Characterization of Bacterial Cellulose Produced by Komagataeibacter maltaceti P285 Isolated from Contaminated Honey Wine
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reference Bacterial Strain and Growth Conditions
2.2. Bacterial Isolation
2.3. Morphological, Biochemical and Physiological Characterization of the Selected Bacterial Isolate
2.4. Genomic DNA Extraction and 16S rRNA Gene Amplification
2.5. Preparation of Inoculum
2.6. Effect of Temperature and Initial Media pH on BC Production
2.7. Effect of Carbon and Nitrogen Sources on BC Production
2.8. Effect of Sugarcane and Honey on BC Production by the Selected Bacterial Isolates
2.9. Sugarcane and Honey Optimization of BC Production by Selected Bacterial Strains
2.10. Harvesting and Weighing of BC
2.11. Physical Property Determination of BC
2.11.1. Scanning Electron Microscopy
2.11.2. Fourier Transform Infrared (FTIR) Spectroscopy
2.11.3. X-ray Diffractometry
2.11.4. Differential Scanning Calorimetry (DSC) Analysis
2.11.5. Tensile Strength Determination
2.12. Statistical Analysis
3. Results
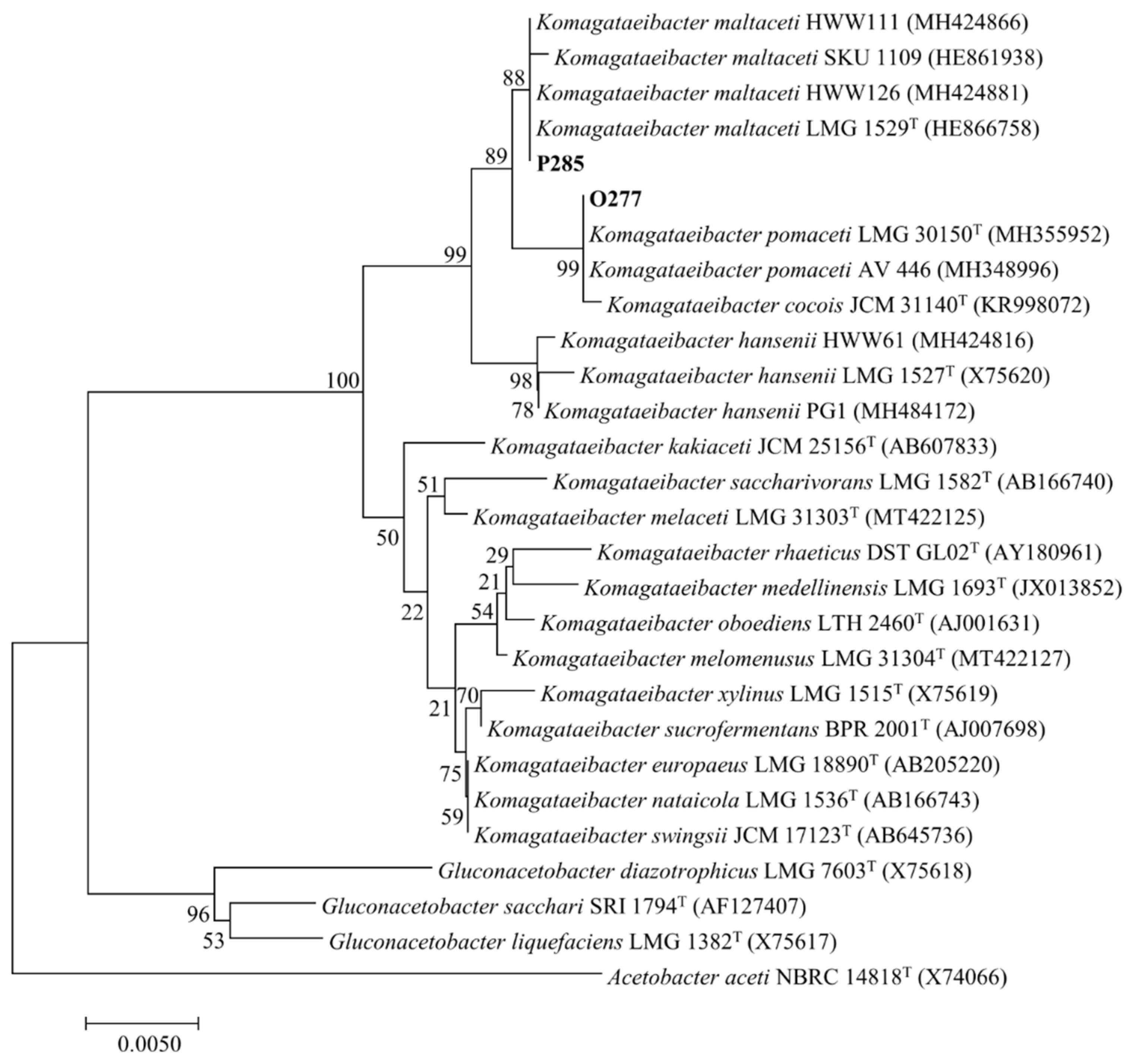
3.1. Bacterial Isolation and Identification
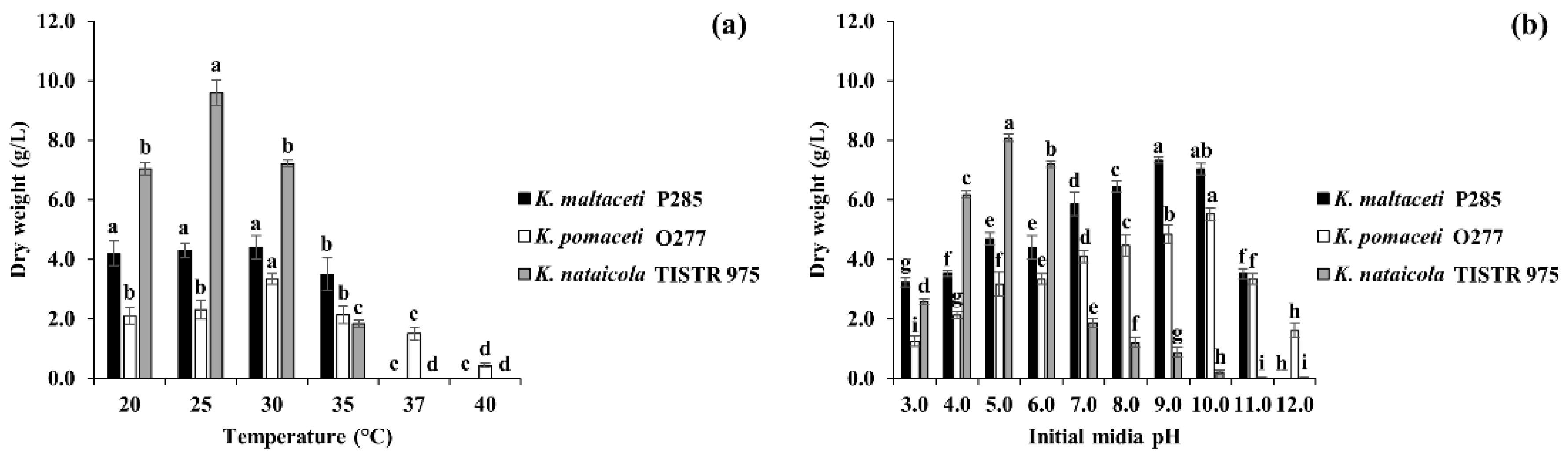
3.2. Effect of Temperatures and Initial Media pH on BC Production by Selected Bacterial Isolates
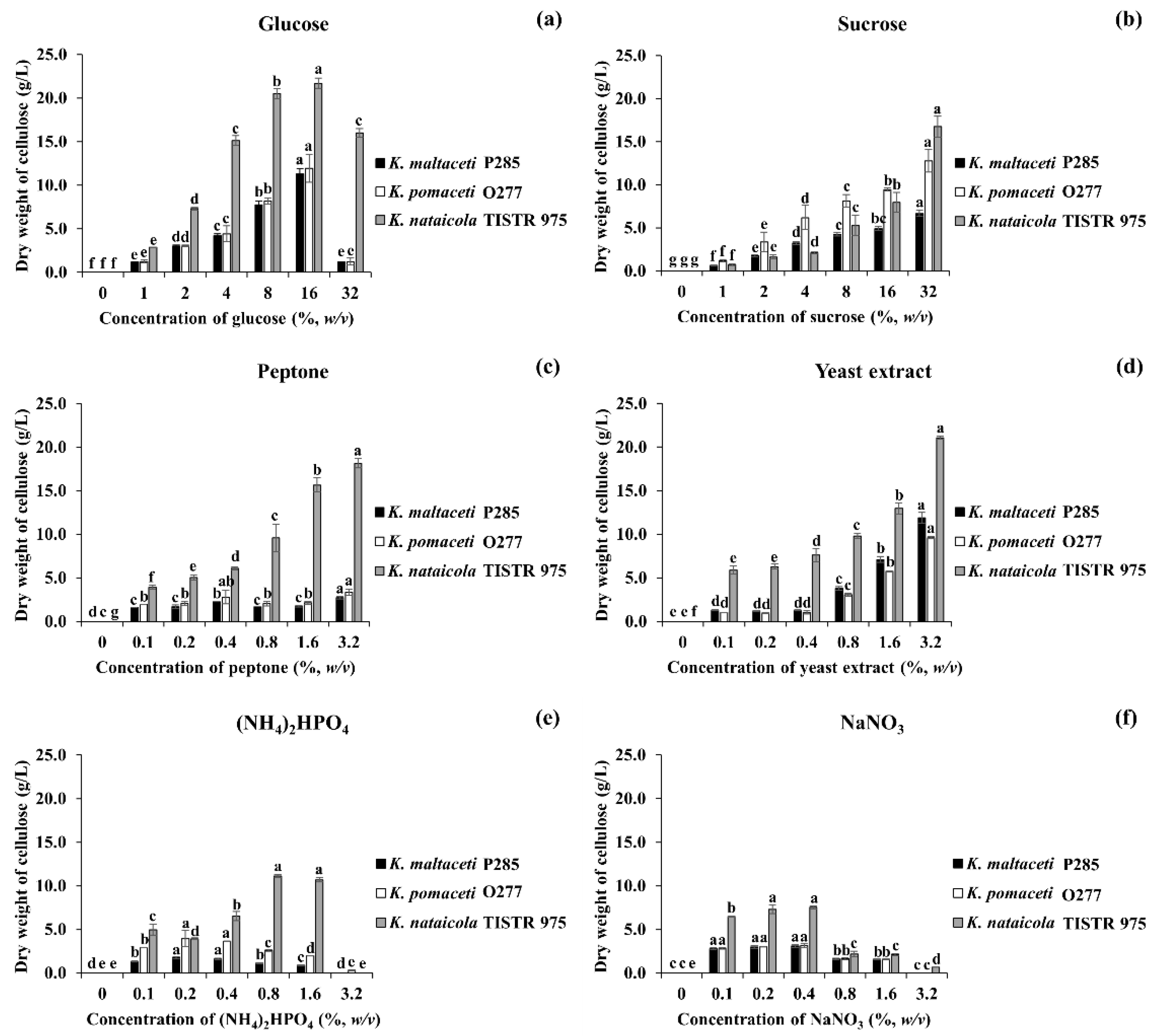
3.3. Effect of Carbon and Nitrogen Sources on BC Production
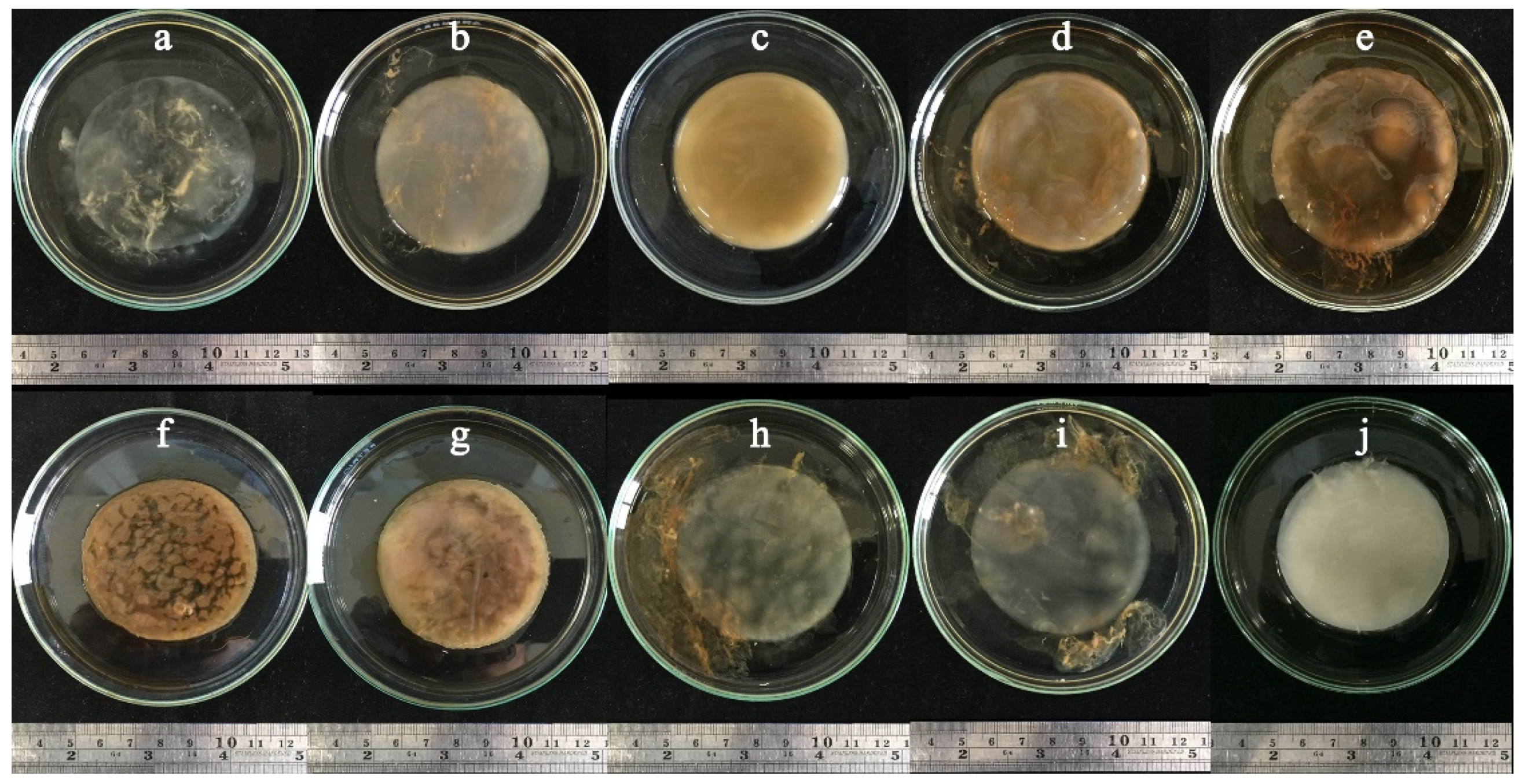
3.4. BC Production Using Sugarcane and Honey by Strains P285 and O277
3.5. BC Production Optimization of Sugarcane and Honey by Selected Bacterial Strains
3.6. Scanning Electron Microscopy (SEM)
3.7. Fourier Transform Infrared (FTIR) Spectroscopy
3.8. X-ray Diffraction Analysis
3.9. Differential Scanning Calorimetry (DSC) Analysis
3.10. Tensile Strength Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Slapšak, N.; Cleenwerck, I.; de Vos, P.; Trček, J. Gluconacetobacter maltaceti sp. nov., a novel vinegar producing acetic acid bacterium. Syst. Appl. Microbiol. 2013, 36, 17–21. [Google Scholar] [CrossRef]
- Yamada, Y. Transfer of Gluconacetobacter kakiaceti, Gluconacetobacter medellinensis and Gluconacetobacter maltaceti to the genus Komagataeibacter as Komagataeibacter kakiaceti comb. nov., Komagataeibacter medellinensis comb. nov. and Komagataeibacter maltaceti comb. nov. Int. J. Syst. Evol. Microbiol. 2014, 64, 1670–1672. [Google Scholar] [CrossRef] [PubMed]
- Vigentini, I.; Fabrizio, V.; Dellacà, F.; Rossi, S.; Azario, I.; Mondin, C.; Benaglia, M.; Foschino, R. Set-up of bacterial cellulose production from the genus Komagataeibacter and its use in a gluten-free bakery product as a case study. Front. Microbiol. 2019, 10, 1953. [Google Scholar] [CrossRef] [PubMed]
