In Vitro Pharmacodynamics and Bactericidal Mechanism of Fungal Defensin-Derived Peptides NZX and P2 against Streptococcus agalactiae
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacteria and Reagents
2.2. Pharmacodynamics Evaluation of NZX and P2
2.2.1. Minimal Inhibitory Concentration (MIC), Mutant Prevention Concentration (MPC), and Selection Index (SI) Determination
2.2.2. Time-Kill Curves Measurement
2.2.3. Synergism and Post-Antibiotic Effect (PAE) of NZX and P2
2.3. Effect of Peptides on the Cell Membrane
2.3.1. FACS Analysis of Cell Membrane Integrity
2.3.2. K+ Leakage
2.3.3. ATP Release Reaction
2.3.4. Membrane Potential
2.4. Effect of Peptides on Cell Wall
2.4.1. Scanning Electron Microscopy (SEM) Observations
2.4.2. Transmission Electron Microscopy (TEM) Observations
2.4.3. Super-Resolution Microscopy (SRM) Observations
2.4.4. Bactericidal Effect of NZX and P2 on Quiescence/Division Period Bacteria
2.4.5. Interaction with Lipid II
3. Results
3.1. Pharmacodynamics Evaluation
3.1.1. MIC, MPC, and SI of NZX and P2 against S. agalactiae ACCC 61733
3.1.2. Time-Killing Curves of NZX and P2
3.1.3. Synergism and PAE of NZX and P2
3.2. Effect of Peptides on the Cell Membrane
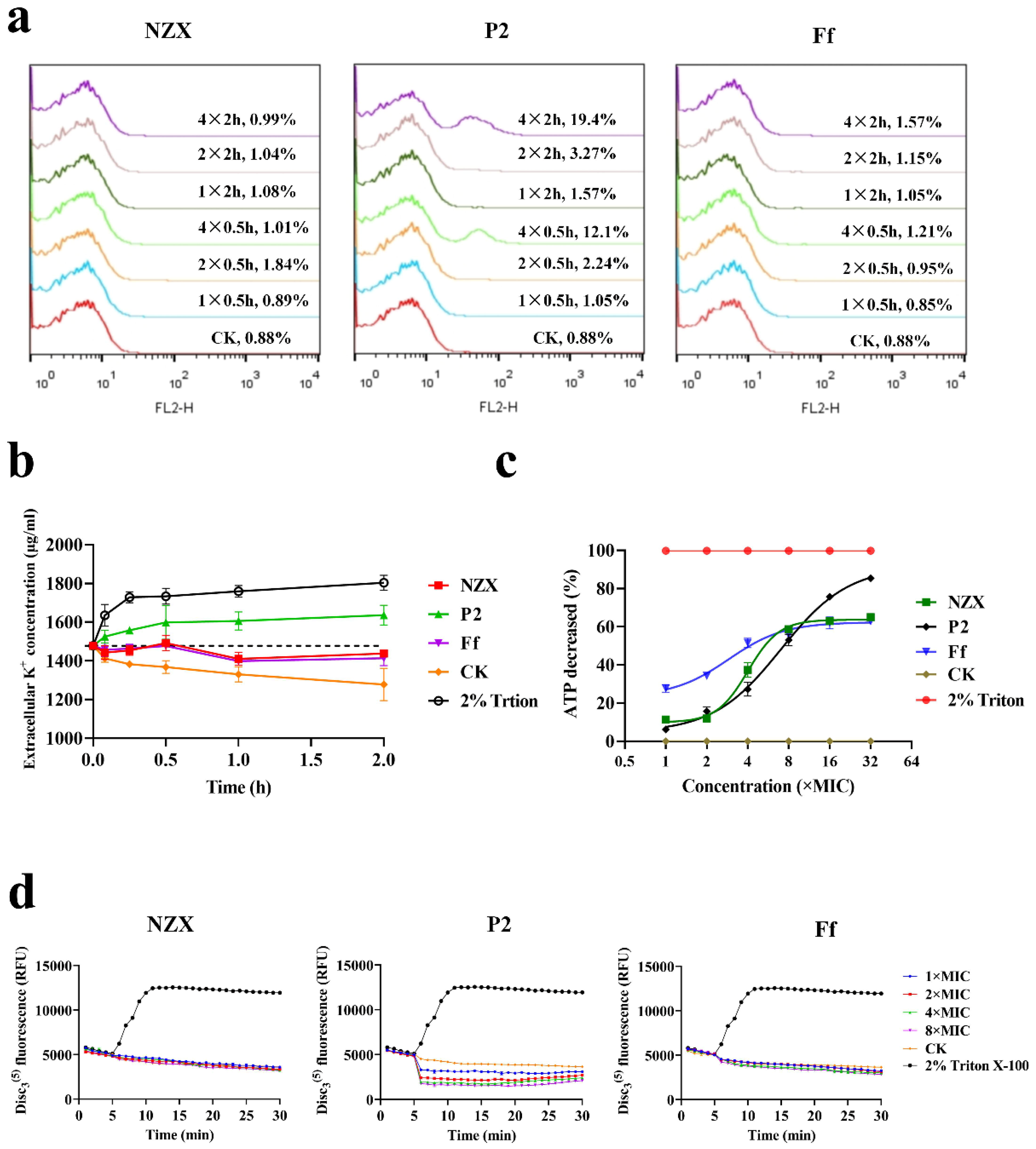
3.2.1. Integrity of Bacterial Membrane
3.2.2. K+ Leakage
3.2.3. ATP Release Reaction
3.2.4. Membrane Potential
3.3. Effect of Peptides on Cell Wall
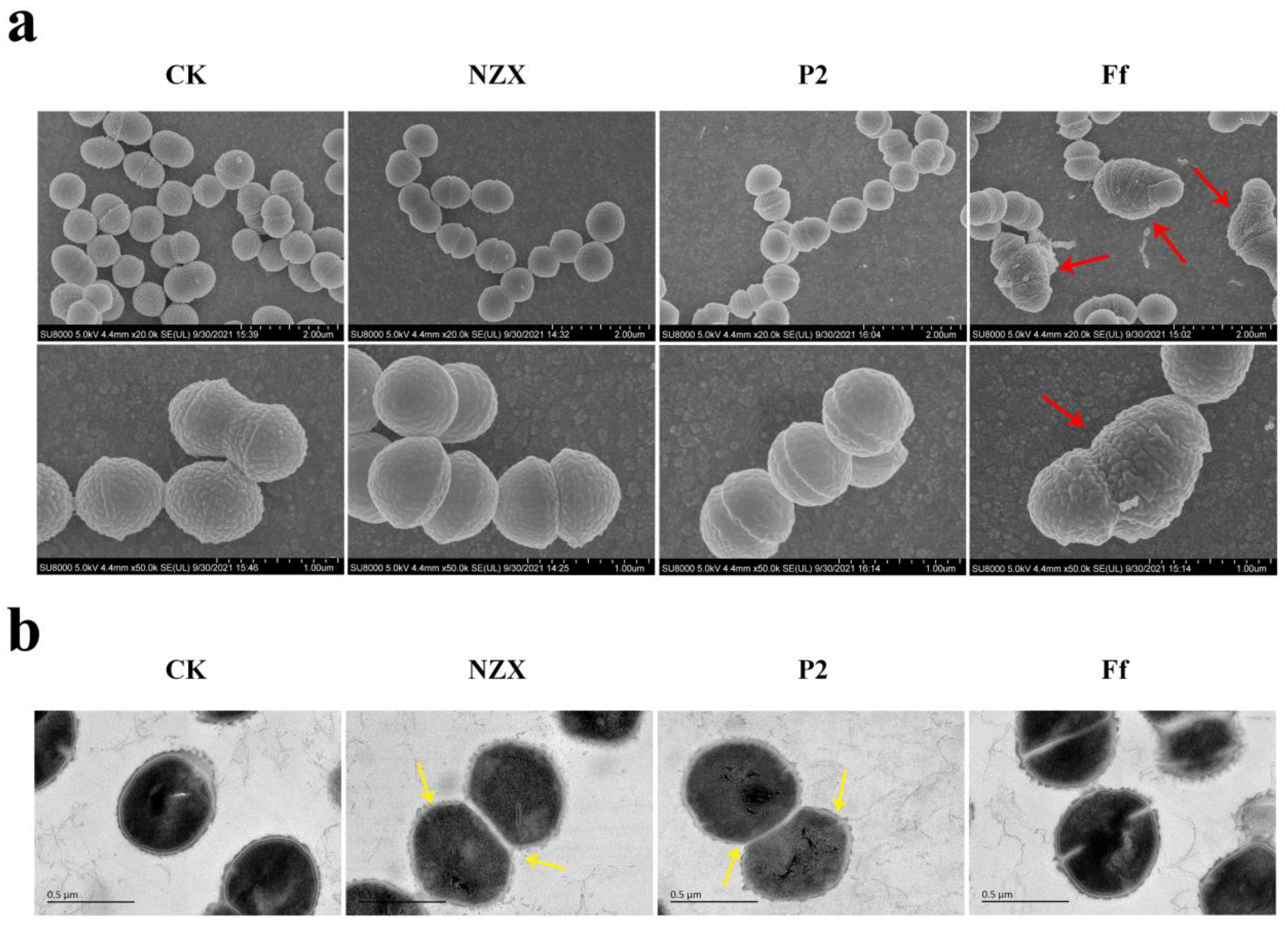
3.3.1. SEM Observations
3.3.2. TEM Observations
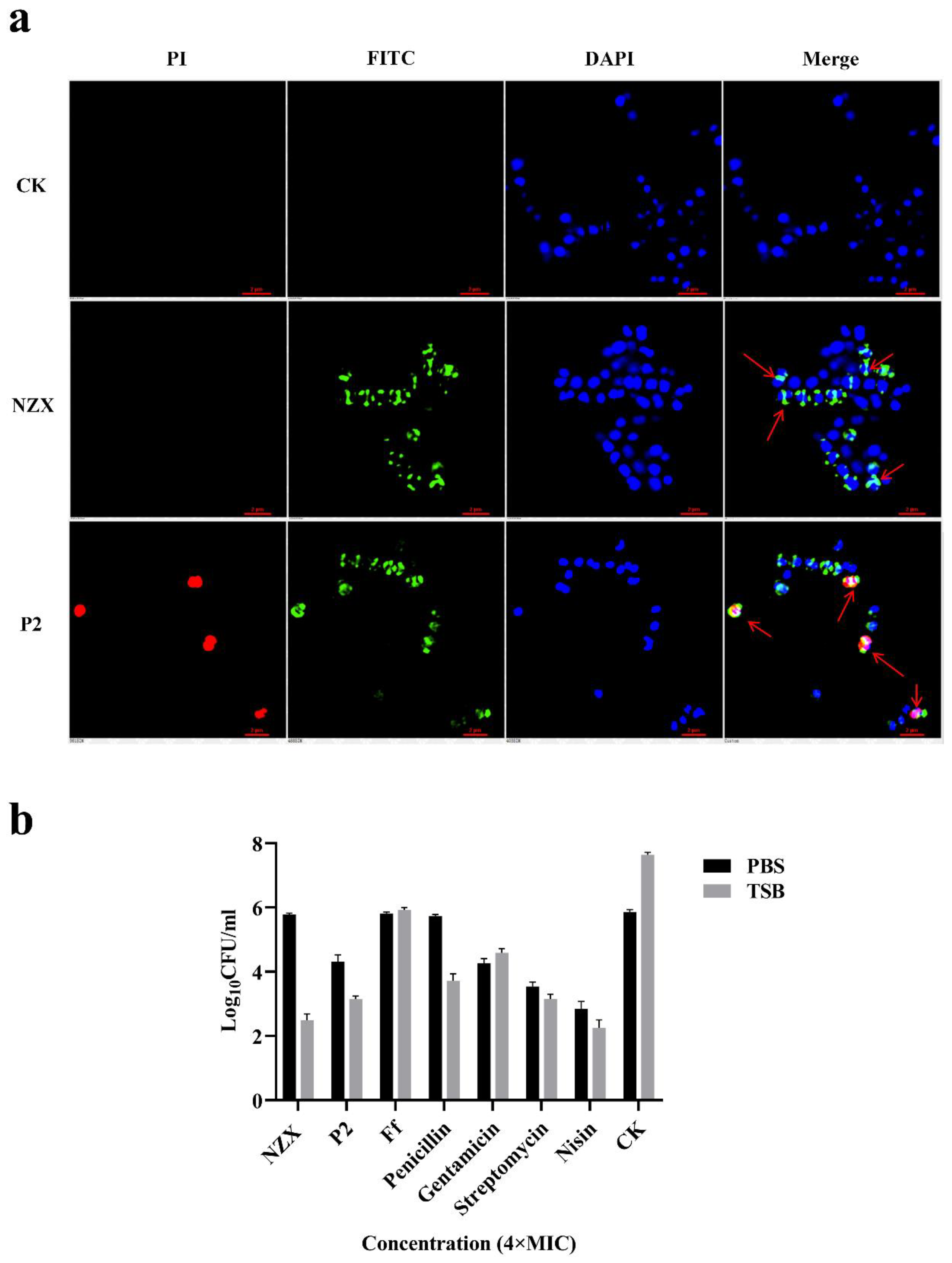
3.3.3. SRM Observations
3.3.4. Bactericidal Effect of NZX and P2 on Quiescence/Division Period Bacteria
3.3.5. Interaction with Lipid II
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barony, G.M.; Tavares, G.C.; Pereira, F.L.; Carvalho, A.F.; Dorella, F.A.; Leal, C.A.G.; Figueiredo, H.C.P. Large-scale genomic analyses reveal the population structure and evolutionary trends of Streptococcus agalactiae strains in Brazilian fish farms. Sci. Rep. 2017, 7, 13538. [Google Scholar] [CrossRef]
- Hernandez, L.; Bottini, E.; Cadona, J.; Cacciato, C.; Monteavaro, C.; Bustamante, A.; Sanso, A.M. Multidrug resistance and molecular characterization of Streptococcus agalactiae isolates from dairy cattle with mastitis. Front. Cell Infect. Microbiol. 2021, 11, 647324. [Google Scholar] [CrossRef]
