 2020, 56, 105978. [Google Scholar] [CrossRef]
- Hu, R.; Zhang, Q.; Gu, Z. Whole-genome analysis of the potentially zoonotic Elizabethkingia miricola FL160902 with two new chromosomal MBL gene variants. J. Antimicrob. Chemother. 2020, 75, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Perrin, A.; Larsonneur, E.; Nicholson, A.C.; Edwards, D.J.; Gundlach, K.M.; Whitney, A.M.; Gulvik, C.A.; Bell, M.E.; Rendueles, O.; Cury, J.; et al. Evolutionary dynamics and genomic features of the Elizabethkingia anophelis 2015 to 2016 Wisconsin outbreak strain. Nat. Commun. 2017, 8, 15483. [Google Scholar] [CrossRef]
- Lin, J.-N.; Lai, C.-H.; Yang, C.-H.; Huang, Y.-H. Elizabethkingia infections in humans: From genomics to clinics. Microorganisms 2019, 7, 295. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.-Y.; Yang, C.-H.; Lai, C.-H.; Huang, Y.-H.; Lin, J.-N. Comparative Genomics of 86 Whole-Genome Sequences in the Six Species of the Elizabethkingia Genus Reveals Intraspecific and Interspecific Divergence. Sci. Rep. 2019, 9, 19167. [Google Scholar] [CrossRef] [PubMed]
- Bellais, S.; Poirel, L.; Naas, T.; Girlich, D.; Nordmann, P. Genetic-Biochemical Analysis and Distribution of the Ambler Class A β-Lactamase CME-2, Responsible for Extended-Spectrum Cephalosporin Resistance in Chryseobacterium (Flavobacterium) meningosepticum. Antimicrob. Agents Chemother. 2000, 44, 1–9. [Google Scholar] [CrossRef]
- Chang, J.; Hsueh, P.; Wu, J.; Ho, S.; Hsieh, W.; Luh, K. Antimicrobial susceptibility of flavobacteria as determined by agar dilution and disk diffusion methods. Antimicrob. Agents Chemother. 1997, 41, 1301–1306. [Google Scholar] [CrossRef]
- Moulin, V.; Freney, J.; Hansen, W.; Philippon, A. Comportement phénotypique des Flavobacterium vis-à-vis de 39 antibiotiques. Méd. Mal. Infect. 1992, 22, 902–907. [Google Scholar] [CrossRef]
- Kwambana-Adams, B.; Laxton, C.; Foster-Nyarko, E.; Weinstock, G.; Antonio, M. Isolation of Methicillin-resistant Staphylococcus aureus and Multidrug-resistant Elizabethkingia meningoseptica from Neonates within Minutes of Birth. Pediatric Infect. Dis. J. 2017, 36, 123–124. [Google Scholar] [CrossRef]
- Huang, Y.; Huang, Y.; Lin, Y.; Wang, F.; Chan, Y.; Yang, T. Risk factors and outcome of levofloxacin-resistant Elizabethkingia meningoseptica bacteraemia in adult patients in Taiwan. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1–8. [Google Scholar] [CrossRef]
- Jean, S.-S.; Hsieh, T.-C.; Ning, Y.-Z.; Hsueh, P.-R. Role of vancomycin in the treatment of bacteraemia and meningitis caused by Elizabethkingia meningoseptica. Int. J. Antimicrob. Agents 2017, 50, 507–511. [Google Scholar] [CrossRef]
- Lee, E.; Robinson, M.; Thong, M.; Puthucheary, S.; Ong, T.; Ng, K. Intraventricular chemotherapy in neonatal meningitis. J. Pediatr. 1977, 91, 991–995. [Google Scholar] [CrossRef]
- Lim, V.; Halijah, M. A comparative study of the in-vitro activity of cefepime and other cephalosporins. Malays. J. Pathol. 1993, 15, 65–68. [Google Scholar] [PubMed]