- Kongruang, S. Bacterial cellulose production by Acetobacter xylinum strains from agricultural waste products. Appl. Biochem. Biotechnol. 2008, 148, 245–256. [Google Scholar] [CrossRef] [PubMed]
- Gorgieva, S.; Trček, J. Bacterial cellulose: Production, modification and perspectives in biomedical applications. Nanomaterials 2019, 9, 1352. [Google Scholar] [CrossRef] [Green Version]
- Marič, L.; Cleenwerck, I.; Accetto, T.; Vandamme, P.; Trček, J. Description of Komagataeibacter melaceti sp. nov. and Komagataeibacter melomenusus sp. nov. isolated from apple cider vinegar. Microorganisms 2020, 8, 1178. [Google Scholar] [CrossRef]
- Cielecka, I.; Ryngajłło, M.; Maniukiewicz, W.; Bielecki, S. Highly stretchable bacterial cellulose produced by Komagataeibacter hansenii SI1. Polymers 2021, 13, 4455. [Google Scholar] [CrossRef]
- Jacek, P.; da Silva, F.A.G.S.; Dourado, F.; Bielecki, S.; Gama, M. Optimization and characterization of bacterial nanocellulose produced by Komagataeibacter rhaeticus K3. Carbohydr. Polym. Technol. Appl. 2021, 2, 100022. [Google Scholar] [CrossRef]
- Gullo, M.; La China, S.; Falcone, P.M.; Giudici, P. Biotechnological production of cellulose by acetic acid bacteria: Current state and perspectives. Appl. Microbiol. Biotechnol. 2018, 102, 6885–6898. [Google Scholar] [CrossRef]
- Rangaswamy, B.E.; Vanitha, K.P.; Hungund, B.S. Microbial cellulose production from bacteria isolated from rotten fruit. Int. J. Polym. Sci. 2015, 2015, 280784. [Google Scholar] [CrossRef] [Green Version]
- La China, S.; De Vero, L.; Anguluri, K.; Brugnoli, M.; Mamlouk, D.; Gullo, M. Kombucha tea as a reservoir of cellulose producing bacteria: Assessing diversity among Komagataeibacter isolates. Appl. Sci. 2021, 11, 1595. [Google Scholar] [CrossRef]
- Costa, A.F.S.; Almeida, F.C.G.; Vinhas, G.M.; Sarubbo, L.A. Production of bacterial cellulose by Gluconacetobacter hansenii using corn steep liquor as nutrient sources. Front. Microbiol. 2017, 8, 2027. [Google Scholar] [CrossRef] [PubMed]
- Esa, F.; Tasirin, S.M.; Rahman, N.A. Overview of bacterial cellulose production and application. Agric. Agric. Sci. Proc. 2014, 2, 113–119. [Google Scholar] [CrossRef] [Green Version]
- Azeredo, H.M.C.; Barud, H.; Farinas, C.S.; Vasconcellos, V.M.; Claro, A.M. Bacterial cellulose as a raw material for food and food packaging applications. Front. Sustain. Food Syst. 2019, 3, 7. [Google Scholar] [CrossRef] [Green Version]
- Ullah, H.; Santos, H.A.; Khan, T. Applications of bacterial cellulose in food, cosmetics and drug delivery. Cellulose 2016, 23, 2291–2314. [Google Scholar] [CrossRef]