- Gao, J.; Yu, F.Q.; Luo, L.P.; He, J.Z.; Hou, R.G.; Zhang, H.Q.; Li, S.M.; Su, J.L.; Han, B. Antibiotic resistance of Streptococcus agalactiae from cows with mastitis. Vet. J. 2012, 194, 423–424. [Google Scholar] [CrossRef]
- Maisey, H.C.; Doran, K.S.; Nizet, V. Recent advances in understanding the molecular basis of group B Streptococcus virulence. Expert Rev. Mol. Med. 2008, 10, e27. [Google Scholar] [CrossRef] [Green Version]
- Campbell, J.R.; Hillier, S.L.; Krohn, M.A.; Ferrieri, P.; Zaleznik, D.F.; Baker, C.J. Group B streptococcal colonization and serotype-specific immunity in pregnant women at delivery. Obs. Gynecol. 2000, 96, 498–503. [Google Scholar] [CrossRef]
- Chideroli, R.T.; Amoroso, N.; Mainardi, R.M.; Suphoronski, S.A.; de Padua, S.B.; Alfieri, A.F.; Alfieri, A.A.; Mosela, M.; Moralez, A.T.P.; de Oliveira, A.G.; et al. Emergence of a new multidrug-resistant and highly virulent serotype of Streptococcus agalactiae in fish farms from Brazil. Aquaculture 2017, 479, 45–51. [Google Scholar] [CrossRef]
- Chu, C.; Huang, P.Y.; Chen, H.M.; Wang, Y.H.; Tsai, I.A.; Lu, C.C.; Chen, C.C. Genetic and pathogenic difference between Streptococcus agalactiae serotype Ia fish and human isolates. BMC Microbiol. 2016, 16, 175. [Google Scholar] [CrossRef] [Green Version]
- Joshi, R.; Skaaurd, A.; Tola Alvarez, A. Experimental validation of genetic selection for resistance against Streptococcus agalactiae via different routes of infection in the commercial Nile tilapia breeding programme. J. Anim. Breed. Genet. 2021, 138, 338–348. [Google Scholar] [CrossRef]
- Kaminska, D.; Ratajczak, M.; Szumala-Kakol, A.; Dlugaszewska, J.; Nowak-Malczewska, D.M.; Gajecka, M. Increasing resistance and changes in distribution of serotypes of Streptococcus agalactiae in Poland. Pathogens 2020, 9, 526. [Google Scholar] [CrossRef]
- Boman, H.G. Antibacterial peptides: Key components needed in immunity. Cell 1991, 65, 205–207. [Google Scholar] [CrossRef]
- Powers, J.P.; Hancock, R.E. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Zhang, Y.; Mao, R.; Teng, D.; Wang, X.; Wang, J. Design and recombination expression of a novel plectasin-derived peptide MP1106 and its properties against Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2015, 99, 2649–2662. [Google Scholar] [CrossRef] [PubMed]
