Comparative Genome Analysis of the Photosynthetic Betaproteobacteria of the Genus Rhodocyclus: Heterogeneity within Strains Assigned to Rhodocyclus tenuis and Description of Rhodocyclus gracilis sp. nov. as a New Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Origin of the Strains of Rhodocyclus Tenuis
2.2. Genome Sequencing
2.3. Whole Genome Comparison
3. Results and Discussion
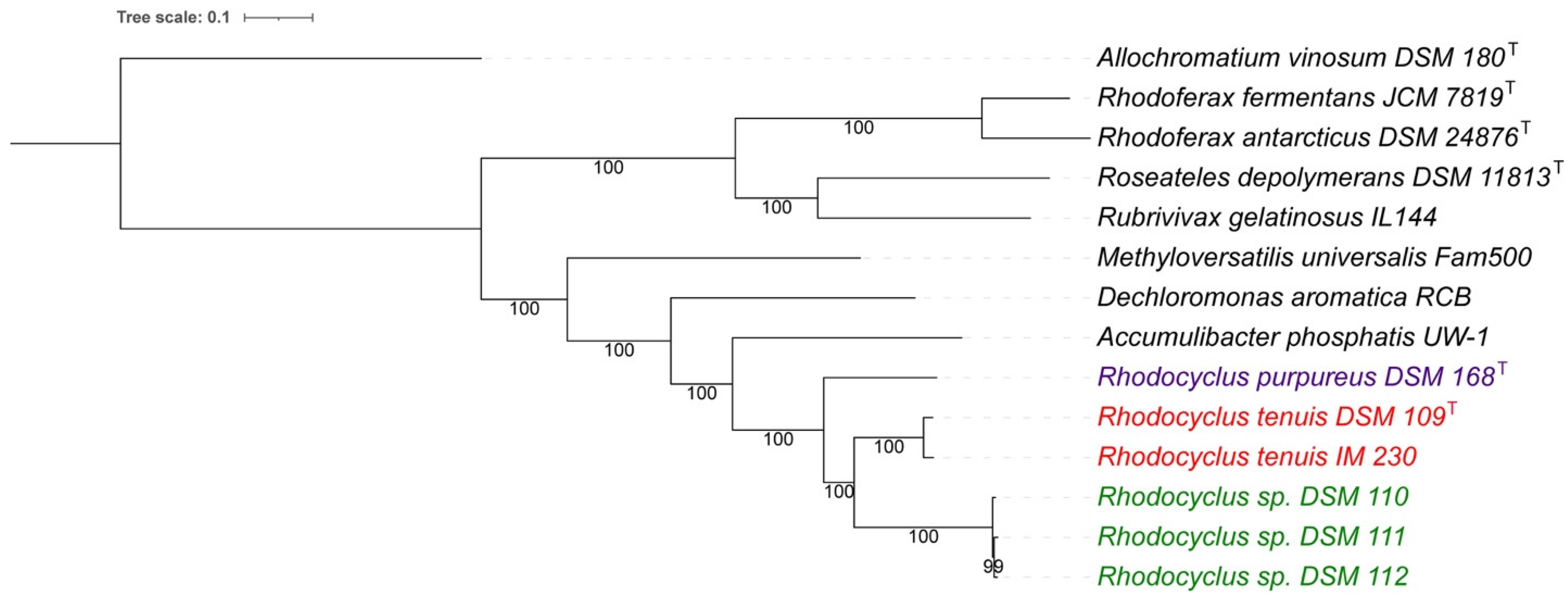
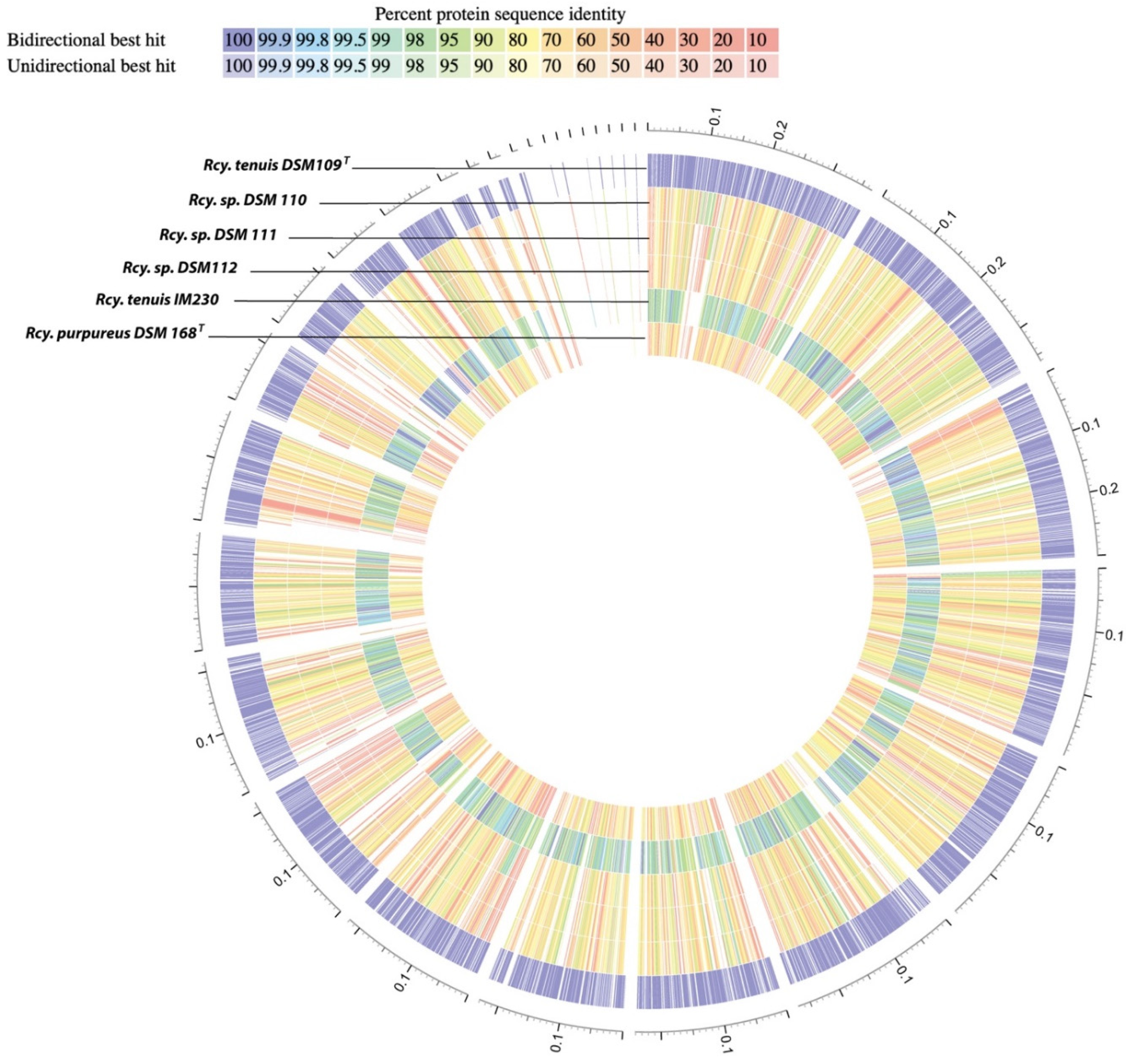
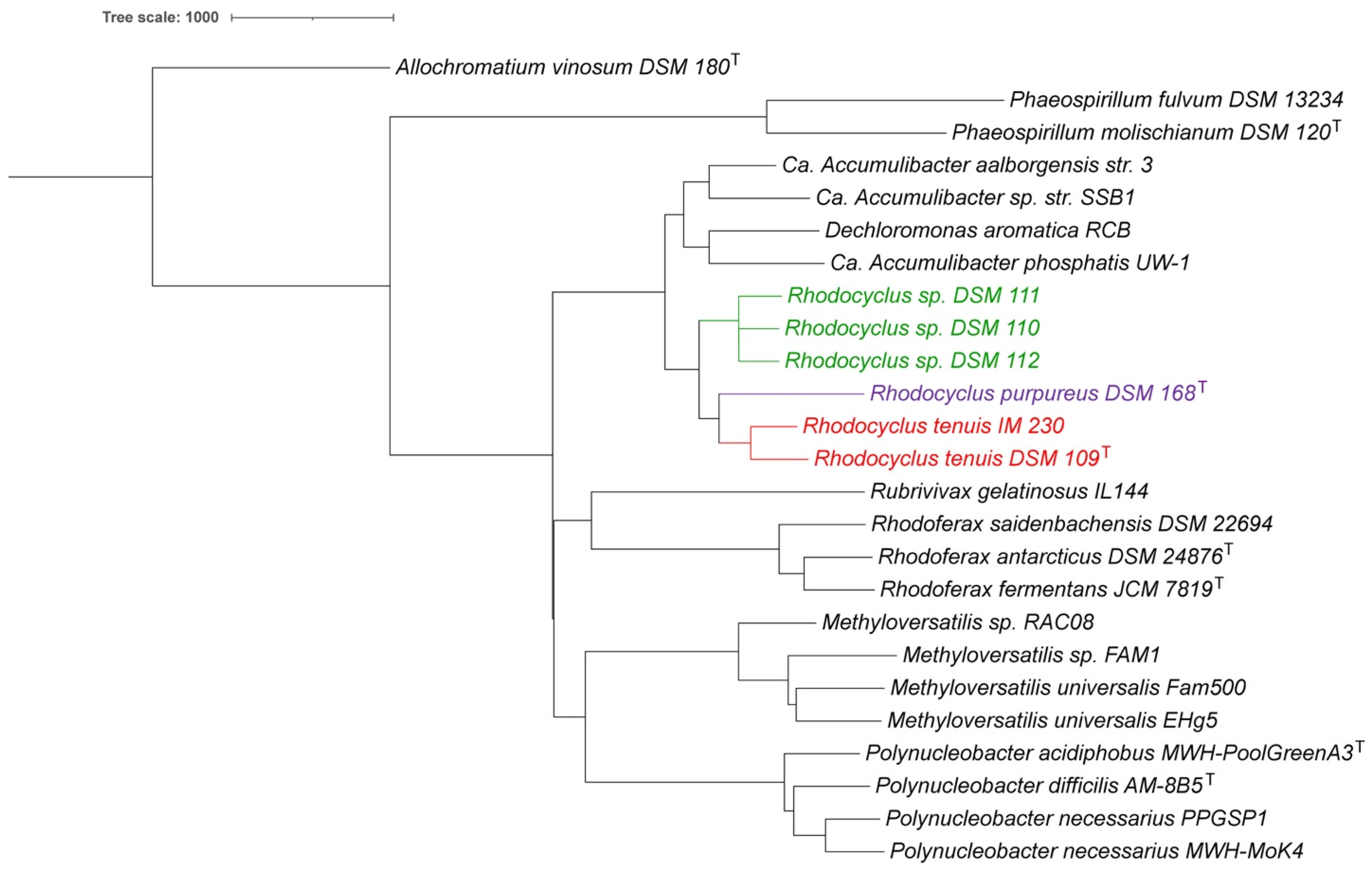
3.1. Whole Genome Analysis
3.2. Cytochrome and High Potential Iron Protein HiPIP Analysis
3.3. Nitrogen Metabolism
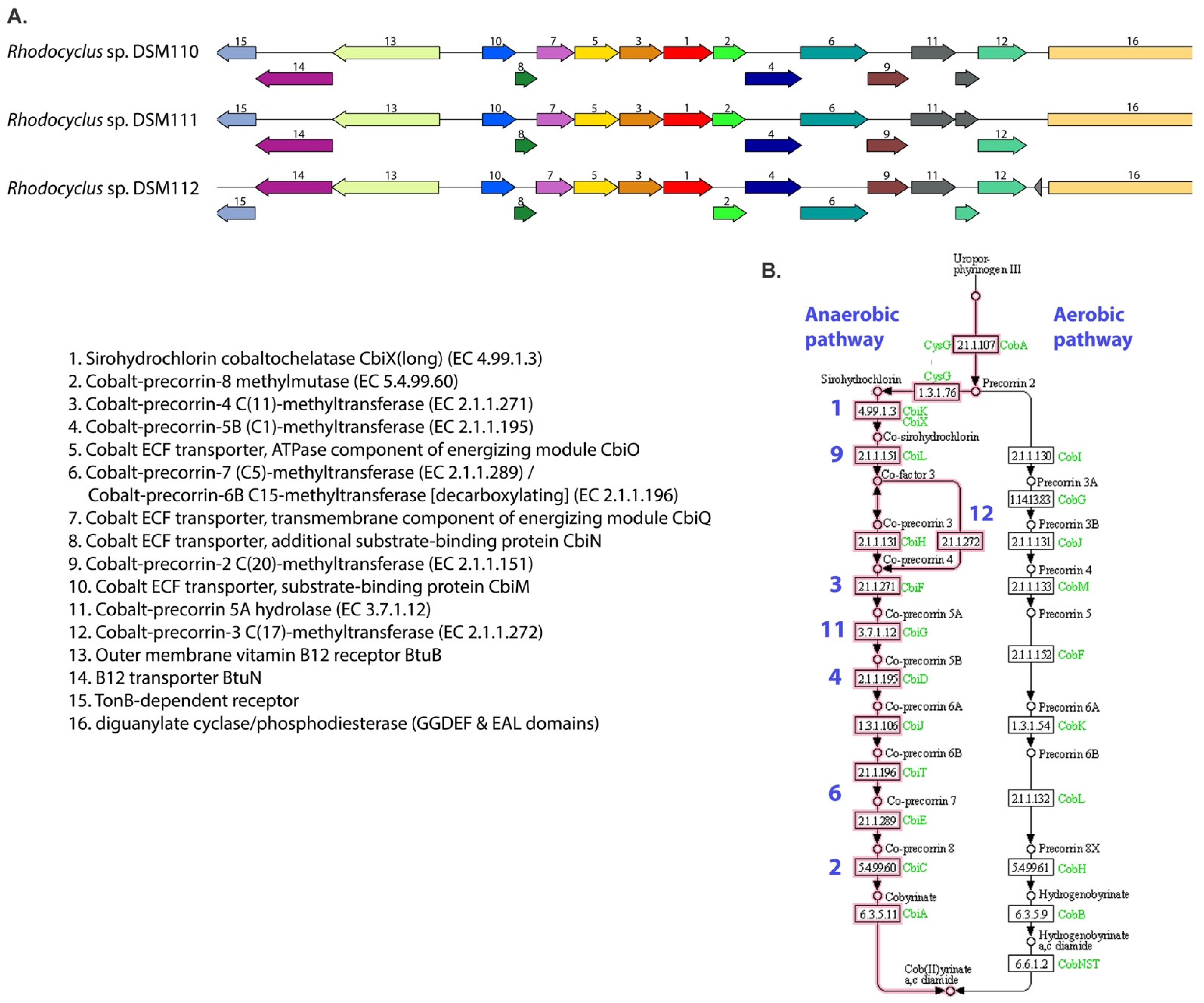
3.4. Cobalamin Metabolism
3.5. Chemotaxis and Motility
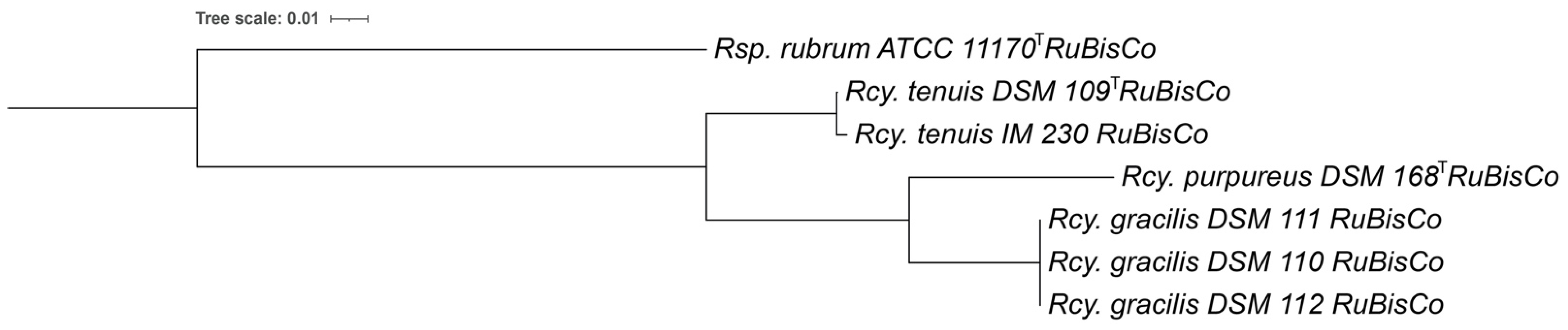
3.6. RuBisCo
3.7. Taxonomic Considerations
3.7.1. Description of Rhodocyclus gracilis sp. nov
Rhodocyclus gracilis. gra’ci.lis M.L. neut. adj. gracilis Slender
3.7.2. Emended Description of Rhodocyclus tenuis Imhoff, Trüper and Pfennig 1984, 341.VP (Rhodospirillum tenue Pfennig 1969, 619.AL)
te’nu.is. L. masc. adj. tenuis Slender, Thin