- Portela, R.; Leal, C.R.; Almeida, P.L.; Sobral, R.G. Bacterial cellulose: A versatile biopolymer for wound dressing applications. Microb. Biotechnol. 2019, 12, 586–610. [Google Scholar] [CrossRef]
- Zang, S.; Zhang, R.; Chen, H.; Lu, Y.; Zhou, J.; Chang, X.; Qiu, G.; Wu, Z.; Yang, G. Investigation on artificial blood vessels prepared from bacterial cellulose. Mater. Sci. Eng. C 2015, 46, 111–117. [Google Scholar] [CrossRef]
- Canilha, L.; Kumar Chandel, A.; dos Santos Milessi, T.S.; Fernandes Antunes, F.A.; da Costa Freitas, W.L.; das Graças Almeida Felipe, M.; da Silva, S.S. Bioconversion of sugarcane biomass into ethanol: An overview about composition, pretreatment methods, detoxification of hydrolysates, enzymatic saccharification, and ethanol fermentation. J. Biomed Biotechnol. 2012, 2012, 989572. [Google Scholar] [CrossRef]
- Kim, M.; Day, D.F. Composition of sugar cane, energy cane, and sweet sorghum suitable for ethanol production at Louisiana sugar mills. J. Ind. Microbiol. Biotechnol. 2011, 38, 803–807. [Google Scholar] [CrossRef]
- Formann, S.; Hahn, A.; Janke, L.; Stinner, W.; Sträuber, H.; Logroño, W.; Nikolausz, M. Beyond sugar and ethanol production: Value generation opportunities through sugarcane residues. Front. Energy Res. 2020, 8, 579577. [Google Scholar] [CrossRef]
- De La Fuente, E.; Ruiz-Matute, A.I.; Valencia-Barrera, R.M.; Sanz, J.; Martínez Castro, I. Carbohydrate composition of Spanish unifloral honeys. Food Chem. 2011, 129, 1483–1489. [Google Scholar] [CrossRef] [Green Version]
- Kečkeš, J.; Trifković, J.; Andrić, F.; Jovetić, M.; Tešić, Z.; Milojković-Opsenica, D. Amino acids profile of Serbian unifloral honeys. J. Sci. Food Agric. 2013, 93, 3368–3376. [Google Scholar] [CrossRef] [PubMed]
- Cherchi, A.; Spanedda, L.; Tuberoso, C.; Cabras, P. Solid-phase extraction and high-performance liquid chromatographic determination of organic acids in honey. J. Chromatogr. A 1994, 669, 59–64. [Google Scholar] [CrossRef]
- Bonté, F.; Desmoulière, A. Honey: Origin and composition. Actual. Pharm. 2013, 52, 18–21. [Google Scholar] [CrossRef]
- Alqarni, A.S.; Owayss, A.A.; Mahmoud, A.A.; Hannan, M.A. Mineral content and physical properties of local and imported honeys in Saudi Arabia. J. Saudi Chem. Soc. 2014, 18, 618–625. [Google Scholar] [CrossRef] [Green Version]
- Da Silva, P.M.; Gauche, C.; Gonzaga, L.V.; Costa, A.C.; Fett, R. Honey: Chemical composition, stability and authenticity. Food Chem. 2016, 196, 309–323. [Google Scholar] [CrossRef]