- Brogden, K.A. Antimicrobial peptides: Pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 2005, 3, 238–250. [Google Scholar] [CrossRef] [PubMed]
- Guilhelmelli, F.; Vilela, N.; Albuquerque, P.; Derengowski Lda, S.; Silva-Pereira, I.; Kyaw, C.M. Antibiotic development challenges: The various mechanisms of action of antimicrobial peptides and of bacterial resistance. Front. Microbiol. 2013, 4, 353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bechinger, B.; Gorr, S.U. Antimicrobial peptides: Mechanisms of action and resistance. J. Dent. Res. 2017, 96, 254–260. [Google Scholar] [CrossRef] [Green Version]
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Sass, V.; Pag, U.; Jansen, A.; Nielsen, A.K.; Mygind, P.H.; Raventós, D.S.; et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor Lipid II. Science 2010, 328, 1168–1172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, X.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Hu, F.; Wang, J. A study on fungal defensin against multidrug-resistant Clostridium perfringens and its treatment on infected poultry. Appl. Microbiol. Biotechnol. 2021, 105, 7265–7282. [Google Scholar] [CrossRef]
- Wang, J.; Hao, Y.; Fuente-Nunez, C.d.l.; Franco, O.L. Editorial: Antimicrobial peptides: Molecular design, structure-function relationship, and biosynthesis optimization. Front. Microbiol. 2022. [Google Scholar] [CrossRef]
- Ahn, K.B.; Kim, A.R.; Kum, K.Y.; Yun, C.H.; Han, S.H. The synthetic human beta-defensin-3 C15 peptide exhibits antimicrobial activity against Streptococcus mutans, both alone and in combination with dental disinfectants. J. Microbiol. 2017, 55, 830–836. [Google Scholar] [CrossRef]
- Lindhauer, N.S.; Bertrams, W.; Poppel, A.; Herkt, C.E.; Wesener, A.; Hoffmann, K.; Greene, B.; Van Der Linden, M.; Vilcinskas, A.; Seidel, K.; et al. Antibacterial activity of a Tribolium castaneum defensin in an in vitro infection model of Streptococcus pneumoniae. Virulence 2019, 10, 902–909. [Google Scholar] [CrossRef] [Green Version]