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, P.; Lang, J.; Wawrousek, K.; Yu, J.; Maness, P.-C.; Chen, J. Draft Genome Sequence of Rubrivivax gelatinosus CBS. J. Bacteriol. 2012, 194, 3262. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Nagashima, S.; Kamimura, A.; Shimizu, T.; Nakamura-Isaki, S.; Aono, E.; Sakamoto, K.; Ichikawa, N.; Nakazawa, H.; Sekine, M.; Yamazaki, S.; et al. Complete genome sequence of phototrophic betaproteobacterium Rubrivivax gelatinosus IL144. J. Bacteriol. 2012, 194, 3541–3542. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, M.; Isukapatla, A.; Mekala, L.P.; Eedara Veera Venkata, R.P.; Chintalapati, S.; Chintalapati, V.R. Genome sequence of the phototrophic betaproteobacterium Rubrivivax benzoatilyticus strain JA2T. J. Bacteriol. 2011, 193, 2898–2899. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Farh, M.E.A.; Yang, D. Genome sequencing of Rhodoferax fermentans JCM 7819. Genbank. Unpublished Work. 2017. [Google Scholar]
- Baker, J.M.; Riester, C.J.; Skinner, B.M.; Newell, A.W.; Swingley, W.D.; Madigan, M.T.; Jung, D.; Asao, M.; Chen, M.; Loughlin, P.C.; et al. Genome Sequence of Rhodoferax antarcticus ANT.BRT; A Psychrophilic Purple Nonsulfur Bacterium from an Antarctic Microbial Mat. Microorganisms 2017, 5, 8. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F.; Meyer, T.E.; Kyndt, J.A. The genome sequence of the giant phototrophic gammaproteobacterium Thiospirillum jenense gives insight into its physiological properties and phylogenetic relationships. Arch. Microbiol. 2021, 203, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Choe, H.; Kim, S.G.; Park, D.S.; Nasir, A.; Kim, B.K.; Kim, K.M. Complete genome of biodegradable plastics-decomposing Roseateles depolymerans KCTC 42856(T) (=61A(T)). J. Biotechnol. 2016, 220, 47–48. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, W.; Li, H.; Zheng, W.; Guo, F. Phylogenomics of Rhodocyclales and its distribution in wastewater treatment systems. Sci. Rep. 2020, 10, 3883. [Google Scholar] [CrossRef] [PubMed]
