Identification and Molecular Characterization of the Operon Required for L-Asparagine Utilization in Corynebacterium glutamicum
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains, Plasmids, and Culture Conditions
2.2. Construction of Plasmids and Mutants
2.3. RNA Extraction
2.4. Determination of Transcriptional Start Site (TSS)
2.5. Quantitative Reverse-Transcription Polymerase Chain Reaction (qRT-PCR)
2.6. Overexpression and Purification of His-Tagged AnsR
2.7. Electrophoretic Mobility Shift Assay (EMSA)
3. Results
3.1. Conservation of the ans Gene Cluster
3.2. Role of Genes in the Operon for L-Asn Utilization
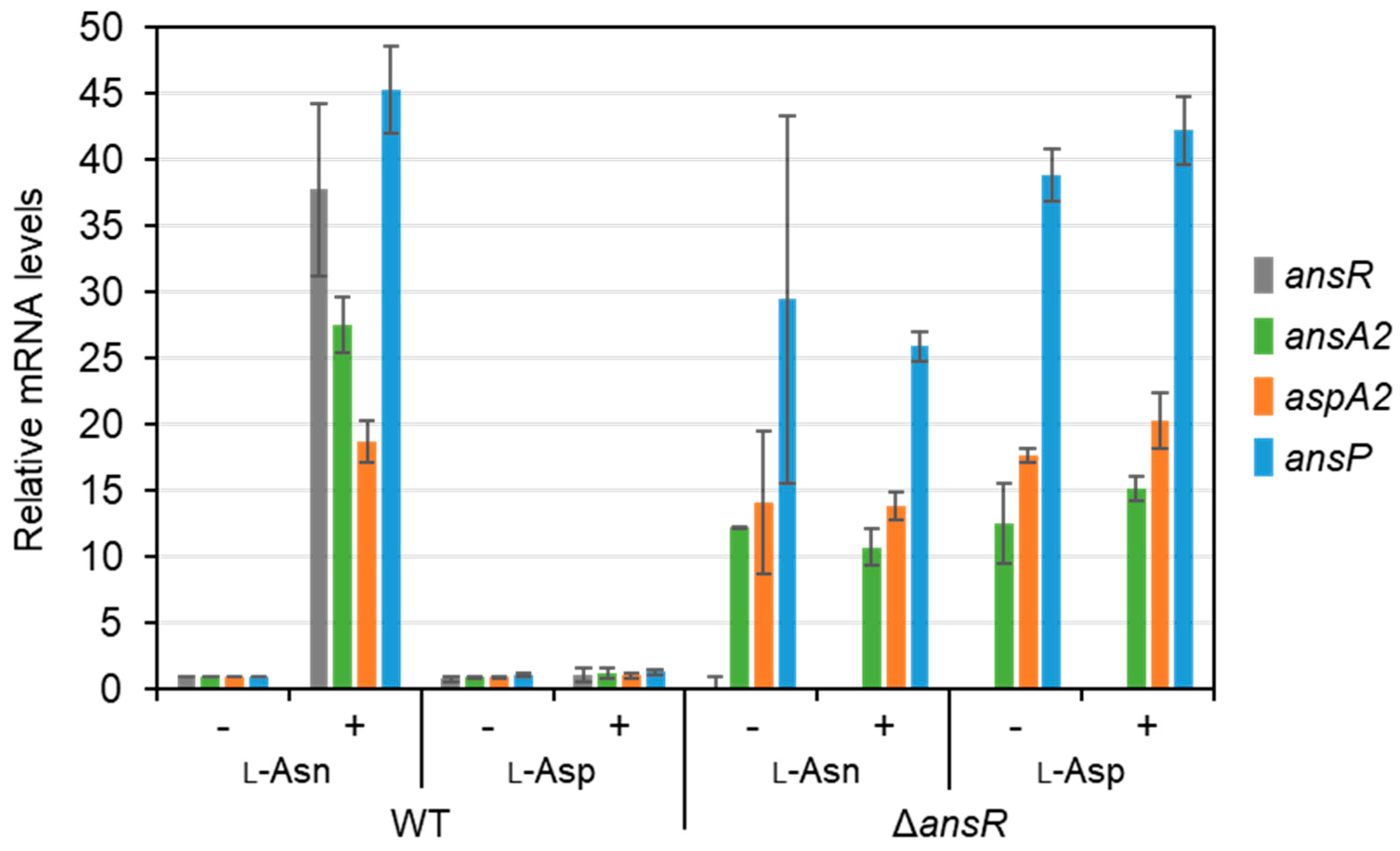
3.3. ans Operon Induced upon L-Asn Supplementation
3.4. ans Operon Repressed by AnsR
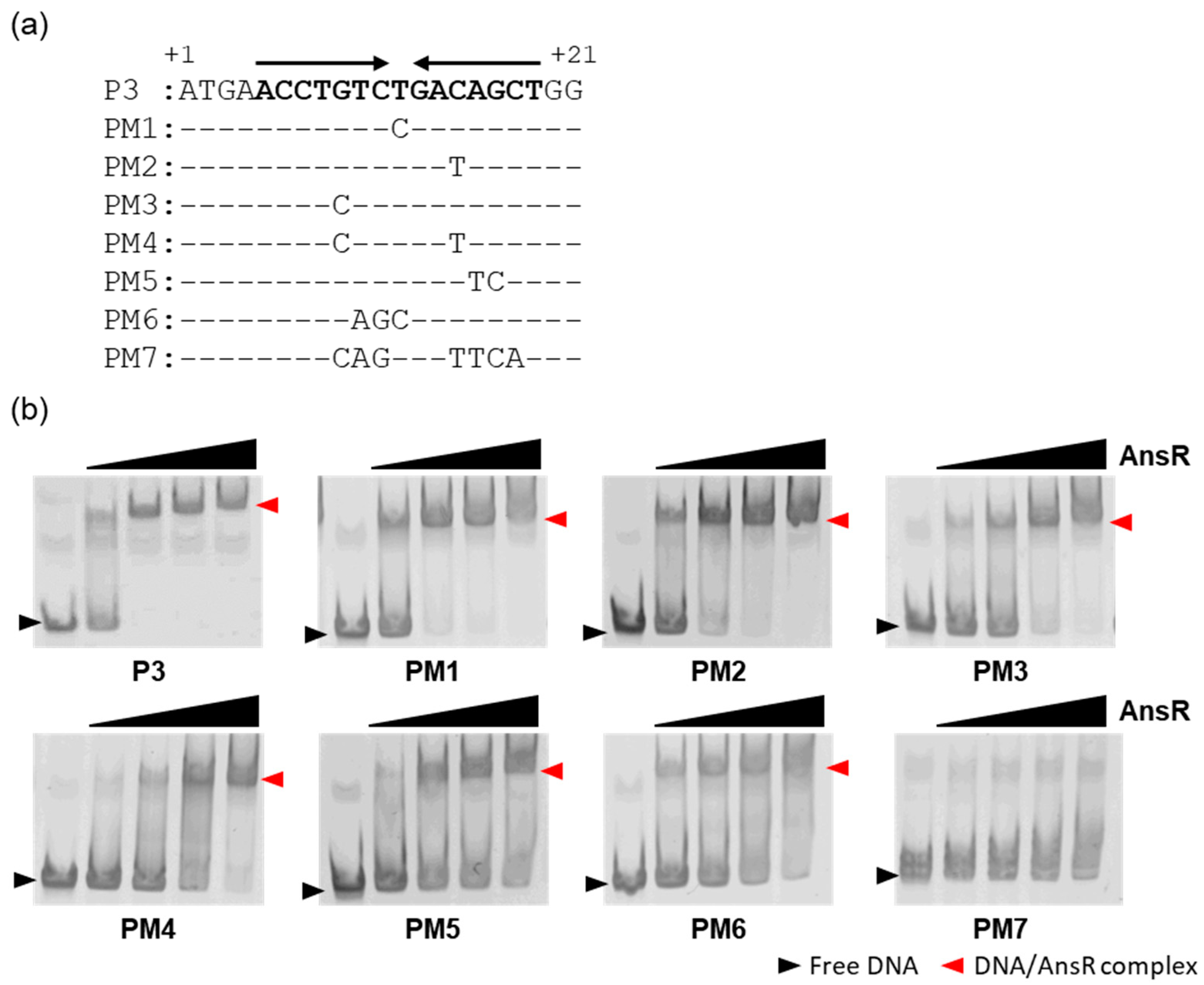
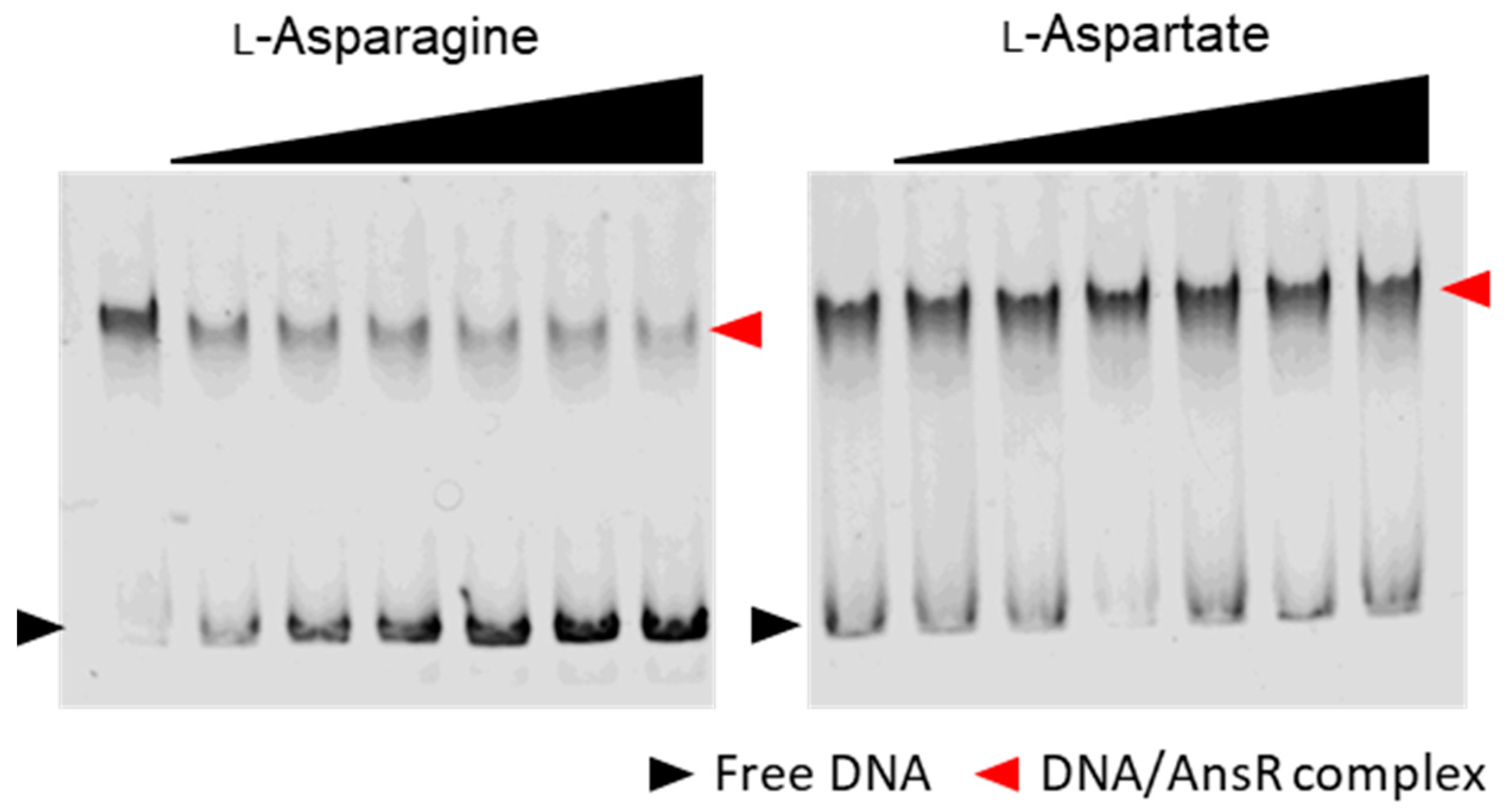
3.5. AnsR Directly Repressed the ans Operon
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Koffas, M.; Stephanopoulos, G. Strain improvement by metabolic engineering: Lysine production as a case study for systems biology. Curr. Opin. Biotechnol. 2005, 16, 361–366. [Google Scholar] [CrossRef] [PubMed]
- Hirasawa, T.; Kim, J.; Shirai, T.; Furusawa, C.; Shimizu, H. Molecular mechanisms and metabolic engineering of glutamate overproduction in Corynebacterium glutamicum. Subcell. Biochem. 2012, 64, 261–281. [Google Scholar] [CrossRef]
- Ikeda, M. Lysine Fermentation: History and Genome Breeding. Adv. Biochem. Eng. Biotechnol. 2017, 159, 73–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikeda, M.; Takeno, S. Recent advances in amino acid production. In Corynebacterium glutamicum: Biology and Biotechnology; Microbiology Monographs; Inui, M., Toyoda, K., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 175–226. ISBN 978-3-030-39267-3. [Google Scholar]