- Chang, T.-Y.; Chen, H.-Y.; Chou, Y.-C.; Cheng, Y.-H.; Sun, J.-R. In vitro activities of imipenem, vancomycin, and rifampicin against clinical Elizabethkingia species producing BlaB and GOB metallo-beta-lactamases. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Santona, A.; Paglietti, B.; Al-Qahtani, A.A.; Bohol, M.F.F.; Senok, A.; Deligios, M.; Rubino, S.; Al-Ahdal, M.N. Novel type of VanB2 teicoplanin-resistant hospital-associated Enterococcus faecium. Int. J. Antimicrob. Agents 2014, 44, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Sader, H.S.; Jones, R.N.; Pfaller, M.A. Relapse of catheter-related Flavobacterium meningosepticum bacteremia demonstrated by DNA macrorestriction analysis. Clin. Infect. Dis. 1995, 21, 997–1000. [Google Scholar] [CrossRef] [PubMed]
- Ozkalay, N.; Anil, M.; Agus, N.; Helvaci, M.; Sirti, S. Community-acquired meningitis and sepsis caused by Chryseobacterium meningosepticum in a patient diagnosed with thalassemia major. J. Clin. Microbiol. 2006, 44, 3037–3039. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Soehnlen, M.; Blom, J.; Terrapon, N.; Henrissat, B.; Walker, E.D. Comparative genomic analyses reveal diverse virulence factors and antimicrobial resistance mechanisms in clinical Elizabethkingia meningoseptica strains. PLoS ONE 2019, 14, e0222648. [Google Scholar] [CrossRef]
- Lin, J.-N.; Lai, C.-H.; Yang, C.-H.; Huang, Y.-H.; Lin, H.-H. Genomic features, phylogenetic relationships, and comparative genomics of Elizabethkingia anophelis strain EM361-97 isolated in Taiwan. Sci. Rep. 2017, 7, 14317. [Google Scholar] [CrossRef]
- Li, Y.; Liu, Y.; Chew, S.C.; Tay, M.; Salido, M.M.S.; Teo, J.; Lauro, F.M.; Givskov, M.; Yang, L. Complete genome sequence and transcriptomic analysis of the novel pathogen Elizabethkingia anophelis in response to oxidative stress. Genome Biol. Evol. 2015, 7, 1676–1685. [Google Scholar] [CrossRef]
- Lau, S.K.; Wu, A.K.; Teng, J.L.; Tse, H.; Curreem, S.O.; Tsui, S.K.; Huang, Y.; Chen, J.H.; Lee, R.A.; Yuen, K.-Y.; et al. Evidence for Elizabethkingia anophelis transmission from mother to infant, Hong Kong. Emerg. Infect. Dis. 2015, 21, 232. [Google Scholar] [CrossRef]
- Jacobs, A.; Chenia, H.Y. Biofilm formation and adherence characteristics of an Elizabethkingia meningoseptica isolate from Oreochromis mossambicus. Ann. Clin. Microbiol. Antimicrob. 2011, 10, 16. [Google Scholar] [CrossRef] [PubMed]
- Alav, I.; Sutton, J.M.; Rahman, K.M. Role of bacterial efflux pumps in biofilm formation. J. Antimicrob. Chemother. 2018, 73, 2003–2020. [Google Scholar] [CrossRef] [PubMed]
- Puah, S.M.; Fong, S.P.; Kee, B.P.; Puthucheary, S.; Chua, K.H. Molecular identification and biofilm-forming ability of Elizabethkingia species. Microb. Pathog. 2022, 162, 105345. [Google Scholar] [CrossRef] [PubMed]
- Dunn, M.; Ramirez-Trujillo, J.; Hernández-Lucas, I. Major roles of isocitrate lyase and malate synthase in bacterial and fungal pathogenesis. Microbiology 2009, 155, 3166–3175. [Google Scholar] [CrossRef]
- McKinney, J.D.; Zu Bentrup, K.H.; Muñoz-Elías, E.J.; Miczak, A.; Chen, B.; Chan, W.-T.; Swenson, D.; Sacchettini, J.C.; Jacobs, W.R.; Russell, D.G. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 2000, 406, 735–738. [Google Scholar] [CrossRef]