- Pfennig, N. Rhodocyclus purpureus gen. nov. and sp. nov., a Ring-Shaped, Vitamin B12-Requiring Member of the Family Rhodospirillaceae. Int. J. Syst. Bacteriol. 1978, 28, 283–288. [Google Scholar] [CrossRef]
- Pfennig, N. Rhodospirillum tenue sp. n., a New Species of the Purple Nonsulfur Bacteria. J. Bacteriol. 1969, 99, 619–620. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F.; Truper, H.G.; Pfennig, N. Rearrangement of the Species and Genera of the Phototrophic “Purple Nonsulfur Bacteria”. Int. J. Syst. Bacteriol. 1984, 34, 340–343. [Google Scholar] [CrossRef]
- Biebl, H. Ökologie einiger Seen in Norddeutschland: Phototrophe Nichtschwefel-Purpurbakterien. Ph.D. Thesis, University Freiburg, Freiburg im Breisgau, Germany, 1973. [Google Scholar]
- Schmidt, K. Biosynthesis of Carotenoids. In The Photosynthetic Bacteria; Clayton, R.K., Sistrom, W.R., Eds.; Plenum Press: New York, NY, USA, 1978; Chapter 39; pp. 729–750. [Google Scholar]
- Drews, G.; Weckesser, J.; Mayer, H. Cell Envelopes. In The Photosynthetic Bacteria; Clayton, R.K., Sistrom, W.R., Eds.; Plenum Press: New York, NY, USA, 1978; Chapter 4; pp. 61–77. [Google Scholar]
