Abstract
The Chlorophyll d-producing cyanobacterium Acaryochloris marina is widely distributed in marine environments enriched in far-red light, but our understanding of its genomic and functional diversity is limited. Here, we take an integrative approach to investigate A. marina diversity for 37 strains, which includes twelve newly isolated strains from previously unsampled locations in Europe and the Pacific Northwest of North America. A genome-wide phylogeny revealed both that closely related A. marina have migrated within geographic regions and that distantly related A. marina lineages can co-occur. The distribution of traits mapped onto the phylogeny provided evidence of a dynamic evolutionary history of gene gain and loss during A. marina diversification. Ancestral genes that were differentially retained or lost by strains include plasmid-encoded sodium-transporting ATPase and bidirectional NiFe-hydrogenase genes that may be involved in salt tolerance and redox balance under fermentative conditions, respectively. The acquisition of genes by horizontal transfer has also played an important role in the evolution of new functions, such as nitrogen fixation. Together, our results resolve examples in which genome content and ecotypic variation for nutrient metabolism and environmental tolerance have diversified during the evolutionary history of this unusual photosynthetic bacterium.
1. Introduction
The marine cyanobacterium Acaryochloris marina is unique in its use of Chlorophyll d (Chl d) as its major photosynthetic pigment [1]. Although Chl d is nearly identical in structure to the ubiquitous Chl a of other cyanobacteria, algae, and plants, this pigment absorbs strongly at far-red wavelengths that are inaccessible to most oxygenic phototrophs. Because these wavelengths attenuate rapidly in water, A. marina appears to be restricted to shallow environments enriched in filtered far-red light, and its abundance has been observed to decrease with depth [2]. Nonetheless, Chl d and its derivatives can be abundant in marine coastal ecosystems [3], and A. marina is widely distributed in temperate and tropical saline environments, often in association with red algae or animals [1,4,5,6,7,8]. These cyanobacteria have also been confirmed, through either laboratory cultivation or environmental DNA sequencing, to occur in marine stromatolites [9], a saline lake epilithic biofilm [10,11], a microbial mat from a high-elevation brackish lake [12], and the aquatic plant rhizosphere of a heavily canopied freshwater stream [13].
Despite its broad distribution, the genomic diversity of A. marina remains poorly understood. This is because, until recently [8], genome data were available for only two laboratory strains [14,15]: the type strain MBIC110017 and strain CCMEE 5410, which was isolated from the Salton Sea, California. Similarly, we are largely ignorant regarding the extent of functional variation for traits that are expected to be important for A. marina life history, including nitrogen metabolism and salinity tolerance. Improving our understanding of this variation is essential for inferring the different ways that this bacterium may contribute to ecosystem processes in far-red enriched environments. We investigated this issue for our collection of 37 A. marina laboratory strains, including twelve newly isolated strains (Table S1). The collection consists of epiphytes of red algae, strains associated with tunicates (MBIC11017, MU11, MU12, and MU13), and an epilithic strain (CCMEE 5410). We identified roles for both plasmid maintenance and loss, along with horizontal gene transfer (HGT), in the origins of metabolic diversity during A. marina evolution. We also report ecotypic variation for phenotypic traits, including salinity tolerance and nitrogen sources, particularly nitrogen fixation. Finally, we identified differences among strains with respect to the presence of an intact CRISPR-Cas system, which may be selectively maintained when foreign DNA-induced mortality is high but otherwise lost.
2. Materials and Methods
2.1. Laboratory Strain Isolation
Twelve new laboratory strains of A. marina were isolated as part of this study from the following sample collections (Table S1): red algae on intertidal rocks at Hug Point State Park, Oregon (28 June 2017); red algae on a low tide sand flat at Wreck Beach, Vancouver, British Columbia (7 August 2018); red algae on intertidal rocks at Praia de Carcavelos, Portugal (24 October 2018); red algae from dock tires at Friday Harbor Laboratories, Washington (5 January 2019); and a red alga on an intertidal rock at Queroianella, Italy (30 December 2019). Strains were isolated from these environmental samples as previously described [8] and are available upon request. Briefly, algal samples were incubated at 20 °C in screw-cap vials containing 15 mL of IOBG-11 medium under low far-red light (~1.25 μmol photons m−2 s−1 from LED diodes with 710 nm peak emission). Clonal laboratory isolates were obtained from successful enrichments through repeated streaks and transfers on IOBG-11 agar plates.
2.2. Genomics
DNA was extracted using the DNeasy PowerBiofilm DNA extraction kit (Qiagen) following manufacturer instructions. Sample libraries were prepared and sequenced on an Illumina NextSeq 550 platform (150-bp paired-end) at the University of Pittsburgh Microbial Genome Sequencing Center. Sequence reads were trimmed of trailing low-quality bases and filtered based on read length and sequence quality with Trimmomatic version 0.36 [16], and draft genome assemblies were obtained with SPAdes version 3.12.0 [17] using manually optimized parameter settings to maximize the N50. Following the removal of contaminant contigs with Bandage version 0.8.1 [18] and Kraken 2 [19], assemblies were annotated by RAST [20]. Genome completeness was estimated with CheckM version 1.0.18 [21] and BUSCO version 4.1.2 [22]. Genome sequence data for strains described in this study are available at NCBI BioProject PRJNA649288.
2.3. Phylogenetics
A genome-wide phylogeny was reconstructed for the A. marina strains A. thomasi RCC1774 (NCBI accession NZ_PQWO00000000) and Cyanothece sp. PCC 7425 (NCBI accession GCA_000022045.1). A total of 1369 single copy groups of orthologous CDS were identified by OrthoFinder v2.2.7 [23] to create a concatenated alignment of protein sequences. We constructed a maximum likelihood tree with 1000 ultrafast bootstrap replicates [24] using IQ-TREE version 2.0 [25] according to the JTT+F+R5 model of sequence evolution selected by AIC in ModelFinder [26]. Additionally, a 16S rRNA gene phylogeny was reconstructed by maximum likelihood according to the TIM3+F+R2 model. For the nif dataset, maximum likelihood trees were reconstructed for nifHDK and 16S rRNA genes with a GTR+I+G model and 1000 bootstrap replicates, and a SH test [27] was used to test for topological congruence of trees.
2.4. Genomics and Bioinformatics
To identify A. marina core genes, we performed two local BLAST [28] approaches for each genome: (1) a blastn search of each CDS (protein coding sequence) in the A. marina reference strain MBIC11017 against each target genome assembly; and (2) a tblastn search of each protein sequence in a genome against the reference strain MBIC11017 genome. For (2), hits with <50% overlap between query and target and/or <50% sequence identity were removed. We used an E-value cut-off of 0.01 for both approaches. For each strain’s genome, BLAST outputs were merged, and a nonredundant gene set was then obtained with a custom Python script. Gene sets for all genomes were then combined, and a custom Python script was used to count how many genomes possessed a given gene.
KEGG analysis was performed with eggNOG [29]. Of CDS sequences in the inferred A. marina core genome, 2642 out of 2757 met the criteria of a minimum hit E-value of 0.001, a minimum sequence identity of 40%, and minimum query and subject coverages of 20%. FeGenie was used with default settings to identify iron metabolism genes [30], and a dendrogram was generated with Pvlcust [31], which hierarchically clustered normalized gene counts by Euclidian distance. Putative siderophore synthesis gene clusters were confirmed using AntiSMASH [32]. CRISPR arrays were obtained from RAST, and CrisprFinder [33] was used to extract individual spacers, which were queried by BLAST against plasmid and virus databases with CRISPRTarget [34].