- Wendisch, V.F.; Lee, J.-H. Metabolic engineering in Corynebacterium glutamicum. In Corynebacterium glutamicum: Biology and Biotechnology; Microbiology Monographs; Inui, M., Toyoda, K., Eds.; Springer: Cham, Switzerland, 2020; pp. 287–322. ISBN 978-3-030-39267-3. [Google Scholar]
- Wendisch, V.F. Metabolic engineering advances and prospects for amino acid production. Metab. Eng. 2020, 58, 17–34. [Google Scholar] [CrossRef] [PubMed]
- Park, J.H.; Lee, S.Y. Metabolic pathways and fermentative production of L-aspartate family amino acids. Biotechnol. J. 2010, 5, 560–577. [Google Scholar] [CrossRef]
- Mesas, J.M.; Gil, J.A.; Martin, J.F. Characterization and partial purification of L-asparaginase from Corynebacterium glutamicum. J. Gen. Microbiol. 1990, 136, 515–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menkel, E.; Thierbach, G.; Eggeling, L.; Sahm, H. Influence of increased aspartate availability on lysine formation by a recombinant strain of Corynebacterium glutamicum and utilization of fumarate. Appl. Environ. Microbiol. 1989, 55, 684–688. [Google Scholar] [CrossRef] [Green Version]
- Willis, R.C.; Woolfolk, C.A. Asparagine utilization in Escherichia coli. J. Bacteriol. 1974, 118, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Del Casale, T.; Sollitti, P.; Chesney, R.H. Cytoplasmic L-asparaginase: Isolation of a defective strain and mapping of ansA. J. Bacteriol. 1983, 154, 513–515. [Google Scholar] [CrossRef] [Green Version]
- Srikhanta, Y.N.; Atack, J.M.; Beacham, I.R.; Jennings, M.P. Distinct physiological roles for the two L-asparaginase isozymes of Escherichia coli. Biochem. Biophys. Res. Commun. 2013, 436, 362–365. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.; Setlow, P. Cloning and nucleotide sequence of the Bacillus subtilis ansR gene, which encodes a repressor of the ans operon coding for L-asparaginase and L-aspartase. J. Bacteriol. 1993, 175, 2501–2506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fisher, S.H.; Wray, L.V. Bacillus subtilis 168 contains two differentially regulated genes encoding L-asparaginase. J. Bacteriol. 