- Mygind, P.H.; Fischer, R.L.; Schnorr, K.M.; Hansen, M.T.; Sonksen, C.P.; Ludvigsen, S.; Raventos, D.; Buskov, S.; Christensen, B.; De Maria, L.; et al. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature 2005, 437, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, Y.; Teng, D.; Tian, Z.; Wang, S.; Wang, J. Expression of plectasin in Pichia pastoris and its characterization as a new antimicrobial peptide against Staphyloccocus and Streptococcus. Protein Expr. Purif. 2011, 78, 189–196. [Google Scholar] [CrossRef] [PubMed]
- Tenland, E.; Krishnan, N.; Ronnholm, A.; Kalsum, S.; Puthia, M.; Morgelin, M.; Davoudi, M.; Otrocka, M.; Alaridah, N.; Glegola-Madejska, I.; et al. A novel derivative of the fungal antimicrobial peptide plectasin is active against Mycobacterium tuberculosis. Tuberculosis 2018, 113, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, N.; Mao, R.; Teng, D.; Hao, Y.; Wang, X.; Wang, J. A new high-yielding antimicrobial peptide NZX and its antibacterial activity against Staphylococcus hyicus in vitro/vivo. Appl. Microbiol. Biotechnol. 2020, 104, 1555–1568. [Google Scholar] [CrossRef]
- Yang, N.; Teng, D.; Mao, R.; Hao, Y.; Wang, X.; Wang, Z.; Wang, X.; Wang, J. A recombinant fungal defensin-like peptide-P2 combats multidrug-resistant Staphylococcus aureus and biofilms. Appl. Microbiol. Biotechnol. 2019, 103, 5193–5213. [Google Scholar] [CrossRef]
- Zhang, Q.; Yang, N.; Mao, R.; Hao, Y.; Ma, X.; Teng, D.; Fan, H.; Wang, J. A recombinant fungal defensin-like peptide-P2 combats Streptococcus dysgalactiae and biofilms. Appl. Microbiol. Biotechnol. 2021, 105, 1489–1504. [Google Scholar] [CrossRef]
- Shan, Y.; Yang, N.; Teng, D.; Wang, X.; Mao, R.; Hao, Y.; Ma, X.; Fan, H.; Wang, J. Recombinant of the staphylococcal bacteriophage lysin CHAPk and its elimination against Streptococcus agalactiae biofilms. Microorganisms 2020, 8, 216. [Google Scholar] [CrossRef] [Green Version]
- Cunha, E.; Janela, R.; Costa, M.; Tavares, L.; Veiga, A.S.; Oliveira, M. Nisin influence on the antimicrobial resistance ability of canine oral Enterococci. Antibiotics 2020, 9, 890. [Google Scholar] [CrossRef]
- Jiang, L.; Xie, N.; Chen, M.; Liu, Y.; Wang, S.; Mao, J.; Li, J.; Huang, X. Synergistic combination of linezolid and fosfomycin closing each other’s mutant selection window to prevent enterococcal resistance. Front. Microbiol. 2020, 11, 605962. [Google Scholar] [CrossRef]