2.5. Nitrogen Fixation and Salt Tolerance Experiments
Stock cultures were grown in 125 mL flasks containing 75 mL of ASN-III medium (https://www.atcc.org, accessed on 20 July 2020), which contains 0.43 M NaCl. Flasks were incubated at 25 °C under a 12:12 h light:dark cycle with a mean light intensity of ~25 μmol photons m−2 s−1 of cool white fluorescent light. For the salt tolerance experiment, each stock of growing cells was inoculated to an optical density at 750 nm (OD750) value of 0.01 in duplicate 125 mL flasks for each experimental salinity treatment (75 mL of ASN-III medium modified to contain final NaCl concentrations of 0, 0.2, 0.43, 0.5, 0.62, 0.8, or 1.6 M). A similar approach was taken for the nitrogen fixation experiment. For the +N control treatment, stock cells were inoculated into fresh ASN-III; for the -N treatment, cells were inoculated into ASN-III-N medium (ASN-III without sodium nitrate added) after rinsing with ASN-III -N. For the -N rinse, cells were first pelleted in a 2 mL microfuge tube and the supernatant removed; then, 1 mL of ASN III -N media was added to the tube, the tube was vortexed, and the process repeated until cells had been rinsed three times. Experimental flasks were incubated at randomized positions in a growth chamber under the same environmental conditions as above. Every 48 h, the OD750 of a 2 mL homogenized culture sub-sample was measured for each flask with a Beckman Coulter DU 530 spectrophotometer (Indianapolis, IN, USA), and flasks were randomly re-positioned in the chamber. Growth was measured as the increase OD750 over time. Exponential growth rates were estimated for cultures that sustained growth for at least three population doublings. All statistical models were analyzed with JMP version 14.2 (SAS Institute Inc., Cary, NC, USA).
3. Results and Discussion
3.1. A. marina Phylogeny and Core Genome
We took a phylogenomics approach to develop a better understanding of A. marina genome variation and functional diversity. Our data set consisted of genome data for 37 A. marina laboratory strains from diverse geographic locations (Figure 1; Tables S1 and S2). Together with previously available data [8,14,15], this included genome assemblies for twelve newly isolated strains. Among these are the first A. marina strains from the Salish Sea (San Juan Island, WA, USA and Vancouver, BC, Canada), the eastern Atlantic Ocean (Carcavelos, Portugal), and the Mediterranean Sea (Queroianella, Italy). Most are draft assemblies constructed from Illumina sequence data but are virtually complete (Table S2).
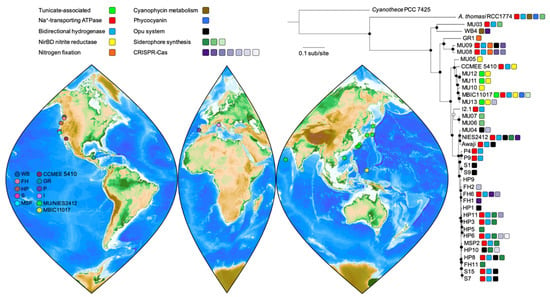
Figure 1.
Sampling locations for A. marina strains and their evolutionary relationships. The genome-wide maximum likelihood phylogeny was reconstructed for a concatenation of 1369 protein sequences from single-copy orthologs according to the JTT+F+R5 model of sequence evolution and outgroup-rooted with Cyanothece sp. PCC 7425. Branch lengths are in units of the expected number of amino acid substitutions per site. Bootstrap support values of 100% (closed circles) and >90% (open circle) are shown for 1000 ultrafast bootstrap replicates. Selected traits discussed in the text are color coded as indicated. For the map of collection sites, note that the markers for the S. China Sea and Arabian Sea strains are approximate, as the exact sampling locations are unknown.
Our phylogeny provides fresh insights on A. marina diversification (Figure 1). As we previously reported [8], A. thomasi strain RCC1774, which produces Chl b instead of Chl d, was sister to the A. marina clade. A. marina strains isolated from the west coast of North America (sites FH, HP, S, MSP) were generally very closely related but did not cluster exclusively by geographic location (Figure 1), which implies a history of migration along the coast. New strains from Portugal and Italy provided a first look at European A. marina diversity, which revealed close connections with strains from both the western and eastern Pacific. Notably, Portugal strains P4 and P9 belonged to the Pacific West Coast clade (and were identical in 16S rRNA gene sequence to these strains). By contrast, the Italy strain I2.1 was sister to strains isolated from Awaji Island, Japan. Finally, although more data will be required to better resolve A. marina global biogeography, it is already clear that distantly related lineages can co-occur (for example, MU03 and MU04 from Hokkaido, Japan; Figure 1).
Tunicate-associated strains (Figure 1) formed part of a clade isolated from Palau (MBIC11017) and Okinawa (MU11, MU12, MU13). All were derived from the surface of didemnid tunicates that have a symbiotic association with the cyanobacterium Prochloron, although members of this clade can also be algal epiphytes (e.g., MU10). A. marina DNA has also been detected from a sponge sample collected in Palau [35]. Whether animal-associated A. marina are restricted to subtropical/tropical environments is not currently known.
Our data set also informs our understanding of the A. marina core genome. We considered a gene to belong to the core if it was detected in at least 36 of the 37 genomes. 2757 protein-coding genes belonged to the core (Figure 2; Table S3; N = 2258 genes for a strict core of all 37 genomes). Few plasmid genes (N = 6) in the A. marina MBIC11017 reference genome are in this core, although they make up ~22% of its genome [14]. The majority of these were annotated as hypothetical proteins, with the exception of an RNA-binding protein and a phage integrase. This observation corroborates the earlier proposal that gene content of A. marina plasmids is highly dynamic and diverges rapidly between genomes [15]. 1502 core genes were assigned a KEGG Orthology ID, with 48 complete KEGG modules (Table S4; 35 additional modules are missing only one block).
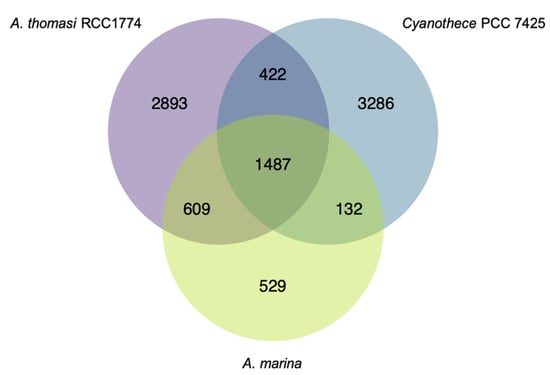
Figure 2.
Venn diagram of shared and idiosyncratic protein-coding gene content among the A. marina core genome, the Acaryochloris thomasi RCC1774 genome, and Cyanothece PCC 7425 genomes. The diagram was generated with the R package VennDiagram.