2002, 184, 2148–2154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Lu, C.-D. Molecular characterization and regulation of operons for asparagine and aspartate uptake and utilization in Pseudomonas aeruginosa. Microbiology 2018, 164, 205–216. [Google Scholar] [CrossRef] [PubMed]
- Gouzy, A.; Larrouy-Maumus, G.; Bottai, D.; Levillain, F.; Dumas, A.; Wallach, J.B.; Caire-Brandli, I.; de Chastellier, C.; Wu, T.-D.; Poincloux, R.; et al. Mycobacterium tuberculosis exploits asparagine to assimilate nitrogen and resist acid stress during infection. PLoS Pathog. 2014, 10, e1003928. [Google Scholar] [CrossRef] [Green Version]
- Hu, Y.; Lu, P.; Zhang, Y.; Li, L.; Chen, S. Characterization of an aspartate-dependent acid survival system in Yersinia pseudotuberculosis. FEBS Lett. 2010, 584, 2311–2314. [Google Scholar] [CrossRef] [Green Version]
- Inui, M.; Suda, M.; Okino, S.; Nonaka, H.; Puskás, L.G.; Vertès, A.A.; Yukawa, H. Transcriptional profiling of Corynebacterium glutamicum metabolism during organic acid production under oxygen deprivation conditions. Microbiology 2007, 153, 2491–2504. [Google Scholar] [CrossRef] [Green Version]
- Teramoto, H.; Suda, M.; Inui, M.; Yukawa, H. Regulation of the expression of genes involved in NAD de novo biosynthesis in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2010, 76, 5488–5495. [Google Scholar] [CrossRef] [Green Version]
- Kubota, T.; Watanabe, A.; Suda, M.; Kogure, T.; Hiraga, K.; Inui, M. Production of para-aminobenzoate by genetically engineered Corynebacterium glutamicum and non-biological formation of an N-glucosyl byproduct. Metab. Eng. 2016, 38, 322–330. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning: A Laboratory Manual, 2nd ed.; Cold Spring Harbor Laboratory: Cold Spring Harbor, NY, USA, 1989. [Google Scholar]