- Flamm, R.K.; Rhomberg, P.R.; Lindley, J.M.; Sweeney, K.; Ellis-Grosse, E.J.; Shortridge, D. Evaluation of the bactericidal activity of fosfomycin in combination with selected antimicrobial comparison agents tested against Gram-negative bacterial strains by using time-kill curves. Antimicrob. Agents Chemother. 2019, 63, e02549-18. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Teng, D.; Wang, X.; Mao, R.; Cao, X.; Hu, X.; Zong, L.; Wang, J. In vitro and in vivo characterization of a new recombinant antimicrobial peptide, MP1102, against methicillin-resistant Staphylococcus aureus. Appl. Microbiol. Biotechnol. 2015, 99, 6255–6266. [Google Scholar] [CrossRef] [PubMed]
- Giguere, S.; Lee, E.A.; Guldbech, K.M.; Berghaus, L.J. In vitro synergy, pharmacodynamics, and postantibiotic effect of 11 antimicrobial agents against Rhodococcus equi. Vet. Microbiol. 2012, 160, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Wang, Z.; Han, H.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Wang, X.; Wang, J. Dual antibacterial activities and biofilm eradication of a marine peptide-N6NH2 and its analogs against multidrug-resistant Aeromonas veronii. Int. J. Mol. Sci. 2020, 21, 9637. [Google Scholar] [CrossRef] [PubMed]
- Han, H.; Teng, D.; Mao, R.; Hao, Y.; Yang, N.; Wang, Z.; Li, T.; Wang, X.; Wang, J. Marine peptide-N6NH2 and its derivative-GUON6NH2 have potent antimicrobial activity against intracellular Edwardsiella tarda in vitro and in vivo. Front. Microbiol. 2021, 12, 637427. [Google Scholar] [CrossRef]
- Elliott, A.G.; Huang, J.X.; Neve, S.; Zuegg, J.; Edwards, I.A.; Cain, A.K.; Boinett, C.J.; Barquist, L.; Lundberg, C.V.; Steen, J.; et al. An amphipathic peptide with antibiotic activity against multidrug-resistant Gram-negative bacteria. Nat. Commun. 2020, 11, 3184. [Google Scholar] [CrossRef]
- Song, J.; Wang, J.; Zhan, N.; Sun, T.; Yu, W.; Zhang, L.; Shan, A.; Zhang, A. Therapeutic potential of Trp-rich engineered amphiphiles by single hydrophobic amino acid end-tagging. ACS Appl. Mater. Interfaces 2019, 11, 43820–43834. [Google Scholar] [CrossRef]