- Roberts, M.F.; Khan, H.M.; Goldstein, R.; Reuter, N.; Gershenson, A. Search and subvert: Minimalist bacterial phosphatidylinositol-specific phospholipase C enzymes. Chem. Rev. 2018, 118, 8435–8473. [Google Scholar] [CrossRef]
- Monturiol-Gross, L.; Villalta-Romero, F.; Flores-Díaz, M.; Alape-Girón, A. Bacterial phospholipases C with dual activity: Phosphatidylcholinesterase and sphingomyelinase. FEBS Open Bio 2021, 11, 3262–3275. [Google Scholar] [CrossRef]
- Vicente, C.S.; Nascimento, F.X.; Ikuyo, Y.; Cock, P.J.; Mota, M.; Hasegawa, K. The genome and genetics of a high oxidative stress tolerant Serratia sp. LCN16 isolated from the plant parasitic nematode Bursaphelenchus xylophilus. BMC Genom. 2016, 17, 301. [Google Scholar] [CrossRef]
- Hassett, D.J.; Alsabbagh, E.; Parvatiyar, K.; Howell, M.L.; Wilmott, R.W.; Ochsner, U.A. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J. Bacteriol. 2000, 182, 4557–4563. [Google Scholar] [CrossRef]
- Wang, M.; Gao, H.; Lin, N.; Zhang, Y.; Huang, N.; Walker, E.D.; Ming, D.; Chen, S.; Hu, S. The antibiotic resistance and pathogenicity of a multidrug-resistant Elizabethkingia anophelis isolate. Microbiologyopen 2019, 8, e804. [Google Scholar] [CrossRef]
- Yang, C.; Liu, Z.; Yu, S.; Ye, K.; Li, X.; Shen, D. Comparison of Whole-Genome Sequences for Three Species of the Elizabethkingia Genus. 2020. Available online: https://www.researchsquare.com/article/rs-61004/v1 (accessed on 21 April 2022).
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [PubMed]
- Laith, A.; Mazlan, A.; Ambak, M.; Jabar, A.; Najiah, M. Isolation and Identification of Elizabethkingia meningoseptica from Diseased African Catfish Clarias gariepinus. J. Microbiol. Biotechnol. Food Sci. 2017, 6, 1070–1076. [Google Scholar] [CrossRef]
- Tew, L.-S.; She, L.-Y.; Chew, C.-H. Isolation, Antimicrobial Susceptibility Profile and Detection of Sul1, blaTEM, and blaSHV in Amoxicillin-Clavulanate-Resistant Bacteria Isolated From Retail Sausages in Kampar, Malaysia. Jundishapur J. Microbiol. 2016, 9, e37897. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, Y.; Wang, F. Risk factors of healthcare-associated Elizabethkingia meningoseptica infections in Taiwan medical center. Int. J. Antimicrob. Agents 2017, 50, S141. [Google Scholar]
- Khan, I.; Lall, M.; Sen, S.; Ninawe, S.; Chandola, P. Multiresistant Elizabethkingia meningoseptica infections in tertiary care. Med. J. Armed Forces India 2015, 71, 282. [Google Scholar] [CrossRef]
- Kämpfer, P.; Matthews, H.; Glaeser, S.P.; Martin, K.; Lodders, N.; Faye, I. Elizabethkingia anophelis sp. nov., isolated from the midgut of the mosquito Anopheles gambiae. Int. J. Syst. Evol. Microbiol. 2011, 61, 2670–2675. [Google Scholar] [CrossRef]
- Chen, S.; Johnson, B.K.; Yu, T.; Nelson, B.N.; Walker, E.D. Elizabethkingia anophelis: Physiologic and transcriptomic responses to iron stress. Front. Microbiol. 2020, 11, 804. [Google Scholar] [CrossRef]
- Onyango, M.; Payne, A.; Stout, J.; Dieme, C.; Kuo, L.; Kramer, L.; Ciota, A. Potential for transmission of Elizabethkingia anophelis by Aedes albopictus and the role of microbial interactions in Zika virus competence. bioRxiv 2020, 702464. [Google Scholar] [CrossRef]