- Ciecholewska-Juśko, D.; Broda, M.; Żywicka, A.; Styburski, D.; Sobolewski, P.; Gorący, K.; Migdał, P.; Junka, A.; Fijałkowski, K. Potato juice, a starch industry waste, as a cost-effective medium for the biosynthesis of bacterial cellulose. Int. J. Mol. Sci. 2021, 22, 10807. [Google Scholar] [CrossRef]
- Hestrin, S.; Schramm, M. Synthesis of cellulose by Acetobacter xylinum. Preparation of freeze-dried cells capable of polymerizing glucose to cellulose. Biochem. J. 1954, 58, 345–352. [Google Scholar] [CrossRef] [Green Version]
- Arroyo, E.; Enríquez, L.; Sánchez, A.; Ovalle, M.; Olivas, A. Scanning electron microscopy of bacteria Tetrasphaera duodecadis. Scanning 2014, 36, 547–550. [Google Scholar] [CrossRef]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [Green Version]
- Rungsirivanich, P.; Inta, A.; Tragoolpua, Y.; Thongwai, N. Partial rpoB gene sequencing identification and probiotic potential of Floricoccus penangensis ML061-4 isolated from Assam tea (Camellia sinensis var. assamica). Sci. Rep. 2019, 9, 16561. [Google Scholar] [CrossRef] [PubMed]
- Rungsirivanich, P.; Supandee, W.; Futui, W.; Chumsai-Na-Ayudhya, V.; Yodsombat, C.; Thongwai, N. Culturable bacterial community on leaves of Assam tea (Camellia sinensis var. assamica) in Thailand and human probiotic potential of isolated Bacillus spp. Microorganisms 2020, 8, 1585. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger dataset. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Zajic, J.E.; Supplisson, B. Emulsification and degradation of “Bunker C” fuel oil by microorganisms. Biotechnol. Bioeng. 1972, 14, 331–343. [Google Scholar] [CrossRef] [PubMed]
- Atykyan, N.; Revin, V.; Shutova, V. Raman and FT-IR Spectroscopy investigation the cellulose structural differences from bacteria Gluconacetobacter sucrofermentans during the different regimes of cultivation on a molasses media. AMB Expr. 2020, 10, 84. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Saha, N.; Saha, P. Characterization of bacterial cellulose produced using media containing waste apple juice. Appl. Biochem. Microbiol. 2018, 54, 649–657. [Google Scholar] [CrossRef]
- Kiziltas, E.E.; Kiziltas, A.; Gardner, D.J. Synthesis of bacterial cellulose using hot water extracted wood sugars. Carbohydr. Polym. 2015, 124, 131–138. [Google Scholar] [CrossRef]
- De Oliveira, J.P.; Bruni, G.P.; Lima, K.O.; El Halal, S.L.M.; Da Rosa, G.S.; Dias, A.R.G.; da Rosa Zavareze, E. Cellulose fibers extracted from rice and oat husks and their application in hydrogel. Food Chem. 2017, 221, 153–160. [Google Scholar] [CrossRef]