- Lunde, C.S.; Hartouni, S.R.; Janc, J.W.; Mammen, M.; Humphrey, P.P.; Benton, B.M. Telavancin disrupts the functional integrity of the bacterial membrane through targeted interaction with the cell wall precursor lipid II. Antimicrob. Agents Chemother. 2009, 53, 3375–3383. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K.; Sugishita, K.; Harada, M.; Fujii, N.; Miyajima, K. Interactions of an antimicrobial peptide, magainin 2, with outer and inner membranes of Gram-negative bacteria. Biochim. Biophys Acta 1997, 1327, 119–130. [Google Scholar] [CrossRef] [Green Version]
- Lapage, G. Mode of action of penicillin. Nature 1945, 155, 403–404. [Google Scholar] [CrossRef] [Green Version]
- Munch, D.; Sahl, H.G. Structural variations of the cell wall precursor lipid II in Gram-positive bacteria-Impact on binding and efficacy of antimicrobial peptides. Biochim. Biophys Acta 2015, 1848, 3062–3071. [Google Scholar] [CrossRef] [Green Version]
- De Francesco, M.A.; Caracciolo, S.; Gargiulo, F.; Manca, N. Phenotypes, genotypes, serotypes and molecular epidemiology of erythromycin-resistant Streptococcus agalactiae in Italy. Eur. J. Clin. Microbiol. Infect. Dis. 2012, 31, 1741–1747. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Kan, Y.; Zhang, Z.; Lu, Z.; Li, Y.; Leng, C.; Ji, J.; Song, S.; Shi, H. New mutations of penicillin-binding proteins in Streptococcus agalactiae isolates from cattle with decreased susceptibility to penicillin. Microb. Drug Resist. 2018, 24, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Hara, S.; Mukae, H.; Sakamoto, N.; Ishimoto, H.; Amenomori, M.; Fujita, H.; Ishimatsu, Y.; Yanagihara, K.; Kohno, S. Plectasin has antibacterial activity and no affect on cell viability or IL-8 production. Biochem. Biophys Res. Commun. 2008, 374, 709–713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drlica, K.; Zhao, X. Mutant selection window hypothesis updated. Clin. Infect. Dis 2007, 44, 681–688. [Google Scholar] [CrossRef]
- Marshall, S.H. Antimicrobial peptides: A natural alternative to chemical antibiotics and a potential for applied biotechnology. Electron. J. Biotechnol. 2003, 6, 271–284. [Google Scholar]
- Ma, Z.; Yang, J.; Han, J.; Gao, L.; Liu, H.; Lu, Z.; Zhao, H.; Bie, X. Insights into the antimicrobial activity and cytotoxicity of engineered alpha-helical peptide amphiphiles. J. Med. Chem. 2016, 59, 10946–10962. [Google Scholar] [CrossRef]