- Akhouayri, I.; Habtewold, T.; Christophides, G. Melanotic pathology and vertical transmission of the gut commensal Elizabethkingia meningoseptica in the major malaria vector Anopheles gambiae. PLoS ONE 2013, 8, e77619. [Google Scholar] [CrossRef]
- Rani, A.; Sharma, A.; Rajagopal, R.; Adak, T.; Bhatnagar, R.K. Bacterial diversity analysis of larvae and adult midgut microflora using culture-dependent and culture-independent methods in lab-reared and field-collected Anopheles stephensi-an Asian malarial vector. BMC Microbiol. 2009, 9, 96. [Google Scholar] [CrossRef]
- Ngwa, C.; Glöckner, V.; Abdelmohsen, U.R.; Scheuermayer, M.; Fischer, R.; Hentschel, U.; Pradel, G. 16S rRNA gene-based identification of Elizabethkingia meningoseptica (Flavobacteriales: Flavobacteriaceae) as a dominant midgut bacterium of the Asian malaria vector Anopheles stephensi (Dipteria: Culicidae) with antimicrobial activities. J. Med. Entomol. 2013, 50, 404–414. [Google Scholar] [CrossRef] [PubMed]
- Lindh, J.; Borg-Karlson, A.; Faye, I. Transstadial and horizontal transfer of bacteria within a colony of Anopheles gambiae (Diptera: Culicidae) and oviposition response to bacteria-containing water. Acta Trop. 2008, 107, 242–250. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gilbreath, T.M., III; Kukutla, P.; Yan, G.; Xu, J. Dynamic gut microbiome across life history of the malaria mosquito Anopheles gambiae in Kenya. PLoS ONE 2011, 6, e24767. [Google Scholar] [CrossRef] [PubMed]
- Boissière, A.; Tchioffo, M.; Bachar, D.; Abate, L.; Marie, A.; Nsango, S.; Shahbazkia, H.; Awono-Ambene, P.; Levashina, E.; Christen, R.; et al. Midgut microbiota of the malaria mosquito vector Anopheles gambiae and interactions with Plasmodium falciparum infection. PLoS Pathog. 2012, 8, e1002742. [Google Scholar] [CrossRef]
- Wang, S.; Ghosh, A.K.; Bongio, N.; Stebbings, K.A.; Lampe, D.J.; Jacobs-Lorena, M. Fighting malaria with engineered symbiotic bacteria from vector mosquitoes. Proc. Natl. Acad. Sci. USA 2012, 109, 12734–12739. [Google Scholar] [CrossRef]
- Dong, Y.; Manfredini, F.; Dimopoulos, G. Implication of the mosquito midgut microbiota in the defense against malaria parasites. PLoS Pathog. 2009, 5, e1000423. [Google Scholar] [CrossRef]
- Tainchum, K.; Dupont, C.; Chareonviriyaphap, T.; Jumas-Bilak, E.; Bangs, M.J.; Manguin, S. Bacterial microbiome in wild-caught Anopheles mosquitoes in western Thailand. Front. Microbiol. 2020, 11, 965. [Google Scholar] [CrossRef]
- Thong, M.; Puthucheary, S.; Lee, E. Flavobacterium meningosepticum infection: An epidemiological study in a newborn nursery. J. Clin. Pathol. 1981, 34, 429–433. [Google Scholar] [CrossRef]
- Chew, K.L.; Cheng, B.; Lin, R.T.; Teo, J.W. Elizabethkingia anophelis is the dominant Elizabethkingia species found in blood cultures in Singapore. J. Clin. Microbiol. 2018, 56, e01445-17. [Google Scholar] [CrossRef]
- Yung, C.-F.; Maiwald, M.; Loo, L.H.; Soong, H.Y.; Tan, C.B.; Lim, P.K.; Li, L.; Tan, N.W.; Chong, C.-Y.; Tee, N.; et al. Elizabethkingia anophelis and association with tap water and handwashing, Singapore. Emerg. Infect. Dis. 2018, 24, 1730. [Google Scholar] [CrossRef]
