Snf1 Kinase Differentially Regulates Botrytis cinerea Pathogenicity according to the Plant Host
Abstract
:1. Introduction
2. Materials and Methods
2.1. Fungal Strains and Growth Conditions
2.2. Construction of Deletion Cassettes by PCR Fusion
| Fungus | Cochliobolus carbonum | Fusarium oxysporum | Magnaporthe oryzae | Alternaria brassicola | Fusarium graminearum | Ustilago maydis | Verticillium dahliae | Penicillium digitatum | Beauveria bassiana | Metarhizium acridum | Magnaporthe oryzae | Leptosphaeria maculans | Trichoderma harzianum | Fusarium virguliforme | Pestalotiopsis microspora | Colletotrichum fructicola | Alternaria alternata | Podospora anserina | Cordyceps militaris | Botrytis cinerea | |||
| Acidic pH | Neutral pH | ||||||||||||||||||||||
| Sporulation | 0 | 3 | 2 | 0 | 3 | 2 | 2 | 3 | 1 | 0 | 3 | 3 | 2 | 3 | 3 | 3 | 3 | ||||||
| Pathogenicity | 2 | 3 | 3 | 0 | 2 | 1 | 3 | 2 | 1 | 3 | 3 | 3 | 2 | 3 | 3 | 2 | 3 | 2 | 3 | ||||
| In vitro radial growth on different carbon sources | Simple sugars | Glucose | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 0 | 1 | 1 | 1 | 2 | ||