- Porto, W.F.; Irazazabal, L.; Alves, E.S.F.; Ribeiro, S.M.; Matos, C.O.; Pires, A.S.; Fensterseifer, I.C.M.; Miranda, V.J.; Haney, E.F.; Humblot, V.; et al. In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design. Nat. Commun. 2018, 9, 1490. [Google Scholar] [CrossRef]
- Li, W.; O’Brien-Simpson, N.M.; Tailhades, J.; Pantarat, N.; Dawson, R.M.; Otvos, L., Jr.; Reynolds, E.C.; Separovic, F.; Hossain, M.A.; Wade, J.D. Multimerization of a proline-rich antimicrobial peptide, Chex-Arg20, alters its mechanism of interaction with the Escherichia coli membrane. Chem. Biol. 2015, 22, 1250–1258. [Google Scholar] [CrossRef]
- Higgins, D.L.; Chang, R.; Debabov, D.V.; Leung, J.; Wu, T.; Krause, K.M.; Sandvik, E.; Hubbard, J.M.; Kaniga, K.; Schmidt, D.E., Jr.; et al. Telavancin, a multifunctional lipoglycopeptide, disrupts both cell wall synthesis and cell membrane integrity in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2005, 49, 1127–1134. [Google Scholar] [CrossRef] [Green Version]
- Lv, X.; Miao, L.; Ma, H.; Bai, F.; Lin, Y.; Sun, M.; Li, J. Purification, characterization and action mechanism of plantaricin JY22, a novel bacteriocin against Bacillus cereus produced by Lactobacillus plantarum JY22 from golden carp intestine. Food Sci. Biotechnol. 2018, 27, 695–703. [Google Scholar] [CrossRef]
- Takahashi, D.; Shukla, S.K.; Prakash, O.; Zhang, G. Structural determinants of host defense peptides for antimicrobial activity and target cell selectivity. Biochimie 2010, 92, 1236–1241. [Google Scholar] [CrossRef] [PubMed]


| Strain | MIC | |||||
|---|---|---|---|---|---|---|
| NZX | P2 | Ff | ||||
| μg/mL | μM | μg/mL | μM | μg/mL | μM | |
| S. agalactiae ATCC13813 | 0.50 | 0.11 | 4 | 0.91 | 1 | 2.80 |
| S. agalactiae CAU-FRI 1 | 0.25 | 0.06 | 2 | 0.46 | 1 | 2.80 |
| S. agalactiae CAU-FRI 2 | 0.50 | 0.11 | 2 | 0.46 | 2 | 5.59 |
| S. agalactiae CAU-FRI 3 | 0.50 | 0.11 | 4 | 0.91 | 2 | 5.59 |
| S. agalactiae CAU-FRI 4 | 0.50 | 0.11 | 4 | 0.91 | 2 | 5.59 |
| S. agalactiae PBSA0903 | 0.50 | 0.11 | 4 | 0.91 | 1 | 2.80 |
| S. dysgalactiae CVCC 3938 | 1 | 0.22 | 2 | 0.46 | 1 | 2.80 |
| Variety | MIC | MPC | SI | ||
|---|---|---|---|---|---|
| μg/mL | μM | μg/mL | μM | ||