- Loo, L.W.; Liew, Y.X.; Choong, H.L.L.; Tan, A.L.; Chlebicki, P. Microbiology and audit of vascular access-associated bloodstream infections in multi-ethnic Asian hemodialysis patients in a tertiary hospital. Infect. Dis. 2015, 47, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Sooklin, L.; Anand, A.J.; Rajadurai, V.S.; Chandran, S. Management of large congenital chylous ascites in a preterm infant: Fetal and neonatal interventions. BMJ Case Rep. CP 2020, 13, e235849. [Google Scholar] [CrossRef]
- Liestiadi, D.E.F.; Azlin, E.; Nafianti, S. A hematologic scoring system and C-reactive protein compared to blood cultures for diagnosing bacterial neonatal sepsis. Paediatr. Indones 2017, 57, 71. [Google Scholar] [CrossRef][Green Version]
- Lee, E.; Robinson, M.; Thong, M.; Puthucheary, S. Rifamycin in Neonatal Flavobacteria meningitis. Arch. Dis. Child. 1976, 51, 209–213. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Raimondi, A.; Moosdeen, F.; Williams, J. Antibiotic resistance pattern of Flavobacterium meningosepticum. Eur. J. Clin. Microbiol. Infect. Dis. 1986, 5, 461–463. [Google Scholar] [CrossRef]
- Zakaria, Z.; Idris, B. Intraoperative Cerebrospinal Fluid Sample from First Ventriculoperitoneal Shunt Operation: Is it Indicated? Malays. J. Med. Sci. MJMS 2013, 20, 102. [Google Scholar]
- Wan Hassan, W.M.N.; Paramasivam, R.P.; Kandasamy, R.; Hassan, M.H.; Zaini, R.H.M. An uncommon Elizabethkingia meningoseptica septicemia in hemorrhagic stroke with septic shock patient during prolonged neuro-intensive care management. Anaesth. Pain Intensive Care 2017, 21, 268–271. [Google Scholar]
- Ali, N.A.M.; Reddy, S.C. Bilateral simultaneous infectious keratitis secondary to contact lens wear: An unusual case report with rare organisms. Eye Contact Lens 2007, 33, 338–340. [Google Scholar] [CrossRef]
- Phoon, H.Y.; Hussin, H.; Hussain, B.M.; Lim, S.Y.; Woon, J.J.; Er, Y.X.; Thong, K.L. Distribution, genetic diversity and antibiotic resistance of clinically important bacteria from the environment of a tertiary hospital. J. Glob. Antimicrob. Resist. 2018, 14, 132–140. [Google Scholar] [CrossRef]
- Haller, L.; Chen, H.; Ng, C.; Le, T.H.; Koh, T.H.; Barkham, T.; Sobsey, M.; Gin, K.Y.-H. Occurrence and characteristics of extended-spectrum β-lactamase-and carbapenemase-producing bacteria from hospital effluents in Singapore. Sci. Total Environ. 2018, 615, 1119–1125. [Google Scholar] [CrossRef]
- Balm, M.; Salmon, S.; Jureen, R.; Teo, C.; Mahdi, R.; Seetoh, T.; Teo, J.; Lin, R.; Fisher, D. Bad design, bad practices, bad bugs: Frustrations in controlling an outbreak of Elizabethkingia meningoseptica in intensive care units. J. Hosp. Infect. 2013, 85, 134–140. [Google Scholar] [CrossRef] [PubMed]
- Venkatachalam, I.; Teo, J.; Balm, M.N.; Fisher, D.A.; Jureen, R.; Lin, R.T. Klebsiella pneumoniae carbapenemase-producing enterobacteria in hospital, Singapore. Emerg. Infect. Dis. 2012, 18, 1381. [Google Scholar] [CrossRef] [PubMed]
- Zainuri, N.; Ransangan, J.; Lal, T.; Jintoni, B.; Chung, V. Identification of Elizabethkingia meningoseptica from American bullfrog (Rana catesbeiana) farmed in Sabah, Malaysia using PCR method and future management of outbreak. Malays. J. Microbiol. 2013, 9, 13–23. [Google Scholar] [CrossRef]