- Imhoff, J.F. Occurrence and evolutionary significance of two sulfate assimilation pathways in the Rhodospirillaceae. Arch. Microbiol. 1982, 132, 197–203. [Google Scholar] [CrossRef]
- Imhoff, J.F.; Bias-Imhoff, U. Lipids, Quinones and Fatty Acids of Anoxygenic Phototrophic Bacteria. In Anoxygenic Photosyn-thetic Bacteria; Blankenship, R.E., Madigan, M.T., Bauer, C.E., Eds.; Kluwer Academic Publ.: Alphen aan den Rijn, The Netherlands, 1995; pp. 179–205. [Google Scholar]
- Imhoff, J.F. Anoxygenic Phototrophic Bacteria. In Methods in Aquatic Bacteriology; Austin, B., Ed.; John Wiley and Sons Ltd.: Hoboken, NJ, USA, 1988; Chapter 9; pp. 207–240. [Google Scholar]
- Imhoff, J.F.; Rahn, T.; Künzel, S.; Neulinger, S.C. New insights into the metabolic potential of the phototrophic purple bacterium Rhodopila globiformis DSM 161T from its draft genome sequence and evidence for a vanadium-dependent nitrogenase. Arch. Microbiol. 2018, 200, 847–857. [Google Scholar] [CrossRef] [PubMed]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling Single-Cell Genomes and Mini-Metagenomes from Chimeric MDA Products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Gorrie, C.L.; Holt, K.E. Unicycler: Resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017, 13, e1005595. [Google Scholar] [CrossRef] [PubMed]
- Wattam, A.R.; Davis, J.J.; Assaf, R.; Boisvert, S.; Brettin, T.; Bun, C.; Conrad, N.; Dietrich, E.M.; Disz, T.; Gabbard, J.L.; et al. Improvements to PATRIC, the all-bacterial Bioinformatics Database and Analysis Resource Center. Nucleic Acids Res. 2017, 45, D535–D542. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Morgulis, A.; Gertz, E.M.; Schäffer, A.A.; Agarwala, R. A Fast and Symmetric DUST Implementation to Mask Low-Complexity DNA Sequences. J. Comput. Biol. 2006, 13, 1028–1040. [Google Scholar] [CrossRef] [PubMed]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar] [CrossRef] [PubMed]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 2015, 3, e1165. [Google Scholar] [CrossRef] [PubMed]
- Bushnell, B. BBMap: A Fast, Accurate, Splice-Aware Aligner. 2014. United States. Available online: https://www.osti.gov/servlets/purl/1241166 (accessed on 29 May 2017).