529 genes in the A. marina core were not present in the outgroup strains (Figure 2; Table S5). This set of “unique core” genes potentially includes the unidentified gene(s) responsible for Chl d synthesis. Although Chl d is produced from Chl a and requires molecular oxygen [36], the identity of this enzyme(s) remains a longstanding puzzle in Acaryochloris biology. Yoneda et al. [37] identified 23 candidates from three oxygenase families, many of which were not co-expressed with other chlorophyll biosynthesis genes. These include the Rieske oxidase (RO; [38]), antibiotic biosynthesis monooxygenase (ABM) [39], and cytochrome P450 [40] families. Our core analysis excludes several of these, which do not belong to the A. marina core: AMB genes AM1_3889 and AM1_4175 (strain MBIC11017 gene designations), RO genes AM1_0159 and AM1_A0067, and P450 genes AM1_3563, AM1_4161, and AM1_5780. In fact, only one of the 23 genes belongs to the A. marina unique core: RO gene AM1_2905, annotated as a pheophorbide a oxygenase (PAO). PAO is involved in Chl a degradation through the oxygenolytic opening of the porphyrin ring [41]; the PAO enzyme in A. marina MBIC11017 is likely AM1_0031, which is the ortholog of PAO in A. thomasi RCC1774 (61% amino acid identity). AM1_2905 appears to have been acquired by an ancient HGT event prior to the diversification of extant A. marina. Whether this PAO homolog has evolved a new function in chlorophyll metabolism remains to be tested.
The estimated total number of genes (and genome size; Table S2) varies greatly among A. marina genomes (~5800–9600) but in all cases greatly exceeds the inferred core. Below, we focus on several examples of variation in gene content among members of the A. marina radiation.
3.2. Differential Retention of Ancestral Plasmid Gene Content Contributes to A. marina Functional Variation
Several notable genes that are observed in A. thomasi RCC1774 and were likely present in the A. marina common ancestor appear to have been differentially retained or lost during A. marina diversification (Figure 1). For example, many A. marina strains possess a second set of ATP synthase (ATPase) genes first reported for strain MBIC11017 [14]. These annotated sodium-transporting ATPase (Na+-ATPase) genes are located on a conserved ~100 kbp block of plasmid sequence in the closed (or nearly closed) genomes of strains MBIC11017 (plasmid pREB4), CCMEE 5410, and S15 (Figure 3A,B). The genes are homologous to and share conserved gene order with the Na+-ATPase operon of the halotolerant cyanobacterium Aphanothece halophytica [42]. The A. halophytica Na+-ATPase is a sodium pump with increased activity at increasing NaCl concentrations [43], and heterologous expression conferred enhanced salt tolerance in the freshwater cyanobacterium Synechococcus PCC 7942 [42]. ATPase selectivity for sodium ions versus protons is controlled by the protein environment in the vicinity of the ion-binding site of the ATPase c-ring (AtpH) [44,45]. In A. marina, the amino acids of this binding site are conserved with A. halophytica [42], which suggests that these genes indeed encode a Na+-ATPase that may be involved in salt tolerance (see below).
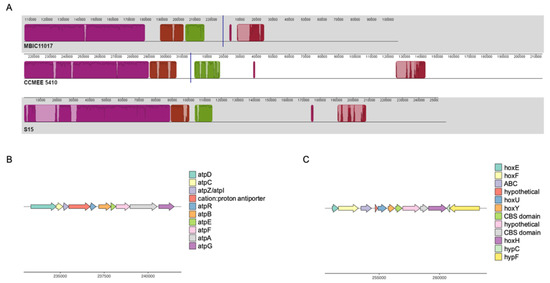
Figure 3.
(A). Multiple alignment of plasmids from A. marina strains MBIC11017, CCMEE 5410, and S15 encoding sodium-transporting ATPase and bidirectional hydrogenase genes. Homologous blocks of aligned sequence share the same color, with missing DNA transparent. Traces within blocks indicate sequence similarity. (B). Gene map of the sodium-transporting ATPase region for A. marina CCMEE 5410. (C). Gene map of the bidirectional hydrogenase region for A. marina CCMEE 5410.
The putative Na+-ATPase completely co-varies with genes encoding a bidirectional NiFe-hydrogenase (hoxEFUYH) and its associated maturation proteins that are found on the same plasmid as the atp genes (Figure 1 and Figure 3A,C). Many cyanobacteria possess a bidirectional hydrogenase, but it is most commonly observed in strains isolated from environments that periodically experience microoxic or anoxic conditions, including microbial mats, salt marshes, and intertidal habitats [46]. The physiological role of this enzyme in cyanobacteria is still debated. Activity is biased toward hydrogen production in Synechocystis PCC 6803 [47], and proposed functions include as an electron sink in fermentation [46] and/or photosynthesis [48], or as a general regulator of the cellular oxidation state [49]. In Synechocystis PCC 6803, electrons for hydrogen production are provided by reduced ferredoxin from photosystem I and pyruvate ferredoxin oxidoreductase (PFOR) activity, respectively, and a functional hydrogenase is essential for growth under anaerobic, nitrate-limiting conditions [50].
Conserved plasmid content also includes a glycogen phosphorylase (MBIC11017 nucleotide positions 164,576–167,203 in Figure 3) and PFOR genes (MBIC11017 positions 209,710–213,333). We propose that the plasmid may therefore provide a module for maintaining redox balance under fermentative conditions similar to the model of Khanna and Lindblad [51], with (1) glycogen catabolism to pyruvate, followed by (2) the oxidation of pyruvate to acetyl-CoA and reduction of ferredoxin by PFOR, and (3) hydrogen production via electron donation from ferredoxin to hydrogenase. This is in accord with the observation that hox expression in A. marina MBIC11017 is induced under anoxic, low light, and far-red light conditions [52,53]. Expression may potentially be controlled by conserved sensory and regulatory genes on the plasmid, which include a RpoD family sigma factor and a PAS domain S-box protein with histidine kinase and response regulator domains. PAS family proteins act as oxygen, redox status, or light sensors [54], and the gene includes a GAF domain with homology to FhlA (E = 4 × 10−11). FhlA plays a role in the transcription of the formate hydrogenlyase complex, which is responsible for fermentative hydrogen production in E. coli [55]. Whether there is a metabolic connection between the hydrogenase and the Na+-ATPase remains to be investigated. In addition to a possible halotolerance function, nitrogen-starved cells of Synechocystis PCC 6803 use ATPase to produce a sodium motive force that maintains sufficient levels of ATP for survival during metabolic dormancy [56]. Both sodium bioenergetics and glycogen catabolism are important during the resumption of growth in Synechocystis PCC 6803 following nitrogen starvation [56], and future work could address if hydrogenase activity also contributes.
The distribution of atp and hox genes (Figure 1) indicates a dynamic history of retention and loss of the entire plasmid that bears these genes over millions of years of A. marina diversification [11]. The persistence of plasmids is a “paradox” [57] that has long puzzled biologists. Since they often impose a fitness cost on the host (e.g., via an expression burden), plasmids are generally expected to be lost over evolutionary time through either segregational instability or by the integration of beneficial plasmid genes into the chromosome [58]. Yet, it is possible for plasmids to be stably maintained not only in selectively favored environments but also in the absence of selection [59,60]. This can occur through compensatory evolution that reduces the fitness cost to the host or by plasmid mechanisms that promote persistence, such as a partitioning system to prevent segregational loss, conjugative transfer, or an addiction module such as a toxin-antitoxin system [61]. While the conserved core of the atp/hox plasmid lacks conjugation machinery and addiction modules, it does include plasmid partition genes parAB.