- Studier, F.W.; Moffatt, B.A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 1986, 189, 113–130. [Google Scholar] [CrossRef]
- Yukawa, H.; Omumasaba, C.A.; Nonaka, H.; Kós, P.; Okai, N.; Suzuki, N.; Suda, M.; Tsuge, Y.; Watanabe, J.; Ikeda, Y.; et al. Comparative analysis of the Corynebacterium glutamicum group and complete genome sequence of strain R. Microbiology 2007, 153, 1042–1058. [Google Scholar] [CrossRef] [Green Version]
- Inui, M.; Kawaguchi, H.; Murakami, S.; Vertès, A.A.; Yukawa, H. Metabolic engineering of Corynebacterium glutamicum for fuel ethanol production under oxygen-deprivation conditions. J. Mol. Microbiol. Biotechnol. 2004, 8, 243–254. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, K.; Teramoto, H.; Gunji, W.; Inui, M.; Yukawa, H. Involvement of regulatory interactions among global regulators GlxR, SugR, and RamA in expression of ramA in Corynebacterium glutamicum. J. Bacteriol. 2013, 195, 1718–1726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toyoda, K.; Teramoto, H.; Inui, M.; Yukawa, H. Expression of the gapA gene encoding glyceraldehyde-3-phosphate dehydrogenase of Corynebacterium glutamicum is regulated by the global regulator SugR. Appl. Microbiol. Biotechnol. 2008, 81, 291–301. [Google Scholar] [CrossRef] [PubMed]
- Pátek, M.; Nešvera, J. Sigma factors and promoters in Corynebacterium glutamicum. J. Biotechnol. 2011, 154, 101–113. [Google Scholar] [CrossRef] [PubMed]
- Ortuño-Olea, L.; Durán-Vargas, S. The L-asparagine operon of Rhizobium etli contains a gene encoding an atypical asparaginase. FEMS Microbiol. Lett. 2000, 189, 177–182. [Google Scholar] [CrossRef]
- González, V.; Santamaría, R.I.; Bustos, P.; Hernández-González, I.; Medrano-Soto, A.; Moreno-Hagelsieb, G.; Janga, S.C.; Ramírez, M.A.; Jiménez-Jacinto, V.; Collado-Vides, J.; et al. The partitioned Rhizobium etli genome: Genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. USA 2006, 103, 3834–3839. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Enríquez, A.; Evangelista-Martínez, Z.; Flores-Carrasco, M.E.; Servín-González, L. Identification of the σ70-dependent promoter controlling expression of the ansPAB operon of the nitrogen-fixing bacterium Rhizobium etli. J. Microbiol. Biotechnol. 2015, 25, 1241–1245. [Google Scholar] [CrossRef]
- Huerta-Zepeda, A.; Durán, S.; Du Pont, G.; Calderón, J. Asparagine degradation in Rhizobium etli. Microbiology 1996, 142, 1071–1076. [Google Scholar] [CrossRef] [Green Version]
- Sun, D.X.; Setlow, P. Cloning, nucleotide sequence, and expression of the Bacillus subtilis ans operon, which codes for L-asparaginase and L-aspartase. J. Bacteriol. 1991, 173, 3831–3845. [Google Scholar] [CrossRef] [Green Version]
- Rigali, S.; Derouaux, A.; Giannotta, F.; Dusart, J. Subdivision of the helix-turn-helix GntR family of bacterial regulators in the FadR, HutC, MocR, and YtrA subfamilies. J. Biol. Chem. 2002, 277, 12507–12515. [Google Scholar] [CrossRef] [Green Version]
- Kotrba, P.; Inui, M.; Yukawa, H. A single V317A or V317M substitution in Enzyme II of a newly identified β-glucoside phosphotransferase and utilization system of Corynebacterium glutamicum R extends its specificity towards cellobiose. Microbiology 2003, 149, 1569–1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, Y.; Teramoto, H.; Inui, M.; Yukawa, H. Identification of a second beta-glucoside phosphoenolpyruvate: Carbohydrate phosphotransferase system in Corynebacterium glutamicum R. Microbiology 2009, 155, 3652–3660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubota, T.; Tanaka, Y.; Takemoto, N.; Hiraga, K.; Yukawa, H.; Inui, M. Identification and expression analysis of a gene encoding a shikimate transporter of Corynebacterium glutamicum. Microbiology 2015, 161, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Kohl, T.A.; Rückert, C.; Rodionov, D.A.; Li, L.-H.; Ding, J.-Y.; Kalinowski, J.; Liu, S.-J. Phenylacetic acid catabolism and its transcriptional regulation in Corynebacterium glutamicum. Appl. Environ. Microbiol. 2012, 78, 5796–5804. [Google Scholar] [CrossRef] [Green Version]