- Tee, L.; Najiah, M. Antibiogram and heavy metal tolerance of bullfrog bacteria in Malaysia. Open Vet. J. 2011, 1, 39–45. [Google Scholar]
- Surat, W.; Mhuantong, W.; Sangsrakru, D.; Chareonviriyaphap, T.; Arunyawat, U.; Kubera, A.; Sittivicharpinyo, T.; Siripan, O.; Pootakham, W. Gut Bacterial Diversity in Plasmodium-infected and Plasmodium-uninfected Anopheles minimus. Chiang Mai J. Sci. 2016, 43, 427–440. [Google Scholar]
- Thi, A.N.T.; Noseda, B.; Samapundo, S.; Nguyen, B.L.; Broekaert, K.; Rasschaert, G.; Heyndrickx, M.; Devlieghere, F. Microbial ecology of Vietnamese Tra fish (Pangasius hypophthalmus) fillets during processing. Int. J. Food Microbiol. 2013, 167, 144–152. [Google Scholar]
- Kim, M.; Singh, D.; Lai-Hoe, A.; Go, R.; Rahim, R.A.; Ainuddin, A.; Chun, J.; Adams, J.M. Distinctive phyllosphere bacterial communities in tropical trees. Microb. Ecol. 2012, 63, 674–681. [Google Scholar] [CrossRef]
- Oh, Y.M.; Kim, M.; Lee-Cruz, L.; Lai-Hoe, A.; Go, R.; Ainuddin, N.; Rahim, R.A.; Shukor, N.; Adams, J.M. Distinctive bacterial communities in the rhizoplane of four tropical tree species. Microb. Ecol. 2012, 64, 1018–1027. [Google Scholar] [CrossRef]
- Boo, N.; Wong, Y.; Lim, V. Pattern of neonatal septicemia in a Malaysian maternity hospital. Med. J. Malays. 1989, 44, 189–193. [Google Scholar]
- Habsah, H.; Zakuan, Z. Profiles of Imipenem Resistance Organism in an Adult ICU in a Teaching Hospital in Malaysia. Int. J. Infect. Dis. 2008, 12, e118–e119. [Google Scholar] [CrossRef][Green Version]
- Lowbridge, C.; Fadhil, S.A.; Krishnan, G.D.; Schimann, E.; Karuppan, R.M.; Sriram, N.; Rajahram, G.S.; Menon, J.; Patel, A.; William, T.; et al. How can gastro-intestinal tuberculosis diagnosis be improved? A prospective cohort study. BMC Infect. Dis. 2020, 20, 255. [Google Scholar] [CrossRef]
- Teo, J.; Tan, S.Y.-Y.; Tay, M.; Ding, Y.; Kjelleberg, S.; Givskov, M.; Lin, R.T.; Yang, L. First case of E anophelis outbreak in an intensive-care unit. Lancet 2013, 382, 855–856. [Google Scholar] [CrossRef]
- Lew, K.Y.; Ng, T.M.; Tan, M.; Tan, S.H.; Lew, E.L.; Ling, L.M.; Ang, B.; Lye, D.; Teng, C.B. Safety and clinical outcomes of carbapenem de-escalation as part of an antimicrobial stewardship programme in an ESBL-endemic setting. J. Antimicrob. Chemother. 2015, 70, 1219–1225. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-L.; Liu, K.-M.; Chang, H.-L.; Liao, Y.-C.; Lin, J.-S.; Kung, F.-Y.; Ho, C.-M.; Lin, K.-H.; Chen, Y.-T. The Evolutionary Trend and Genomic Features of an Emerging Lineage of Elizabethkingia anophelis Strains in Taiwan. Microbiol. Spectr. 2022, 10, e01682-21. [Google Scholar] [CrossRef] [PubMed]
- Santoso, M.S.; Yohan, B.; Denis, D.; Hayati, R.F.; Haryanto, S.; Trianty, L.; Noviyanti, R.; Hibberd, M.L.; Sasmono, R.T. Diagnostic accuracy of 5 different brands of dengue virus non-structural protein 1 (NS1) antigen rapid diagnostic tests (RDT) in Indonesia. Diagn. Microbiol. Infect. Dis. 2020, 98, 115116. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Soehnlen, M.; Downes, F.; Walker, E. Insights from the draft genome into the pathogenicity of a clinical isolate of Elizabethkingia meningoseptica Em3. Stand. Genom. Sci. 2017, 12, 56. [Google Scholar] [CrossRef] [PubMed]