The role of selection in atp/hox plasmid persistence remains to be determined, as does whether plasmid loss is neutral or favored in particular environments. However, it is clear that these ecological and evolutionary dynamics can operate at a local scale. For example, strains with or without the plasmid co-occur at Hug Point, Oregon (Figure 1), where samples were collected at low tide from exposed rock and tide pools. One avenue for future research would be to investigate whether cells that have retained the plasmid are more likely to experience periods of anoxia, salt stress, and/or metabolic dormancy, perhaps due to microenvironmental differences in the probability or duration of tidal exposure.
3.3. HGT and the Evolution of Nitrogen Metabolism
A. marina strains have the coding potential to assimilate a variety of nitrogen (N) sources, including ammonium, nitrate, nitrite, urea, and cyanate. In addition, some A. marina genomes contain idiosyncratic N metabolism genes beyond these core capacities. For example, in addition to an assimilatory ferredoxin nitrite-reductase, one A. marina clade possesses transposase-flanked nirBD genes for nitrite reductase (Figure 1) that were likely acquired by HGT from another cyanobacterium based on sequence similarity. In enterobacteria, NirBD is a dissimilatory enzyme that detoxifies nitrite produced by respiratory nitrate reduction under anoxic conditions [62]. Its role in A. marina is not clear; however, in strain CCMEE 5410, expression of these genes is similar to assimilatory ferredoxin nitrite-reductase in N deficient versus N replete conditions [37], which suggests that it may be an alternative assimilatory nitrite reductase.
N fixation of atmospheric N2 to ammonium is an important source of biologically available N. The ability to fix N has been previously reported for one strain of A. marina (HICR111A) [63]. Like many unicellular, N-fixing cyanobacteria, strain HICR111A fixes N during the dark period [63], when nitrogenase-inhibiting oxygen is not produced by photosynthesis. N fixation genes are absent from the genomes of strains MBIC11017 and CCMEE 5410, and N fixation ability in HICR111A has been proposed to have been acquired by HGT [63]. However, the evolutionary history of this trait, including whether the common ancestor of A. marina possessed nif genes, has not been fully resolved due to limited taxon sampling.
Our new phylogeny provides fresh insights into this history. We have identified a clade of epiphytic A. marina near the base of the phylogeny that also have N fixation genes (Figure 1). Members of this clade can grow with N2 as the sole N source at the same rate as on nitrate (Figure 4A). Although genome data are not publicly available for HICR111A, we believe that this strain likely belongs to this clade as well, based on a 16S rRNA gene tree (Figure S1), the nif gene sequence similarities of these strains, and the conservation of genes flanking the nif region among genomes [63].
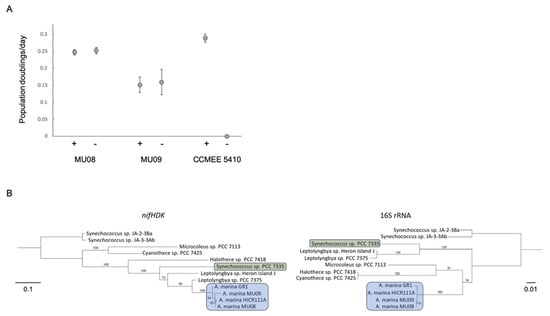
Figure 4.
(A). Population growth rate in the presence (+) or absence (−) of combined nitrogen for nif-containing strains MU08 and MU09 and for nif-lacking strain CCMEE 5410. Error bars are standard errors. (B). Maximum likelihood phylogenies for nifHDK and 16S rRNA genes reconstructed with a GTR+I+G model. Bootstrap support values greater than 50% are shown for 1000 bootstrap replicates. Branch lengths are in units of expected number of nucleotide substitutions per site. The two topologies were significantly different by an SH test (p < 0.0001).
There are two alternative hypotheses for how these strains obtained nif genes. The first is that they were vertically inherited from the A. marina common ancestor, with subsequent gene loss in other descendant strains. If this were the case, then we predict that the nif phylogeny will resemble the species tree inferred with 16S rRNA gene data. The second is that the most recent common ancestor of this clade acquired the nif genes more recently by HGT, in which case the nif and species trees will differ in topology. To test these hypotheses, we inferred maximum likelihood trees for both nifHDK (encoding the nitrogenase complex proteins) and 16S rRNA genes. In the species tree, the sister group of A. marina included Cyanothece PCC 7425 (Figure 4B), as has been observed in genome-wide phylogenies (Figure 1) [8,64]. By contrast, the most closely related sequences of A. marina nifHDK genes were from Leptolyngbya PCC 7375, with Cyanothece PCC 7425 genes more distantly related (Figure 4B). The topologies of the two trees were significantly different from each other, as determined by an SH test (−Δln L = 310.4, p < 0.0001) [27]. This indicates that the A. marina common ancestor did not possess nif genes; rather, these genes were obtained later by the ancestor of the GR1/MU08/MU09 clade (Figure 1) via HGT from another cyanobacterium.
Finally, N storage pools vary among strains. The genes required to produce and degrade cyanophycin, the primary N storage compound of many cyanobacteria [65], is absent in all A. marina strains except strain WB4 (Figure 1). The amino acid sequences of cyanophycin synthetase and cyanophycinase in WB4 have high similarity (83% and 79%) with A. thomasi RCC1774, indicating that these genes were likely present in the A. marina common ancestor but lost by other strains. Phycobilisomes are the primary light-harvesting complexes of most cyanobacteria and can account for approximately 50% of cell soluble protein [66,67]; however, they are quickly degraded during N limitation to provide N for other functions [68]. We have previously reported that the A. marina common ancestor had lost the genes for phycobiliprotein production and degradation but were uniquely reacquired by strain MBIC11017 [8]. Even with the addition here of twelve new A. marina genomes, MBIC11017 is still the only known strain to produce phycobiliproteins (Figure 1).
3.4. Ecotypic Variation in Salt Tolerance
All of the A. marina strains in our collection were isolated from different zones along tidal gradients, from the upper intertidal to brackish or saline subtidal environments (Table S1). Consequently, they are expected to have historically experienced different frequencies and durations of stress associated with tidal activity, including the degree of exposure to both hyper- and hyposaline conditions. The salt tolerance range of marine algae (on which A. marina often grows as epiphytes) is generally broader for upper intertidal taxa than for subtidal species [69,70]. We therefore predicted that A. marina strains isolated from the upper intertidal zone would exhibit broader salinity tolerance than other strains.
To test this, we assayed growth at a range of NaCl concentrations for six A. marina strains. Strains S1 and S15 were isolated from upper intertidal and subtidal habitat, respectively, along a tidal gradient on the northern California coast (Shelter Cove; Table S1).
Like S1, HP9 was isolated from the upper intertidal zone (Hug Point, OR). Strain FH11, which is very closely related to the above strains (Figure 1), was isolated from a submerged estuarine environment (San Juan Island, WA) that has a salinity that is approximately 85% of that of seawater (30 ppt; https://www.bco-dmo.org/dataset/775732, accessed on 26 May 2021). Strains MBIC11017 and CCMEE 5410 are from submerged habitats in the tropical western Pacific [1] and the Salton Sea, CA (34 ppt salinity at time of collection; [11]), respectively.