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.O.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A.; Hoover, P.; Rougemont, J. A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst. Biol. 2008, 57, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 2019, 10, 2182. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.A.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef]
- Skennerton, C.T.; Barr, J.; Slater, F.R.; Bond, P.; Tyson, G.W. Expanding our view of genomic diversity in Candidatus Accumulibacter clades. Environ. Microbiol. 2015, 17, 1574–1585. [Google Scholar] [CrossRef] [PubMed]
- Stackebrandt, E.; Ebers, J. Taxonomic parameters revisited: Tarnished gold standards. Microbiol. Today 2006, 33, 152–155. [Google Scholar]
- Menin, L.; Yoshida, M.; Jaquinod, M.; Nagashima, K.V.P.; Matsuura, K.; Parot, P.; Verméglio, A. Dark Aerobic Growth Conditions Induce the Synthesis of a High Midpoint Potential Cytochrome c8 in the Photosynthetic Bacterium Rubrivivax gelatinosus. Biochemistry 1999, 38, 15238–15244. [Google Scholar] [CrossRef] [PubMed]
- Hochkoeppler, A.; Zannoni, D.; Ciurli, S.; Meyer, T.E.; Cusanovich, M.A.; Tollin, G. Kinetics of photo-induced electron transfer from high-potential iron-sulfur protein to the photosynthetic reaction center of the purple phototroph Rhodoferax fermentans. Proc. Natl. Acad. Sci. USA 1996, 93, 6998–7002. [Google Scholar] [CrossRef] [PubMed]
- Menin, L.; Schoepp, B.; Parot, P.; Verméglio, A. Photoinduced Cyclic Electron Transfer in Rhodocyclus tenuis Cells: Participation of HiPIP or Cyt c8 Depending on the Ambient Redox Potential. Biochemistry 1997, 36, 12183–12188. [Google Scholar] [CrossRef] [PubMed]
- Meyer, T.E.; Kamen, M.D. Structural and Functional Diversity among Bacterial Electron Transport Protein. In Electron Transport and Oxygen Utilization; Ho, C., Ed.; Elsevier North Holland: New York, NY, USA, 1982; pp. 33–41. [Google Scholar]
- Devreese, B.; Brigé, A.; Backers, K.; Van Driessche, G.; Meyer, T.E.; Cusanovich, M.A.; Van Beeumen, J.J. Primary Structure Characterization of a Rhodocyclus tenuis Diheme Cytochrome c Reveals the Existence of Two Different Classes of Low-Potential Diheme Cytochromes c in Purple Phototropic Bacteria. Arch. Biochem. Biophys. 2000, 381, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Hartig, E.; Zumft, W.G. Kinetics of nirS expression (cytochrome cd1 nitrite reductase) in Pseudomonas stutzeri during the tran-sition from aerobic respiration to denitrification: Evidence for a denitrification-specific nitrate- and nitrite-responsive regulatory system. J. Bacteriol. 1999, 181, 161–166. [Google Scholar] [CrossRef] [PubMed]
- Masters, R.A.; Madigan, M. Nitrogen metabolism in the phototrophic bacteria Rhodocyclus purpureus and Rhodospirillum tenue. J. Bacteriol. 1983, 155, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Roth, J.R.; Lawrence, J.G.; Rubenfield, M.; Kieffer-Higgins, S.; Church, G. Characterization of the cobalamin (vitamin B12) biosynthetic genes of Salmonella typhimurium. J. Bacteriol. 1993, 175, 3303–3316. [Google Scholar] [CrossRef] [PubMed]
- Blanche, F.; Thibaut, D.; Debussche, L.; Hertle, R.; Zipfel, F.; Müller, G. Parallels and Decisive Differences in Vitamin B12 Biosyntheses. Angew. Chem. Int. Ed. 1993, 32, 1651–1653. [Google Scholar] [CrossRef]
- Raux, E.; Lanois, A.; Rambach, A.; Warren, M.; Thermes, C. Cobalamin (vitamin B12) biosynthesis: Functional characterization of the Bacillus megaterium cbi genes required to convert uroporphyrinogen III into cobyrinic acid a,c-diamide. Biochem. J. 1998, 335, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Moore, S.J.; Lawrence, A.D.; Biedendieck, R.; Deery, E.; Frank, S.; Howard, M.J.; Rigby, S.E.J.; Warren, M.J. Elucidation of the anaerobic pathway for the corrin component of cobalamin (vitamin B12). Proc. Natl. Acad. Sci. USA 2013, 110, 14906–14911. [Google Scholar] [CrossRef] [PubMed]