| NZX | 0.5 | 0.11 | 8 | 1.82 | 16 |
| P2 | 4 | 0.91 | >128 | >29.2 | >32 |
| Ff | 2 | 5.59 | >128 | >357.5 | >64 |
| Combination | Variety | MICa (μg/mL) | MICc (μg/mL) | FIC | FICI |
|---|---|---|---|---|---|
| NZX-Ff | NZX | 0.5 | 0.5 | 1 | 1.0625 |
| Ff | 2 | 0.125 | 0.0625 | ||
| P2-Ff | P2 | 4 | 0.5 | 1 | 1.0625 |
| Ff | 2 | 0.125 | 0.0625 |
| Variety | None | dKAA(D) | Sodium Pyrophosphate (S) | D-Glutamate (D-G) |
|---|---|---|---|---|
| MICA (μg/mL) | MICA+D (μg/mL) | MICA+S (μg/mL) | MICA+D-G (μg/mL) | |
| NZ2114 | 0.5 | 0.5 | 0.5 | 0.5 |
| NZX | 0.5 | 0.5 | 0.5 | 0.5 |
| P2 | 4 | 4 | 4 | 4 |
| VAN | 2 | 64 | 2 | 2 |
| Peptides | Sequences | Similarity (%) | Length (Amino Acid) | MW (Da) | pI | Charge (+) | GRAVY | Hydrophobicity |
|---|---|---|---|---|---|---|---|---|
| Plectasin | GFGCNGPWDEDDMQCHNHCKSIKGYKGGYCAKGGFVCKCY | 100.0 | 40 | 4407.9 | 7.77 | 1 | −0.695 | 0.353 |
| P2 | GFGCNGPWDEDDMKCHNHCKSIKGYKGGYCASAGFVCKCY | 92 | 40 | 4380.9 | 7.77 | 1 | −0.573 | 0.366 |
| NZX | GFGCNGPWSEDDIQCHNHCKSIKGYKGGYCARGGFVCKCY | 92 | 40 | 4390.0 | 8.3 | 2 | −0.578 | 0.385 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.; Yang, N.; Mao, R.; Hao, Y.; Teng, D.; Wang, J. In Vitro Pharmacodynamics and Bactericidal Mechanism of Fungal Defensin-Derived Peptides NZX and P2 against Streptococcus agalactiae. Microorganisms 2022, 10, 881. https://doi.org/10.3390/microorganisms10050881
Wu Y, Yang N, Mao R, Hao Y, Teng D, Wang J. In Vitro Pharmacodynamics and Bactericidal Mechanism of Fungal Defensin-Derived Peptides NZX and P2 against Streptococcus agalactiae. Microorganisms. 2022; 10(5):881. https://doi.org/10.3390/microorganisms10050881
Chicago/Turabian StyleWu, Yankang, Na Yang, Ruoyu Mao, Ya Hao, Da Teng, and Jianhua Wang. 2022. "In Vitro Pharmacodynamics and Bactericidal Mechanism of Fungal Defensin-Derived Peptides NZX and P2 against Streptococcus agalactiae" Microorganisms 10, no. 5: 881. https://doi.org/10.3390/microorganisms10050881
APA StyleWu, Y., Yang, N., Mao, R., Hao, Y., Teng, D., & Wang, J. (2022). In Vitro Pharmacodynamics and Bactericidal Mechanism of Fungal Defensin-Derived Peptides NZX and P2 against Streptococcus agalactiae. Microorganisms, 10(5), 881. https://doi.org/10.3390/microorganisms10050881