As predicted, upper intertidal strains S1 and HP9 exhibited the broadest range of NaCl concentration that could support sustained growth (0.2–0.8 M; Figure 5; all six strains bleached at 0 M and 1.6 M NaCl). S1 and subtidal strain S15 did not differ significantly in growth rate at 0.2 M and 0.43 M NaCl; however, S1 grew faster than S15 at higher NaCl concentrations (FDR-adjusted p = 0.08 at 0.5 M, p < 0.01 at 0.62 M, and only S1 could grow at 0.8 M). The lower salt tolerance of S15 was expected given its subtidal origin. Estuarine strain FH11 was even less halotolerant, with optimal growth observed at lower NaCl concentrations (p = 0.38 for the comparison between 0.2 M and 0.43 M) and particularly low growth rates at NaCl concentrations greater than 0.43 M (including no sustained growth at 0.8 M). Other strains from less variable salinity environments grew well at intermediate NaCl concentrations but did not grow at either the lowest salinity (MBIC11017) or highest salinity (CCMEE 5410) treatments. We conclude that A. marina halotolerance generally matches local environmental conditions.
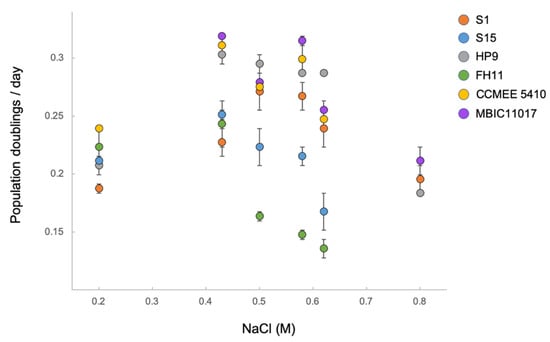
Figure 5.
Population growth rates at different NaCl concentrations for upper intertidal A. marina strains S1 and HP9, subtidal strains S15 and MBIC11017, estuarine strain FH11, and saline lake strain CCMEE 5410. Error bars are standard errors. Note that 0.43 M is the NaCl concentration of standard marine ASN-III medium. Rates were estimated if growth was sustained for at least three generations. Pigments of strains for which growth was not observed at 0.2 M or 0.8 M NaCl did not bleach, whereas all strains bleached at 0 M and 1.6 M.
In cyanobacteria, halotolerance involves both the export of sodium ions and the production of compatible solutes to maintain osmotic balance under conditions of high salinity; however, other key aspects of tolerance, including how cells recognize salt stress signals and regulate salt acclimation, is not well understood [71]. Understanding the observed differences in halotolerance among A. marina strains is therefore not possible from a consideration of gene content alone. Nonetheless, it is instructive to briefly discuss how differences in gene content may contribute to these phenotypic differences.
A. marina strains vary in terms of the types and number of systems for removing sodium ions. For example, strain S1 has three CPA1 family Na+/H+ antiporter genes, compared with one each in the close relatives S15, FH11, and HP9. Most strains also have one copy of the mrpCDEFGAB gene cluster, a multi-subunit CPA3 family Na+/H+ antiporter. However, the mrp cluster is absent in the FH1 genome, whereas strains S15, S7, HP8, and HP11 have two copies. Recent duplicates in A. marina are often found on different genetic elements [15], and we confirmed that duplicated Mrp genes are on a plasmid (contig 10, nucleotide positions 263,524–267,402) for strain S15. The homologous A. halophytica cluster complements a salt-sensitive mutant of E. coli [72]; in Anabaena PCC 7120, mrpA is upregulated in response to increasing NaCl, and its inactivation by transposon mutagenesis results in a salt-sensitive phenotype [73]. Finally, as discussed above, many strains have plasmid-encoded genes for a sodium-transporting ATPase (Figure 1).
Strains also exhibit variation in genes involved in the production or import of compatible solutes. All possess ggpP and ggpS genes required to synthesize glucosylglycerol, a common compatible solute of marine cyanobacteria [71]. In addition, many strains also encode an ABC transporter in the osmoprotectant uptake (Opu) family of compatible solute transporters [74] with unknown substrate specificity (Figure 1).
3.5. Iron Metabolism
Iron is an important nutrient for most organisms but is often limiting in oxic environments, where it is insoluble [75]. Consequently, bacteria have evolved mechanisms for scavenging and storing iron when it is rare. This includes the ability to synthesize and/or import siderophores, low molecular weight compounds that chelate iron with high affinity [76]. The ability to acquire iron may vary among organisms depending on iron availability in the environment. We used FeGenie [30] to identify genes related to iron metabolism in A. marina genomes. Strains clustered into two groups based on iron gene content that do not closely reflect the A. marina phylogeny (Figure 6 and Figure S2). Strains with more genes related to iron metabolism tend to be enriched in genes involved in the regulation of iron assimilation, siderophore transport, and, often, siderophore synthesis (Figure 6; Table S6).
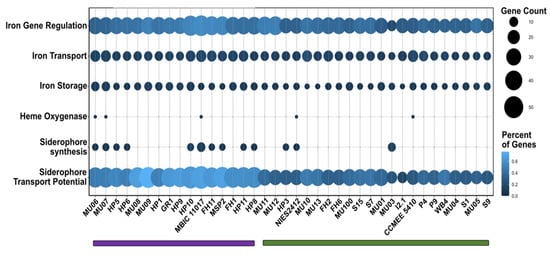
Figure 6.
Distribution of genes involved in the regulation, acquisition, and storage of iron among A. marina strains based on both gene count and percent of genes in the genome. A dendrogram clustered strains into two groups based on high (purple) and low (green) total iron gene content. The approximately unbiased p-value of each cluster was 99%. See Figure S2 for the hierarchical clustering dendrogram.
All strains possess annotated siderophore import genes (though they vary in number). By contrast, genes involved in the synthesis of siderophores are scattered throughout the phylogeny and appear to have been independently acquired at least three times during A. marina diversification (Figure 1). In addition to the MBIC11017 cluster, the most common siderophore gene cluster was observed in strains isolated from the U.S. West Coast (FH, HP, and MSP sites) and in the Japanese strain NIES2412. The closest matches in the antiSMASH database of microbial secondary metabolites [32] were paenibactin and myxochelin. Homologs of these genes were detected by BLAST in an assembly of the cyanobacterium Leptolyngbya from metagenome data obtained for a South African peritidal stromatolite (NCBI BioProject PRJNA612530, assembly SM1_1_3) [77]. It is not clear whether this cluster was inherited from a common ancestor or independently acquired. A third cluster in strains MU06, MU07, and MU03 exhibited the closest similarity to cupriachelin in the antiSMASH database and exhibit homology to genes in A. thomasi RCC1774 (48–68% amino acid identity between the latter and MU06).
Enhanced iron gene content may be selectively favored in low iron environments. For example, the genome of strain MBIC11017, which was isolated from an iron-poor region of the Pacific (1), contains both plasmid-encoded iron transport gene duplicates and a transposase-flanked cluster of genes [78] that is homologous to characterized siderophore synthesis genes in the cyanobacterium Anabaena PCC 7120 [79]. These genes are associated with higher rates of iron acquisition and growth under conditions of low iron availability compared with strain CCMEE 5410 [78], which is a member of the “low iron” gene content cluster (Figure 6).