- Trilokesh, C.; Uppuluri, K.B. Isolation and characterization of cellulose nanocrystals from jackfruit peel. Sci. Rep. 2019, 9, 16709. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Zhang, Y.; Phillips, G.O.; Yang, G. Utilization of bacterial cellulose in food. Food Hydrocoll. 2014, 35, 539–545. [Google Scholar] [CrossRef]
- Lin, D.; Lopez-Sanchez, P.; Li, R.; Li, Z. Production of bacterial cellulose by Gluconacetobacter hansenii CGMCC 3917 using only waste beer yeast as nutrient source. Bioresour. Technol. 2014, 151, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.M.; Liu, R.H. Cost-effective production of bacterial cellulose in static cultures using distillery wastewater. J. Biosci. Bioeng. 2013, 115, 284–290. [Google Scholar] [CrossRef] [PubMed]
- Alga, I.; Fernandes, S.C.M.; Mondragon, G.; Castro, C.; Garcia-Astrain, C.; Gabilondo, N.; Retegi, A.; Eceiza, A. Pineapple agroindustrial residues for the production of high value bacterial cellulose with different morphologies. J. Appl. Polym. Sci. 2015, 132, 41237. [Google Scholar] [CrossRef]
- Chen, L.; Hong, F.; Yang, X.X.; Han, S.F. Biotransformation of wheat straw to bacterial cellulose and its mechanism. Bioresour. Technol. 2013, 135, 464–468. [Google Scholar] [CrossRef]
- Basu, A.; Vadanan, S.V.; Lim, S.A. Novel platform for evaluating the environmental impacts on bacterial cellulose production. Sci. Rep. 2018, 8, 5780. [Google Scholar] [CrossRef] [Green Version]
- Kadier, A.; Ilyas, R.A.; Huzaifah, M.R.M.; Harihastuti, N.; Sapuan, S.M.; Harussani, M.M.; Azlin, M.N.M.; Yuliasni, R.; Ibrahim, R.; Atikah, M.S.N.; et al. Use of industrial wastes as sustainable nutrient sources for bacterial cellulose (BC) production: Mechanism, advances, and future perspectives. Polymers 2021, 13, 3365. [Google Scholar] [CrossRef]
- Jagannath, A.; Kalaiselvan, A.; Manjunatha, S.S.; Raju, P.S.; Bawa, A.S. The effect of pH, sucrose and ammonium sulphate concentrations on the production of bacterial cellulose (Nata-de-coco) by Acetobacter xylinum. World J. Microbiol. Biotechnol. 2008, 24, 2593. [Google Scholar] [CrossRef]
- Coban, E.P.; Biyik, H. Evaluation of different pH and temperatures for bacterial cellulose production in HS (hestrin-scharmm) medium and beet molasses medium. Afr. J. Microbiol. Res. 2011, 5, 1037–1045. [Google Scholar] [CrossRef]
- Zahan, K.A.; Pa’e, N.; Muhamad, I.I. Monitoring the effect of pH on bacterial cellulose production and Acetobacter xylinum 0416 growth in a rotary discs reactor. Arab. J. Sci. Eng. 2015, 40, 1881–1885. [Google Scholar] [CrossRef]
- Alemam, A.M.; Shaheen, T.I.; Hassan, S.E.D.; Desouky, S.E.; El-Gamal, M.S. Production enhancement of bacterial cellulose nanofiber using local Komagataeibacter xylinus SB3.1 under static conditions. Egypt. J. Chem. 2021, 64, 2213–2221. [Google Scholar] [CrossRef]