| Sucrose | 0 | 2 | 2 | 0 | 0 | 2 | 1 | 1 | 1 | 3 | 2 | 0 | 1 | 2 | |||||||||
| Fructose | 1 | 2 | 2 | 0 | 1 | 1 | 0 | 2 | 1 | 0 | |||||||||||||
| Galactose | 2 | 2 | 2 | 3 | 0 | 2 | 3 | 2 | |||||||||||||||
| Trehalose | 2 | 2 | 1 | 1 | |||||||||||||||||||
| Maltose | 2 | 2 | 1 | 0 | 1 | 1 | 0 | ||||||||||||||||
| Glycerol | 1 | 1 | 1 | 1 | 1 | 0 | |||||||||||||||||
| GA | 2 | 1 | 2 | ||||||||||||||||||||
| Xylose | 2 | 2 | 2 | 0 | 0 | 1 | 2 | 1 | 1 | 1 | |||||||||||||
| Arabinose | 3 | 1 | 2 | 0 | 1 | 2 | |||||||||||||||||
| Polysaccharides | Cellulose | 0 | 0 | 1 | 2 | ||||||||||||||||||
| PGA or Pectin | 2 | 1 | 2 | 0 | 0 | 2 | 3 | 2 | 0 | 1 | 2 | 1 | 2 | ||||||||||
| Xylan | 2 | 1 | 2 | 0 | 2 | 0 | 1 | 2 | |||||||||||||||
| Plant cell wall | 3 | 2 | |||||||||||||||||||||
| Chitin | 1 | 3 | |||||||||||||||||||||
| Lipids | Tween 80 | 2 | 0 | 2 | 2 | ||||||||||||||||||
| Olive oil | 2 | 1 | 2 | ||||||||||||||||||||
| Triolein | 2 | 1 | 2 | ||||||||||||||||||||
| Acetate | 2 | 1 | 0 | 2 | |||||||||||||||||||