3.6. Complex History of CRISPR-Cas Systems
A. marina genomes also vary with respect to the presence of CRISPR-Cas systems (Figure 1), all of which are either type I or type III class 1 systems (Table 1; Figure 7). The number of CRISPR arrays varied among these genomes (1–8 arrays), as did the total number of spacers (17–140 spacers; Table S7). The majority of spacers were derived from uncharacterized viral sequences, although plasmid sequences were also detected (Table S8).

Table 1.
CRISPR-Cas systems detected in A. marina genomes.
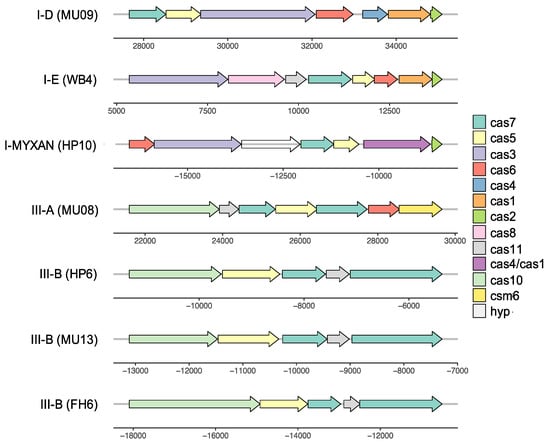
Figure 7.
Representative gene maps for CRISPR-Cas systems detected in A. marina genomes (strain is indicated in parentheses).
The scattered distribution of CRISPR-Cas among extant A. marina strains (Figure 1) suggests a complicated history of gains, retentions, and losses. Two of the systems (the I-E and one of the III-B systems) are observed in strains from geographically distant locations (both cases involve western North America and Japan) among distantly related strains (Figure 1). Both exhibit a highest sequence identity with distantly related cyanobacteria (Romeria gracilis (I-E) and Aphanocapsa montana (III-B), respectively) and therefore clearly have been acquired by HGT.
There is both theoretical and empirical support for the idea that organisms with functional CRISPR-Cas systems obtain protection from phages and plasmids at the cost of a reduction in the ability to acquire potentially beneficial genes by HGT [80,81]. We speculate that such a trade-off may have played out during A. marina diversification. An intact CRISPR-Cas may be selectively favored under conditions of high foreign DNA-induced mortality but otherwise lost; its absence may then result in a greater influx of new genes by HGT.
4. Conclusions
Laboratory strain collections and genomic resources are essential for developing an understanding of the genetic and phenotypic variation of recently discovered microorganisms. Our study illustrates several ways by which genome content and ecotypic variation for nutrient metabolism and environmental tolerance have evolved during A. marina diversification. It further identifies phenotypic variation of interest for future comparative physiology and omics investigations.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10030569/s1, Figure S1: 16S rRNA gene phylogeny showing HICR111A as a member of the same clade as other nitrogen-fixing strains (GR1, MU08, MU09)., Figure S2: Dendrogram of A. marina iron gene content, hierarchically clustered by Euclidean distance. Table S1: Sampling information for A. marina strain collection, Table S2: Genome assembly statistics, Table S3: A. marina core genome, Table S4: Complete KEGG modules in the Acaryocloris marina core genome, Table S5: Genes unique to the A. marina core, Table S6: Summary of FeGenie results, Table S7: A. marina CRISPR arrays, repeat sequences and spacer numbers, Table S8: CRISPRTarget output for A. marina strain FH6.
Author Contributions
S.R.M. conceived the study; H.E.A., J.J.B., A.R.O. and N.J.U. performed experiments and obtained genome data; all authors contributed to data analysis and the writing of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by award NNA15BB04A from the National Aeronautics and Space Administration to S.R.M.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Genome data generated in this study can be found at NCBI BioProject PRJNA649288.
Acknowledgments
We thank the two anonymous reviewers for their helpful comments.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Miyashita, H.; Ikemoto, H.; Kurano, N.; Adachi, K.; Chihara, M.; Miyachi, S. Chlorophyll d as a major pigment. Nature 1996, 383, 402. [Google Scholar] [CrossRef]
- Behrendt, L.; Larkum, A.W.D.; Trampe, E.; Norman, A.; Sørensen, S.J.; Kühl, M. Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. ISME J. 2012, 6, 1222–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kashiyama, Y.; Miyashita, H.; Ohkubo, S.; Ogawa, N.O.; Chikaraishi, Y.; Takano, Y.; Suga, H.; Toyofuku, T.; Nomaki, H.; Kitazato, H.; et al. Evidence of global chlorophyll d. Science 2008, 321, 658. [Google Scholar] [CrossRef] [PubMed]
- Murakami, A.; Miyashita, H.; Iseki, M.; Adachi, K.; Mimuro, M. Chlorophyll d in an epiphytic cyanobacterium of red algae. Science 2004, 303, 1633. [Google Scholar] [CrossRef] [PubMed]
- Mohr, R.; Voß, B.; Schliep, M.; Kurz, T.; Maldener, I.; Adams, D.G.; Larkum, A.D.W.; Chen, M.; Hess, W.R. A new chlorophyll d-containing cyanobacterium: Evidence for niche adaptation in the genus Acaryochloris. ISME J. 2010, 4, 1456–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Larkum, A.W.; Chen, M.; Li, Y.; Schliep, M.; Trampe, E.; West, J.; Salih, A.; Kühl, M. A novel epiphytic Chlorophyll d-containing cyanobacterium isolated from a mangrove-associated red alga. J. Phycol. 2012, 48, 1320–1327. [Google Scholar] [CrossRef]
- Behrendt, L.; Nielsen, J.L.; Sørensen, S.J.; Larkum, A.W.D.; Winther, J.R.; Kühl, M. Rapid TaqMan-based quantification of Chlorophyll d-containing cyanobacteria in the genus Acaryochloris. Appl. Environ. Microbiol. 2014, 80, 3244–3249. [Google Scholar] [CrossRef] [Green Version]
- Ulrich, N.J.; Uchida, H.; Kanesaki, Y.; Hirose, E.; Murakami, A.; Miller, S.R. Reacquisition of light-harvesting genes in a marine cyanobacterium confers a broader solar niche. Curr. Biol. 2021, 31, 1539–1546. [Google Scholar] [CrossRef]
- Goh, F.; Allen, M.A.; Leuko, S.; Kawaguchi, T.; Decho, A.W.; Burns, B.P.; Neilan, B. Determining the specific microbial populations and their spatial distribution within the stromatolite ecosystem of Shark Bay. ISME J. 2009, 3, 383–396. [Google Scholar] [CrossRef] [Green Version]
- Wood, A.M.; Miller, S.R.; Li, W.K.W.; Castenholz, R.W. Preliminary studies of cyanobacteria, picoplankton, and virioplankton in the Salton Sea with special attention to phylogenetic diversity among eight strains of filamentous cyanobacteria. Hydrobiologia 2002, 473, 77–92. [Google Scholar] [CrossRef]
- Miller, S.R.; Augustine, S.; Le Olson, T.; Blankenship, R.E.; Selker, J.; Wood, A.M. Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proc. Natl. Acad. Sci. USA 2005, 102, 850–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleming, E.D.; Prufert-Bebout, L. Characterization of cyanobacterial communities from high-elevation lakes in the Bolivian Andes. J. Geophys. Res. 2010, 115. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.-C.; Li, Z.-K.; Yin, Y.-C.; Li, Y.; Jia, Y.; Chen, M.; Qiu, B.-S. Widespread occurrence and unexpected diversity of red-shifted chlorophyll producing cyanobacteria in humid subtropical forest ecosystems. Environ. Microbiol. 2019, 21, 1497–1510. [Google Scholar] [CrossRef] [PubMed]