- Kumar, V.; Sharma, D.K.; Sandhu, P.P.; Jadaun, J.; Sangwan, R.S.; Yadav, S.K. Sustainable process for the production of cellulose by an Acetobacter pasteurianus RSV-4 (MTCC 25117) on whey medium. Cellulose 2021, 28, 103–116. [Google Scholar] [CrossRef]
- Aswini, K.; Gopal, N.O.; Uthandi, S. Optimized culture conditions for bacterial cellulose production by Acetobacter senegalensis MA1. BMC Biotechnol. 2020, 20, 46. [Google Scholar] [CrossRef] [PubMed]
- Zahan, K.A.; Nordin, K.; Mustapha, M.; Zairi, M.N.M. Effect of incubation temperature on growth of Acetobacter xylinum 0416 and bacterial cellulose production. Appl. Mech. Mater. 2015, 815, 3–8. [Google Scholar] [CrossRef]
- Lin, S.P.; Huang, Y.H.; Hsu, K.D.; Lai, Y.J.; Chen, Y.K.; Cheng, K.C. Isolation and identification of cellulose-producing strain Komagataeibacter intermedius from fermented fruit juice. Carbohydr. Polym. 2016, 151, 827–833. [Google Scholar] [CrossRef]
- Raiszadeh-Jahromi, Y.; Rezazadeh-Bari, M.; Almasi, H.; Amiri, S. Optimization of bacterial cellulose production by Komagataeibacter xylinus PTCC 1734 in a low-cost medium using optimal combined design. J. Food Sci. Technol. 2020, 57, 2524–2533. [Google Scholar] [CrossRef]
- Ratzke, C.; Gore, J. Modifying and reacting to the environmental pH can drive bacterial interactions. PLoS Biol. 2018, 16, e2004248. [Google Scholar] [CrossRef] [Green Version]
- Siedler, S.; Rau, M.H.; Bidstrup, S.; Vento, J.M.; Aunsbjerg, S.D.; Bosma, E.F.; McNair, L.M.; Beisel, C.L.; Neves, A.R. Competitive exclusion is a major bioprotective mechanism of Lactobacilli against fungal spoilage in fermented milk products. Appl. Environ. Microbiol. 2020, 86, e02312-19. [Google Scholar] [CrossRef] [Green Version]
- Ramana, K.; Tomar, A.; Singh, L. Effect of various carbon and nitrogen sources on cellulose synthesis by Acetobacter xylinum. World J. Microbiol. Biotechnol. 2000, 16, 245–248. [Google Scholar] [CrossRef]
- Morgan, J.L.W.; Strumillo, J.; Zimmer, J. Crystallographic snapshot of cellulose synthesis and membrane translocation. Nature 2012, 493, 181–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Römling, U.; Galperin, M.Y. Bacterial cellulose biosynthesis: Diversity of operons, subunits, products, and functions. Trends Microbiol. 2015, 23, 545–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Son, H.J.; Kim, H.G.; Kim, K.K.; Kim, H.S.; Kim, Y.G.; Lee, S.J. Increased production of bacterial cellulose by Acetobacter sp. V6 in synthetic media under shaking culture conditions. Bioresour. Technol. 2003, 86, 215–219. [Google Scholar] [CrossRef]