- Hansmeier, N.; Albersmeier, A.; Tauch, A.; Damberg, T.; Ros, R.; Anselmetti, D.; Pühler, A.; Kalinowski, J. The surface (S)-layer gene cspB of Corynebacterium glutamicum is transcriptionally activated by a LuxR-type regulator and located on a 6 kb genomic island absent from the type strain ATCC 13032. Microbiology 2006, 152, 923–935. [Google Scholar] [CrossRef] [Green Version]



| Strain or Plasmid | Relevant Characteristics | Source or Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| JM109 | recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 Δ(lac-proAB)/F’[traD36 proAB+ lacIq lacZΔM15] | Takara |
| JM110 | dam dcm supE44 hsdR17 tih leu rpsL lacy galK galT ara tonA thr tsx Δ(lac-proAB)/F’[traD36 proAB+ lacIq lacZΔM15] | [21] |
| BL21(DE3) | F− ompT gal dcm lon hsdSB(rB− mB−) λ(DE3) | [22] |
| C. glutamicum | ||
| R (JCM 18229) | Wild-type strain | [23] |
| ATCC 13032 | Wild-type strain | American Type Culture Collection, Manassas, VA, USA |
| ΔaspA1 | R with deletion in aspA1 | This study |
| ΔansA1 | R with deletion in ansA1 | This study |
| ΔansR | R with deletion in ansR | This study |
| ΔansA2 | R with deletion in ansA2 | This study |
| ΔaspA2 | R with deletion in aspA2 | This study |
| ΔansP | R with deletion in ansP | This study |
| ΔaspA1A2 | ΔaspA2 with deletion in aspA1 | This study |
| ΔansA1A2 | ΔansA2 with deletion in ansA1 | This study |
| Plasmids | ||
| pCold | Apr: a vector for cold-inducible expression | TaKaRa |
| pCRA725 | Kmr; the suicide vector containing the B. subtilis sacB gene | [24] |
| pCRC667 | Apr: pColdI with a coding region of the ansR gene | This study |
| pCRC668 | Kmr; pCRA725 with a fragment containing flanking regions of aspA1 | This study |