| Host species and organ tested | Maize leaf | Green cabbage, Arabidopsis seedlings | Rice leaf | Green cabbage leaf | Barley head; Wheat head | Maize seedling | Tomato plant, Eggplant leaf | Citrus fruit | Greater wax moth larvae (insect) | Locust (insect) | Barley leaf, Rice leaf | Canola cotyledons | Fusarium spp. (fungi) | Soybean plant and root | Not pathogenic (endophyte) | Tea-oil tree leaf | Tangerine leaf | Not pathogenic | Silkworm pupae (insect) | Apple fruit | Cucumber cotyledon, Bean leaf | ||
| Reference | [8] Tonukary et al., 2000 | [9] Ospina-Giraldo et al., 2003 | [10] Yi et al., 2008 | [7] Cho et al., 2009 | [11] Lee et al., 2009 | [22] Nadal et al., 2010 | [12] Tzima et al., 2011 | [13] Zhang et al., 2013 | [18] Wang et al., 2014 | [19] Ming et al., 2014 | [25] Zeng et al., 2014 | [14] Feng et al, 2014 | [21] Galarza et al., 2015 | [15] Islam et al., 2017 | [26] Wang et al., 2018 | [16] Zhang et al., 2019 | [17] Tang et al., 2020 | [23] Li et al., 2020 | [20] Wang et al., 2020 | This study | |||
| Mutant defect vs WT | 0 | No | 1 | Mild | 2 | 2 Moderate | 3 | Severe | GA: galacturonic acid PGA: Polygalacturonic acid | ||||||||||||||
2.3. Chemical Transformation of Fungal Protoplasts with PEG/CaCl2