- Chai, J.M.; Adnan, A. Effect of different nitrogen source combinations on microbial cellulose production by Pseudomonas aeruginosa in batch fermentation. IOP Conf. Ser. Mater. Sci. Eng. 2018, 440, 012044. [Google Scholar] [CrossRef]
- Potivara, K.; Phisalaphong, M. Development and characterization of bacterial cellulose reinforced with natural rubber. Materials 2019, 12, 2323. [Google Scholar] [CrossRef] [Green Version]
- Morán, J.I.; Alvarez, V.A.; Cyras, V.P.; Vázquez, A. Extraction of cellulose and preparation of nanocellulose from sisal fibers. Cellulose 2008, 15, 149–159. [Google Scholar] [CrossRef]
- Rosa, S.M.L.; Rehman, N.; de Miranda, M.I.G.; Nachtigall, S.M.B.; Bica, C.I.D. Chlorine-free extraction of cellulose from rice husk and whisker isolation. Carbohydr. Polym. 2012, 87, 1131–1138. [Google Scholar] [CrossRef] [Green Version]
- Auta, R.; Adamus, G.; Kwiecien, M.; Radecka, I.; Hooley, P. Production and characterization of bacterial cellulose before and after enzymatic hydrolysis. Afr. J. Biotechnol. 2017, 10, 470–482. [Google Scholar] [CrossRef]
- Schenzel, K.; Fischer, S.; Brendler, E. New method for determining the degree of cellulose I crystallinity by means of FT Raman spectroscopy. Cellulose 2005, 12, 223–231. [Google Scholar] [CrossRef]
- Sun, X.F.; Sun, R.C.; Su, Y.; Sun, J.X. Comparative study of crude and purified cellulose from wheat straw. J. Agric. Food Chem. 2004, 52, 839–847. [Google Scholar] [CrossRef]
- Treesuppharat, W.; Rojanapanthu, P.; Siangsanoh, C.; Manuspiya, H.; Ummartyotin, S. Synthesis and characterization of bacterial cellulose and gelatin-based hydrogel composites for drug-delivery systems. Biotechnol. Rep. 2017, 15, 84–91. [Google Scholar] [CrossRef] [PubMed]
- George, J.; Ramana, K.V.; Sabapathy, S.N.; Jagannath, J.H.; Bawa, A.S. Characterization of chemically treated bacterial (Acetobacter xylinum) biopolymer: Some thermo-mechanical properties. Int. J. Biol. Macromol. 2005, 37, 189–194. [Google Scholar] [CrossRef]
- Vazquez, A.; Foresti, M.L.; Cerrutti, P.; Galvagno, M. Bacterial cellulose from simple and low cost production media by Gluconacetobacter xylinus. J. Polym. Environ. 2013, 21, 545–554. [Google Scholar] [CrossRef]
- Pérez-Rigueiro, J.; Viney, C.; Llorca, J.; Elices, M. Mechanical properties of single-brin silkworm silk. J. Appl. Polym. Sci. 2000, 75, 1270–1277. [Google Scholar] [CrossRef]