- Amladi, A.; Jacob, J.J.; Anandan, S.; Veeraraghavan, B. Draft genome sequence of carbapenem-resistant Elizabethkingia anophelis strain BP8467 clinical isolate from India. J. Glob. Antimicrob. Resist. 2020, 21, 200–202. [Google Scholar] [CrossRef]
- Yum, J.H.; Lee, E.Y.; Hur, S.-H.; Jeong, S.H.; Lee, H.; Yong, D.; Chong, Y.; Lee, E.-W.; Nordmann, P.; Lee, K. Genetic diversity of chromosomal metallo-β-lactamase genes in clinical isolates of Elizabethkingia meningoseptica from Korea. J. Microbiol. 2010, 48, 358–364. [Google Scholar] [CrossRef]
- Bellais, S.; Aubert, D.; Naas, T.; Nordmann, P. Molecular and biochemical heterogeneity of class B carbapenem-hydrolyzing β-lactamases in Chryseobacterium meningosepticum. Antimicrob. Agents Chemother. 2000, 44, 1878–1886. [Google Scholar] [CrossRef]
- Matyi, S.A.; Hoyt, P.R.; Hosoyama, A.; Yamazoe, A.; Fujita, N.; Gustafson, J.E. Draft genome sequences of Elizabethkingia meningoseptica. Genome Announc. 2013, 1, e00444-13. [Google Scholar] [CrossRef]
- Sun, G.; Wang, L.; Bao, C.; Li, T.; Ma, L.; Chen, L. Complete genome sequence of Elizabethkingia meningoseptica, isolated from a T-cell non-Hodgkin’s lymphoma patient. Genome Announc. 2015, 3, e00673-15. [Google Scholar] [CrossRef] [PubMed]
- Steinberg, J.P.; Burd, E.M. Other gram-negative and gram-variable bacilli. Princ. Pract. Infect. Dis. 2010, 2, 2751–2768. [Google Scholar]
- Rossolini, G.M.; Franceschini, N.; Lauretti, L.; Caravelli, B.; Riccio, M.L.; Galleni, M.; Frère, J.-M.; Amicosante, G. Cloning of a Chryseobacterium (Flavobacterium) meningosepticum chromosomal gene (blaA CME) encoding an extended-spectrum class A β-lactamase related to the Bacteroides cephalosporinases and the VEB-1 and PER β-lactamases. Antimicrob. Agents Chemother. 1999, 43, 2193–2199. [Google Scholar] [CrossRef] [PubMed]
- Ghafoori, S.M.; Robles, A.M.; Arada, A.M.; Shirmast, P.; Dranow, D.M.; Mayclin, S.J.; Lorimer, D.D.; Myler, P.J.; Edwards, T.E.; Kuhn, M.L. Structural characterization of a Type B chloramphenicol acetyltransferase from the emerging pathogen Elizabethkingia anophelis NUHP1. Sci. Rep. 2021, 11, 9453. [Google Scholar] [CrossRef]
- Naidenov, B.; Lim, A.; Willyerd, K.; Torres, N.J.; Johnson, W.L.; Hwang, H.J.; Hoyt, P.; Gustafson, J.E.; Chen, C. Pan-genomic and polymorphic driven prediction of antibiotic resistance in Elizabethkingia. Front. Microbiol. 2019, 10, 1446. [Google Scholar] [CrossRef]
- Lin, J.-N.; Lai, C.-H.; Yang, C.-H.; Huang, Y.-H.; Lin, H.-H. Clinical manifestations, molecular characteristics, antimicrobial susceptibility patterns and contributions of target gene mutation to fluoroquinolone resistance in Elizabethkingia anophelis. J. Antimicrob. Chemother. 2018, 73, 2497–2502. [Google Scholar] [CrossRef]