| pCRC669 | Kmr; pCRA725 with a fragment containing flanking regions of ansA1 | This study |
| pCRC670 | Kmr; pCRA725 with a fragment containing flanking regions of ansR | This study |
| pCRC671 | Kmr; pCRA725 with a fragment containing flanking regions of ansA2 | This study |
| pCRC672 | Kmr; pCRA725 with a fragment containing flanking regions of aspA2 | This study |
| pCRC673 | Kmr; pCRA725 with a fragment containing flanking regions of ansP | This study |
| Genotype | Carbon or Nitrogen Source | Amino Acids As | |
|---|---|---|---|
| Glucose | L-Asparagine | ||
| R (Wild type) | ++ 1 | ++ | Carbon |
| ++ | Nitrogen | ||
| ATCC 13032 | ++ | − 1 | Carbon |
| − | Nitrogen | ||
| ΔaspA1 | ++ | ++ | Carbon |
| ++ | Nitrogen | ||
| ΔaspA2 | ++ | − | Carbon |
| + 1 | Nitrogen | ||
| ΔaspA1ΔaspA2 | ++ | − | Carbon |
| − | Nitrogen | ||
| ΔansA1 | ++ | ++ | Carbon |
| ++ | Nitrogen | ||
| ΔansA2 | ++ | − | Carbon |
| ++ | Nitrogen | ||
| ΔansA1ΔansA2 | ++ | − | Carbon |
| − | Nitrogen | ||
| ΔansP | ++ | − | Carbon |
| + | Nitrogen | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toyoda, K.; Sugaya, R.; Domon, A.; Suda, M.; Hiraga, K.; Inui, M. Identification and Molecular Characterization of the Operon Required for L-Asparagine Utilization in Corynebacterium glutamicum. Microorganisms 2022, 10, 1002. https://doi.org/10.3390/microorganisms10051002
Toyoda K, Sugaya R, Domon A, Suda M, Hiraga K, Inui M. Identification and Molecular Characterization of the Operon Required for L-Asparagine Utilization in Corynebacterium glutamicum. Microorganisms. 2022; 10(5):1002. https://doi.org/10.3390/microorganisms10051002
Chicago/Turabian StyleToyoda, Koichi, Riki Sugaya, Akihiro Domon, Masako Suda, Kazumi Hiraga, and Masayuki Inui. 2022. "Identification and Molecular Characterization of the Operon Required for L-Asparagine Utilization in Corynebacterium glutamicum" Microorganisms 10, no. 5: 1002. https://doi.org/10.3390/microorganisms10051002