- Almeida, L.R.; Martins, A.R.; Fernandes, E.M.; Oliveir, M.B.; Correlo, V.M.; Pashkuleva, I.; Marques, A.P.; Ribeiro, A.S.; Durães, N.F.; Silva, C.J.; et al. New biotextiles for tissue engineering: Development, characterization and in vitro cellular viability. Acta Biomater. 2013, 9, 8167–8181. [Google Scholar] [CrossRef]

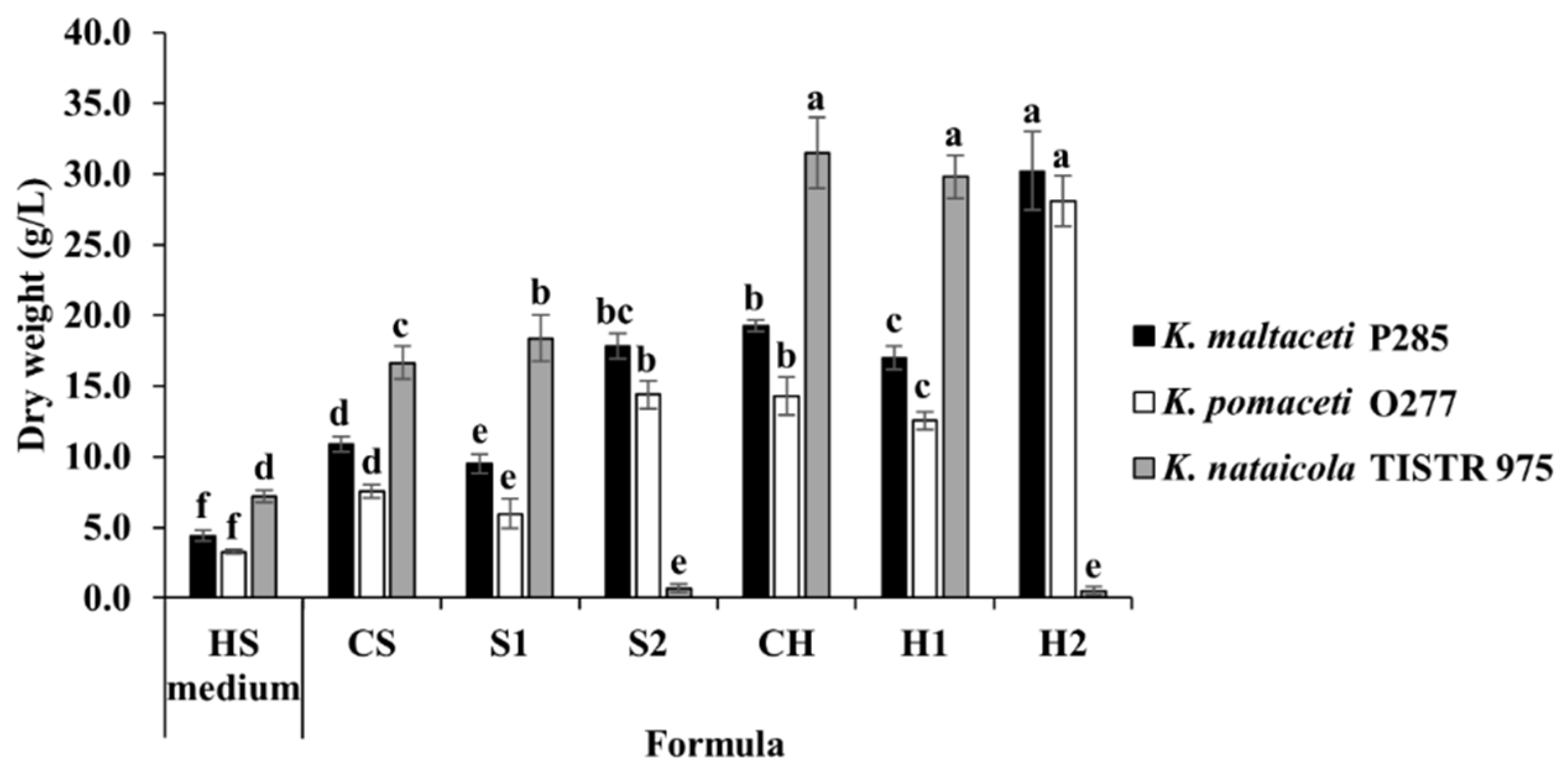



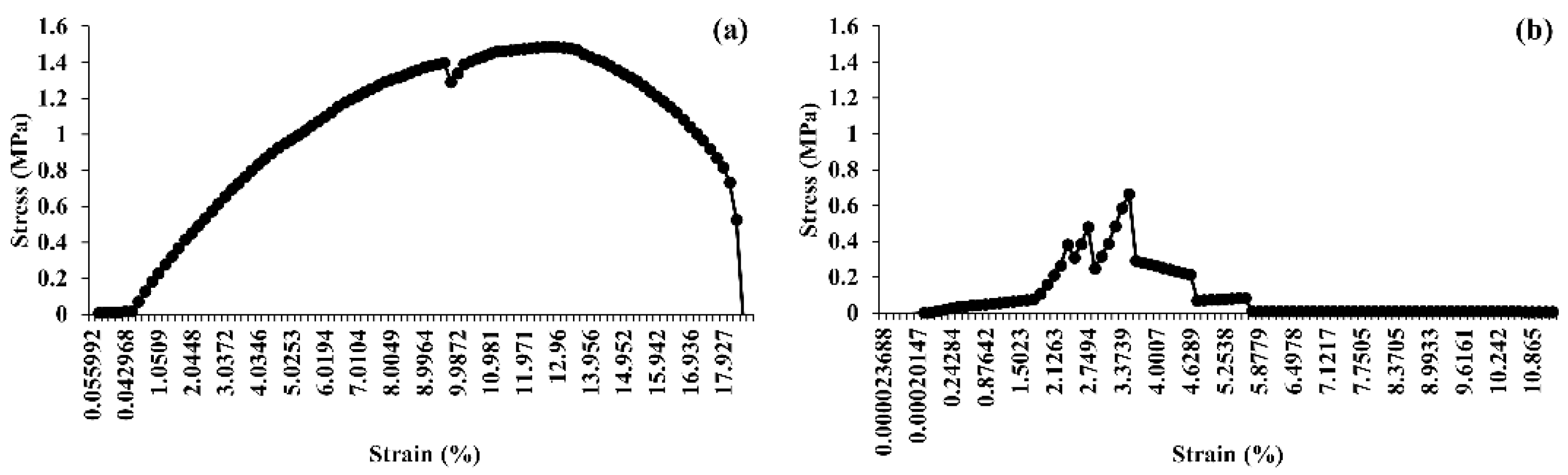
| Component | CS | S1 | S2 | CH | H1 | H2 |
|---|---|---|---|---|---|---|
| Sugarcane (8%, w/v) | + | + | + | − | − | − |
| Honey (1:4 of honey: water ratio) | − | − | − | + | + | + |
| Yeast extract (0.2%, w/v) | + | − | − | + | − | − |
| Yeast extract (3.2%, w/v) | − | + | + | − | + | + |
| pH 5.0 | − | + | − | − | + | − |
| pH 6.0 | + | − | − | + | − | − |
| pH 9.0 | − | − | + | − | − | + |
| Test | Strain P285 | Strain O277 | K. nataicola TISTR 975 |
|---|---|---|---|
| Catalase | + | + | + |
| Citrate | − | + | + |
| Cytochrome oxidase | − | − | − |
| Gelatinase | − | − | − |
| Indole | − | − | − |
| Lysine deaminase | − | − | − |
| Lysine decarboxylase | + | + | + |
| Methyl red | + | + | − |
| Nitrate reduction | − | − | − |
| String | + | + | + |
| Voges-Proskauer | − | − | − |
| Urease | − | − | − |
| Carbohydrate fermentation | |||
| Glucose | + | + | w |
| Galactose | + | + | w |
| Mannitol | + | w | + |
| Mannose | w | − | − |
| Lactose | w | − | − |
| Maltose | w | w | w |
| Rhamnose | w | − | − |
| Sucrose | w | w | w |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thongwai, N.; Futui, W.; Ladpala, N.; Sirichai, B.; Weechan, A.; Kanklai, J.; Rungsirivanich, P. Characterization of Bacterial Cellulose Produced by Komagataeibacter maltaceti P285 Isolated from Contaminated Honey Wine. Microorganisms 2022, 10, 528. https://doi.org/10.3390/microorganisms10030528
Thongwai N, Futui W, Ladpala N, Sirichai B, Weechan A, Kanklai J, Rungsirivanich P. Characterization of Bacterial Cellulose Produced by Komagataeibacter maltaceti P285 Isolated from Contaminated Honey Wine. Microorganisms. 2022; 10(3):528. https://doi.org/10.3390/microorganisms10030528
Chicago/Turabian StyleThongwai, Narumol, Wirapong Futui, Nanthiwa Ladpala, Benjamat Sirichai, Anuwat Weechan, Jirapat Kanklai, and Patthanasak Rungsirivanich. 2022. "Characterization of Bacterial Cellulose Produced by Komagataeibacter maltaceti P285 Isolated from Contaminated Honey Wine" Microorganisms 10, no. 3: 528. https://doi.org/10.3390/microorganisms10030528
APA StyleThongwai, N., Futui, W., Ladpala, N., Sirichai, B., Weechan, A., Kanklai, J., & Rungsirivanich, P. (2022). Characterization of Bacterial Cellulose Produced by Komagataeibacter maltaceti P285 Isolated from Contaminated Honey Wine. Microorganisms, 10(3), 528. https://doi.org/10.3390/microorganisms10030528