- Johnson, W.L.; Ramachandran, A.; Torres, N.J.; Nicholson, A.C.; Whitney, A.M.; Bell, M.; Villarma, A.; Humrighouse, B.W.; Sheth, M.; Dowd, S.E. The draft genomes of Elizabethkingia anophelis of equine origin are genetically similar to three isolates from human clinical specimens. PLoS ONE 2018, 13, e0200731. [Google Scholar] [CrossRef]
- Jiang, X.; Wang, D.; Wang, Y.; Yan, H.; Shi, L.; Zhou, L. Occurrence of antimicrobial resistance genes sul and dfrA 12 in hospital environmental isolates of Elizabethkingia meningoseptica. World J. Microbiol. Biotechnol. 2012, 28, 3097–3102. [Google Scholar] [CrossRef]
| Source of Isolation | Country of Origin | Citation |
|---|---|---|
| Blood | Malaysia, Singapore, Thailand, Indonesia, Cambodia | [8,13,15,23,24,25,26,36,65,91,92,93,94,95,96] |
| Peritoneal fluid | Malaysia | [91] |
| Cerebrospinal fluid (CSF) | Malaysia, Singapore | [15,23,91,97,98,99,100] |
| Contact lens | Malaysia | [101] |
| Hospital environment (aerators, sink drains and traps at ICUs, pediatric wards, surgical wards, orthopedic wards) | Singapore | [8,102,103,104] |
| Catheter tips | Singapore | [8] |
| Respiratory specimens | Singapore Malaysia | [8,65] |
| Rectal swabs | Singapore | [105] |
| Urine | Malaysia | [65] |
| Wound swabs | Malaysia | [65] |
| Nasal swabs | Malaysia | [65] |
| Vitreous culture | Singapore | [8] |
| Frogs (Rana catesbeiana (American bullfrogs) and Theloderma bicolor Chapa bug-eyed frogs, Warty toads (Bombina microdeladigitora), and Northern leopard frogs (Lithobates pipiens) | Malaysia Vietnam | [27,106,107] |
| Mosquitoes (Anopheles minimus, Anopheles dirus, Anopheles maculatus, Anopheles sawadwongporni, and Anopheles dravidicus) | Thailand | [90,108] |
| Fish (Clarias gariepinus (African sharptooth catfish) and Pangasius hypophthalmus (Tra catfish) | Malaysia, Vietnam | [28,75,109] |
| Retail sausages | Malaysia | [76] |
| Gnetum gnemon (Tree) | Malaysia | [110,111] |
| Isolate | Malaysia | Singapore | Thailand | Indonesia | Vietnam | Cambodia |
|---|---|---|---|---|---|---|
| NR | NR | NR | NR | NR | NR | |
| E. meningoseptica (CL) | 12 | 11 | 2 | 4 | - | 1 |
| E. meningoseptica (EN) | 2 | - | 1 | - | - | - |
| E. anophelis (CL) | 1 | 2 | - | - | - | 2 |
| E. anophelis (EN) | - | - | - | - | - | - |
| E. miricola (CL) | 1 | 1 | - | - | - | - |
| E. miricola (EN) | 2 | - | - | - | 2 | - |
| Elizabethkingia spp. (UI) | - | - | 1 | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zajmi, A.; Teo, J.; Yeo, C.C. Epidemiology and Characteristics of Elizabethkingia spp. Infections in Southeast Asia. Microorganisms 2022, 10, 882. https://doi.org/10.3390/microorganisms10050882
Zajmi A, Teo J, Yeo CC. Epidemiology and Characteristics of Elizabethkingia spp. Infections in Southeast Asia. Microorganisms. 2022; 10(5):882. https://doi.org/10.3390/microorganisms10050882
Chicago/Turabian StyleZajmi, Asdren, Jeanette Teo, and Chew Chieng Yeo. 2022. "Epidemiology and Characteristics of Elizabethkingia spp. Infections in Southeast Asia" Microorganisms 10, no. 5: 882. https://doi.org/10.3390/microorganisms10050882
APA StyleZajmi, A., Teo, J., & Yeo, C. C. (2022). Epidemiology and Characteristics of Elizabethkingia spp. Infections in Southeast Asia. Microorganisms, 10(5), 882. https://doi.org/10.3390/microorganisms10050882