- Swingley, W.D.; Chen, M.; Cheung, P.C.; Conrad, A.L.; Dejesa, L.C.; Hao, J.; Honchak, B.M.; Karbach, L.E.; Kurdoglu, A.; Lahiri, S.; et al. Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc. Natl. Acad. Sci. USA 2008, 105, 2005–2010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, S.R.; Wood, A.M.; Blankenship, R.E.; Kim, M.; Ferriera, S. Dynamics of gene duplication in the genomes of chlorophyll d-producing cyanobacteria: Implications for the ecological niche. Genome Biol. Evol. 2011, 3, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comp. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Wick, R.R.; Schultz, M.B.; Zobel, J.; Holt, K.E. Bandage: Interactive visualization of de novo genome assemblies. Bioinformatics 2015, 31, 3350–3352. [Google Scholar] [CrossRef] [Green Version]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef] [Green Version]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [Green Version]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019, 20, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [Green Version]
- Shimodaira, H.; Hasegawa, M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999, 16, 1114–1116. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.V.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. eggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2018, 47, D309–D314. [Google Scholar] [CrossRef] [Green Version]
- Garber, A.I.; Nealson, K.H.; Okamoto, A.; McAllister, S.M.; Chan, C.S.; Barco, R.A.; Merino, N. FeGenie: A comprehensive tool for the identification of iron genes and iron gene neighborhoods in genome and metagenome assemblies. Front Microbiol. 2020, 11, 37. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, R.; Shimodaira, H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 2006, 22, 1540–1542. [Google Scholar] [CrossRef] [PubMed]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grissa, I.; Vergnaud, G.; Pourcel, C. CRISPRFinder: A web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 2007, 35, W52–W57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biswas, A.; Gagnon, J.N.; Brouns, S.J.J.; Fineran, P.C.; Brown, C.M. CRISPRTarget: Bioinformatic prediction and analysis of crRNA targets. RNA Biol. 2013, 10, 817–827. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohkubo, S.; Miyashita, H. Selective detection and phylogenetic diversity of Acaryochloris spp. that exist in association with didemnid ascidians and sponge. Microbes Environ. 2012, 27, 217–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schliep, M.; Crossett, B.; Willows, R.D.; Chen, M. 18O labeling of chlorophyll d in Acaryochloris marina reveals that chlorophyll a and molecular oxygen are precursors. J. Biol. Chem. 2010, 285, 28450–28456. [Google Scholar] [CrossRef] [Green Version]
- Yoneda, A.; Wittmann, B.J.; King, J.D.; Blankenship, R.E.; Dantas, G. Transcriptomic analysis illuminates genes involved in chlorophyll synthesis after nitrogen starvation in Acaryochloris sp. CCMEE 5410. Photosynth. Res. 2016, 129, 171–182. [Google Scholar] [CrossRef]
- Barry, S.M.; Challis, G.L. Mechanism and catalytic diversity of Rieske non-heme iron-dependent oxygenases. ACS Catal. 2013, 3, 2362–2370. [Google Scholar] [CrossRef] [Green Version]
- Sciara, G.; Kendrew, S.G.; Miele, A.E.; Marsh, N.G.; Federici, L.; Malatesta, F. The structure of ActVA-Orf6, a novel type of monooxygenase involved in actinorhodin biosynthesis. EMBO J. 2003, 22, 205–215. [Google Scholar] [CrossRef] [Green Version]
- Denisov, I.G.; Makris, T.M.; Sligar, S.G.; Schlichting, I. Structure and chemistry of cytochrome P450. Chem. Rev. 2005, 105, 2253–2277. [Google Scholar] [CrossRef]
- Pružinská, A.; Tanner, G.; Anders, I.; Roca, M.; Hörtensteiner, S. Chlorophyll breakdown: Pheophorbide a oxygenase is a Rieske-type iron–sulfur protein, encoded by the accelerated cell death 1 gene. Proc. Natl. Acad. Sci. USA 2003, 100, 15259–15264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soontharapirakkul, K.; Promden, W.; Yamada, N.; Kageyama, H.; Incharoensakdi, A.; Iwamoto-Kihara, A.; Takabe, T. Halotolerant cyanobacterium Aphanothece halophytica contains an Na+-dependent F1F0-ATP synthase with a potential role in salt-stress tolerance. J. Biol. Chem. 2011, 286, 10169–10176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soontharapirakkul, K.; Incharoensakdi, A. Na+-stimulated ATPase of alkaliphilic halotolerant cyanobacterium Aphanothece halophytica translocates Na+ into proteoliposomes via Na+ uniport mechanism. BMC Biochem. 2010, 11, 30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulz, S.; Iglesias-Cans, M.; Krah, A.; Yildiz, Ö.; Leone, V.; Matthies, D.; Cook, G.M.; Faraldo-Gómez, J.D.; Meier, T. A New Type of Na+-driven ATP synthase membrane rotor with a two-carboxylate ion-coupling motif. PLoS Biol. 2013, 11, e1001596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leone, V.; Pogoryelov, D.; Meier, T.; Faraldo-Gómez, J.D. On the principle of ion selectivity in Na⁺/H⁺-coupled membrane proteins: Experimental and theoretical studies of an ATP synthase rotor. Proc. Natl. Acad. Sci. USA 2015, 112, E1057–E1066. [Google Scholar] [CrossRef] [Green Version]
- Barz, M.; Beimgraben, C.; Staller, T.; Germer, F.; Opitz, F.; Marquardt, C.; Schwarz, C.; Gutekunst, K.; Vanselow, K.H.; Schmitz, R.; et al. Distribution analysis of hydrogenases in surface waters of marine and freshwater environments. PLoS ONE 2010, 5, e13846. [Google Scholar] [CrossRef]
- McIntosh, C.L.; Germer, F.; Schulz, R.; Appel, J.; Jones, A.K. The [NiFe]-hydrogenase of the cyanobacterium Synechocystis sp. PCC 6803 works bidirectionally with a bias to H2 production. J. Am. Chem. Soc. 2011, 133, 11308–11319. [Google Scholar] [CrossRef]
- Appel, J.; Phunpruch, S.; Steinmüller, K.; Schulz, R. The bidirectional hydrogenase of Synechocystis sp. PCC 6803 works as an electron valve during photosynthesis. Arch. Microbiol. 2000, 173, 333–338. [Google Scholar] [CrossRef]
- Carrieri, D.; Wawrousek, K.; Eckert, C.; Yu, J.; Maness, P.-C. The role of the bidirectional hydrogenase in cyanobacteria. Bioresour. Tech. 2011, 102, 8368–8377. [Google Scholar] [CrossRef]