- Cadieux, N.; Bradbeer, C.; Reeger-Schneider, E.; Köster, W.; Mohanty, A.K.; Wiener, M.C.; Kadner, R.J. Identification of the Periplasmic Cobalamin-Binding Protein BtuF of Escherichia coli. J. Bacteriol. 2002, 184, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Badger, M.R.; Bek, E.J. Multiple Rubisco forms in proteobacteria: Their functional significance in relation to CO2 acquisition by the CBB cycle. J. Exp. Bot. 2008, 59, 1525–1541. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F. Genus Rhodocyclus. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Brenner, D.J., Krieg, N.R., Staley, J.T., Eds.; Springer: New York, NY, USA, 2005; Volume 2, Part C; pp. 887–890. [Google Scholar]

| Species | Genome Size | GC Content | Contigs | Coverage | CDS | tRNAs | Reference | Genbank Accession # |
|---|---|---|---|---|---|---|---|---|
| Rhodocyclus tenuis DSM109 | 3.85 Mb | 64.7 | 30 | 100x | 3605 | 51 | Wang et.al., 2020 | SSSP00000000 |
| Rhodocyclus tenuis IM230 | 3.65 Mb | 64.7 | 28 | 93x | 3359 | 51 | this study | NRRZ00000000 |
| Rhodocyclus sp. DSM110 | 2.93 Mb | 64.5 | 27 | 54x | 2735 | 47 | this study | WIXJ00000000 |
| Rhodocyclus sp. DSM111 | 2.93 Mb | 64.4 | 30 | 55x | 2705 | 47 | this study | WJED00000000 |
| Rhodocyclus sp. DSM112 | 2.98 Mb | 64.5 | 27 | 73x | 2768 | 47 | this study | JAATWB000000000 |
| Rhodocyclus purpureus DSM168 | 3.62 Mb | 66.1 | 69 | 81x | 3600 | 51 | this study | NHRX00000000 |
| Rhodocyclus tenuis DSM 109T | ||||||||
| 97.1 | Rhodocyclus tenuis IM 230 | |||||||
| 80.3 | 80.3 | Rhodocyclus purpureus DSM 168T | ||||||
| 79.7 | 79.4 | 79.9 | Rhodocyclus sp. DSM 111 | |||||
| 79.8 | 79.4 | 77.8 | 98.8 | Rhodocyclus sp. DSM 110 | ||||
| 79.7 | 79.5 | 77.9 | 98.9 | 98.9 | Rhodocyclus sp. DSM 112 | |||
| 73.3 | 72.9 | 73.1 | 72.5 | 72.5 | 72.5 | Ca. Accumulibacter phosphatis UW1 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kyndt, J.A.; Aviles, F.A.; Imhoff, J.F.; Künzel, S.; Neulinger, S.C.; Meyer, T.E. Comparative Genome Analysis of the Photosynthetic Betaproteobacteria of the Genus Rhodocyclus: Heterogeneity within Strains Assigned to Rhodocyclus tenuis and Description of Rhodocyclus gracilis sp. nov. as a New Species. Microorganisms 2022, 10, 649. https://doi.org/10.3390/microorganisms10030649
Kyndt JA, Aviles FA, Imhoff JF, Künzel S, Neulinger SC, Meyer TE. Comparative Genome Analysis of the Photosynthetic Betaproteobacteria of the Genus Rhodocyclus: Heterogeneity within Strains Assigned to Rhodocyclus tenuis and Description of Rhodocyclus gracilis sp. nov. as a New Species. Microorganisms. 2022; 10(3):649. https://doi.org/10.3390/microorganisms10030649
Chicago/Turabian StyleKyndt, John A., Fabiola A. Aviles, Johannes F. Imhoff, Sven Künzel, Sven C. Neulinger, and Terrance E. Meyer. 2022. "Comparative Genome Analysis of the Photosynthetic Betaproteobacteria of the Genus Rhodocyclus: Heterogeneity within Strains Assigned to Rhodocyclus tenuis and Description of Rhodocyclus gracilis sp. nov. as a New Species" Microorganisms 10, no. 3: 649. https://doi.org/10.3390/microorganisms10030649
APA StyleKyndt, J. A., Aviles, F. A., Imhoff, J. F., Künzel, S., Neulinger, S. C., & Meyer, T. E. (2022). Comparative Genome Analysis of the Photosynthetic Betaproteobacteria of the Genus Rhodocyclus: Heterogeneity within Strains Assigned to Rhodocyclus tenuis and Description of Rhodocyclus gracilis sp. nov. as a New Species. Microorganisms, 10(3), 649. https://doi.org/10.3390/microorganisms10030649