- Gutekunst, K. The Bidirectional NiFe-hydrogenase in Synechocystis sp. PCC 6803 is reduced by flavodoxin and ferredoxin and is essential under mixotrophic, nitrate-limiting conditions. J. Biol. Chem. 2014, 289, 1930–1937. [Google Scholar] [CrossRef] [Green Version]
- Khanna, N.; Lindblad, P. Cyanobacterial hydrogenases and hydrogen metabolism revisited: Recent progress and future prospects. Int. J. Mol. Sci. 2015, 16, 10537–10561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiss, É.; Kós, P.B.; Chen, M.; Vass, I. Functioning of the bidirectional hydrogenase in different unicellular cyanobacteria. In Photosynthesis Research for Food, Fuel and the Future; Kuang, T., Lu, C., Zhang, L., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 733–736. [Google Scholar]
- Loughlin, P.; Lin, Y.; Chen, M. Chlorophyll d and Acaryochloris marina: Current status. Photosynth. Res. 2013, 116, 277–293. [Google Scholar] [CrossRef] [PubMed]
- Taylor, B.L.; Zhulin, I.B. PAS domains: Internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 1999, 63, 479–506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sankar, P.; Lee, J.H.; Shanmugam, K.T. Gene-product relationships of fhlA and fdv genes of Escherichia coli. J. Bacteriol. 1988, 170, 5440–5445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doello, S.; Burkhardt, M.; Forchhammer, K. The essential role of sodium bioenergetics and ATP homeostasis in the developmental transitions of a cyanobacterium. Curr. Biol. 2021, 31, 1606–1615. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.; Brockhurst, M.A. Plasmid-mediated horizontal gene transfer is a coevolutionary process. Trends Microbiol. 2012, 20, 262–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San Millan, A.; MacLean, R.C. Fitness costs of plasmids: A limit to plasmid transmission. Microbiol. Spectr. 2017, 5, 65–79. [Google Scholar] [CrossRef] [Green Version]
- Harrison, E.; Guymer, D.; Spiers, A.J.; Paterson, S.; Brockhurst, M.A. Parallel compensatory evolution stabilizes plasmids across the parasitism-mutualism continuum. Curr. Biol. 2015, 25, 2034–2039. [Google Scholar] [CrossRef] [Green Version]
- Wein, T.; Hülter, N.F.; Mizrahi, I.; Dagan, T. Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nature Comm. 2019, 10, 2595. [Google Scholar] [CrossRef] [Green Version]
- Carroll, A.C.; Wong, A. Plasmid persistence: Costs, benefits, and the plasmid paradox. Can. J. Microbiol. 2018, 64, 293–304. [Google Scholar] [CrossRef]
- Stewart, V. Regulation of nitrate and nitrite reductase synthesis in enterobacteria. Antonie Leeuwenhoek 1994, 66, 37–45. [Google Scholar] [CrossRef] [PubMed]
- Pfreundt, U.; Stal, L.J.; Voß, B.; Hess, W.R. Dinitrogen fixation in a unicellular chlorophyll d-containing cyanobacterium. ISME J. 2012, 6, 1367–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, P.M.; Wu, D.; Latifi, A.; Axen, S.; Fewer, D.; Talla, E.; Calteau, A.; Cai, F.; de Marsac, N.T.; Rippka, R.; et al. Improving the coverage of the cyanobacterial phylum using diversity-driven genome sequencing. Proc. Natl. Acad. Sci. USA 2013, 110, 1053–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watzer, B.; Forchhammer, K. Cyanophycin synthesis optimizes nitrogen utilization in the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2018, 84, e01298-18. [Google Scholar] [CrossRef] [Green Version]
- Boussiba, S.; Richmond, A.E. C-phycocyanin as a storage protein in the blue-green alga Spirulina platensis. Arch. Microbiol. 1980, 125, 143–147. [Google Scholar] [CrossRef]
- Glazer, A.N. Phycobiliproteins. Methods Enzymol. 1988, 167, 291–303. [Google Scholar]
- Allen, M.M.; Smith, A.J. Nitrogen chlorosis in blue-green algae. Arch. Microbiol. 1969, 69, 114–120. [Google Scholar] [CrossRef]
- Russell, G. Salinity and seaweed vegetation. In Plant Life in Aquatic and Amphibious Habitats; Crawford, R.M.M., Ed.; Blackwell: Oxford, UK, 1987; pp. 35–52. [Google Scholar]
- Karsten, U. Seaweed acclimation to salinity and desiccation stress. In Seaweed Biology; Wiencke, C., Bischof, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 87–107. [Google Scholar]
- Hagemann, M. Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol. Rev. 2011, 35, 87–123. [Google Scholar] [CrossRef]
- Fukaya, F.; Promden, W.; Hibino, T.; Tanaka, Y.; Nakamura, T.; Takabe, T. An Mrp-like cluster in the halotolerant cyanobacterium Aphanothece halophytica functions as a Na+/H+ antiporter. Appl. Environ. Microbiol. 2009, 75, 6626–6629. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Rivero, A.; Leganes, F.; Fernandez-Valiente, E.; Calle, P.; Fernandez-Piñas, F. mrpA, a gene with roles in resistance to Na+ and adaptation to alkaline pH in the cyanobacterium Anabaena sp. PCC7120. Microbiology 2005, 151, 1671–1682. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, T.; Bremer, E. Guardians in a stressful world: The Opu family of compatible solute transporters from Bacillus subtilis. Biol. Chem. 2017, 398, 193–214. [Google Scholar] [CrossRef] [PubMed]
- Emerson, D. The irony of iron–biogenic iron oxides as an iron source to the ocean. Front. Microbiol. 2016, 6, 1502. [Google Scholar] [CrossRef] [PubMed]
- Kranzler, C.; Rudolf, M.; Keren, N.; Schleiff, E. Iron in cyanobacteria. Adv. Bot. Res. 2013, 65, 57–105. [Google Scholar]
- Waterworth, S.C.; Isemonger, E.W.; Rees, E.R.; Dorrington, R.A.; Kwan, J.C. Conserved bacterial genomes from two geographically isolated peritidal stromatolite formations shed light on potential functional guilds. Environ. Microbiol. Rep. 2021, 13, 126–137. [Google Scholar] [CrossRef]
- Gallagher, A.L.; Miller, S.R. Expression of novel gene content drives adaptation to low iron in the cyanobacterium Acaryochloris. Genome Biol. Evol. 2018, 10, 1484–1492. [Google Scholar] [CrossRef] [Green Version]
- Jeanjean, R.; Talla, E.; Latifi, A.; Havaux, M.; Janicki, A.; Zhang, C.C. A large gene cluster encoding peptide synthetases and polyketide synthases is involved in production of siderophores and oxidative stress response in the cyanobacterium Anabaena sp. strain PCC 7120. Environ. Microbiol. 2008, 10, 2574–2585. [Google Scholar] [CrossRef]
- Wheatley, R.M.; MacLean, R.C. CRISPR-Cas systems restrict horizontal gene transfer in Pseudomonas aeruginosa. ISME J. 2021, 15, 1420–1433. [Google Scholar] [CrossRef]
- Jiang, W.; Maniv, I.; Arain, F.; Wang, Y.; Levin, B.R.; Marraffini, L.A. Dealing with the evolutionary downside of CRISPR immunity: Bacteria and beneficial plasmids. PLoS Genet. 2013, 9, e1003844. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).