2.4. Molecular Validation of Gene Deletion by PCR and Southern Blot
2.5. Functional Complementation
2.6. Pathogenicity Assays on Plants
2.7. In Vitro Radial Growth Tests
2.8. Xylanase Enzymatic Assay
2.9. Monitoring Ambient pH Changes in Liquid Culture
2.10. Quantitative Real-Time RT-PCR
2.11. Statistical Analysis
3. Results
3.1. Targeted Gene Deletion of Bcsnf1 Gene in Botrytis Cinerea
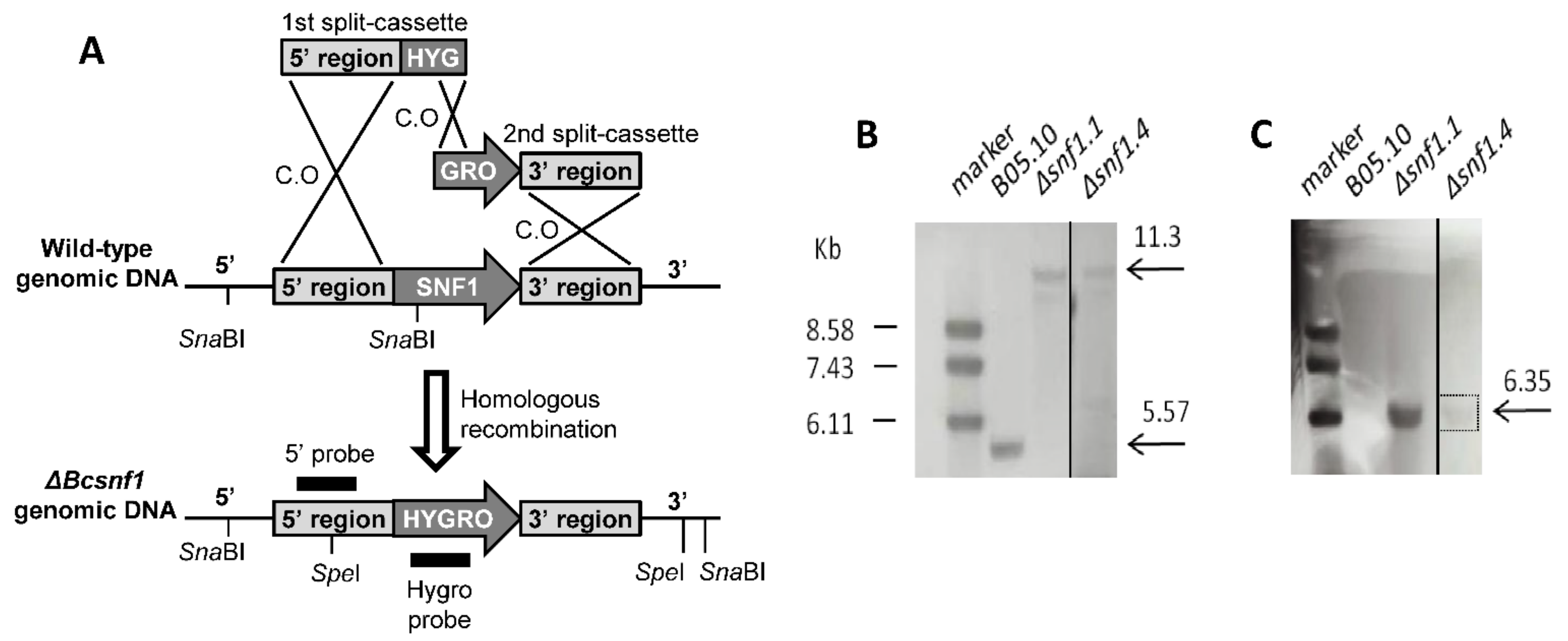
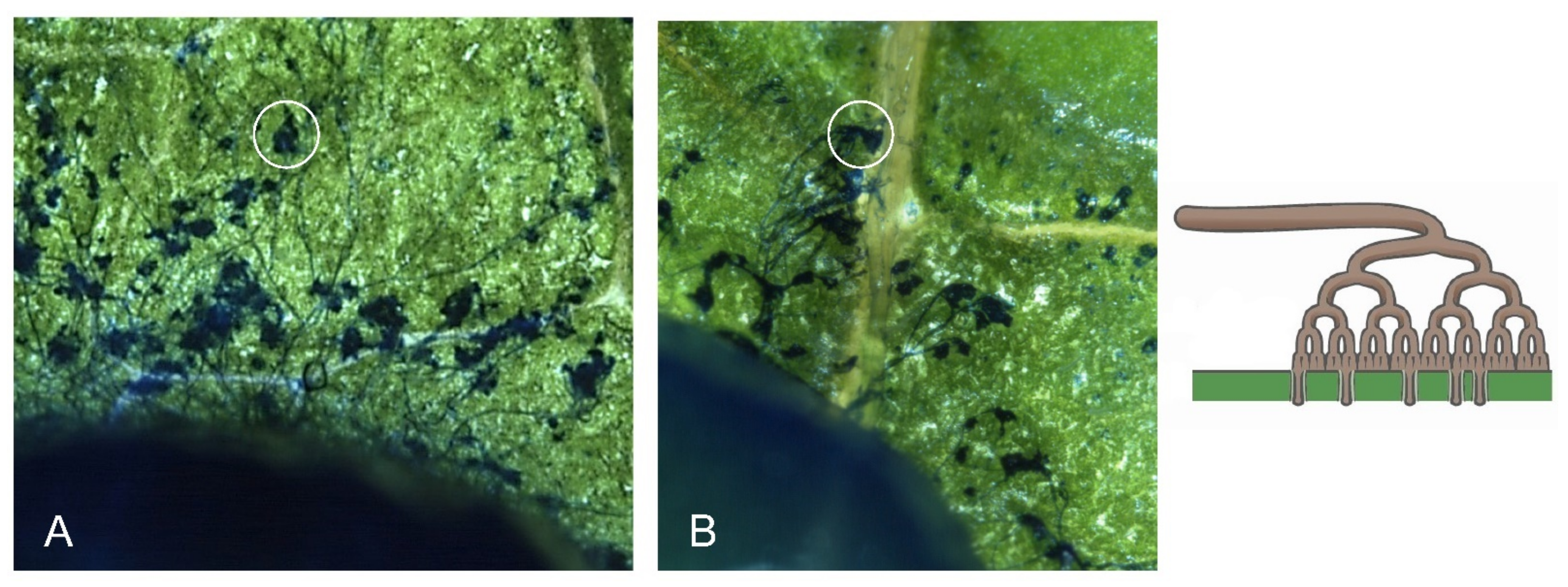
3.2. Bcsnf1 Gene Deletion Abolishes Asexual Sporulation and Production of Macroconidia
3.3. Bcsnf1 Gene Deletion Does Not Affect in Planta Penetration by the Fungus, but Alters Virulence According to the Host
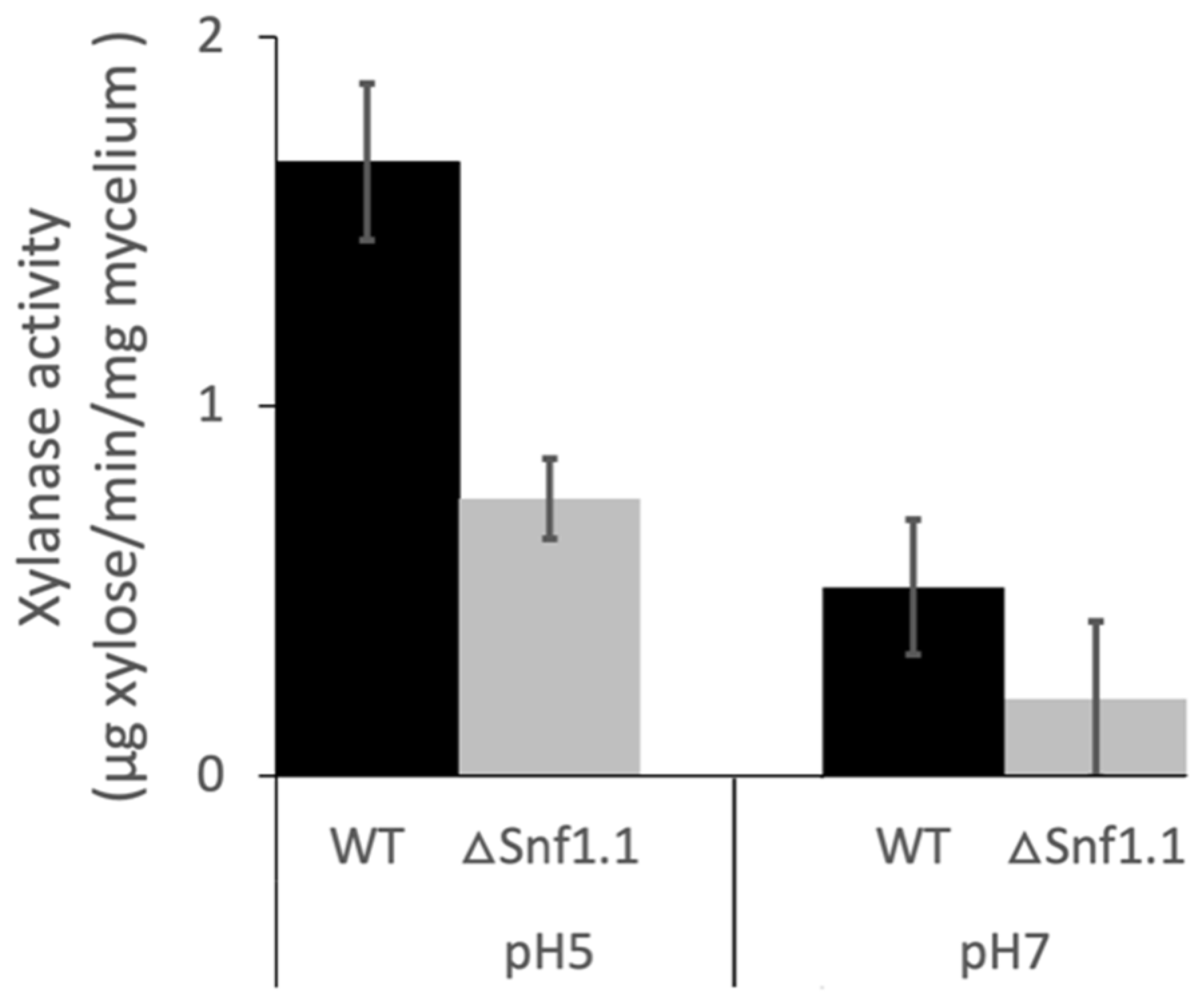
3.4. Bcsnf1 Gene Deletion Alters Xylanase Secretion and Carbon Nutrition
3.5. Bcsnf1 Deletion Alters the Ability of the Fungus to Modulate the Alkaline pH
4. Discussion
4.1. Role of Snf1 in Conidiation of B. cinerea
4.2. Role of Snf1 in Pathogenicity of B. cinerea
4.3. Role of Snf1 in B. cinerea Growth on Different Carbon Sources
4.4. A Suggested Role of Snf1 on Alkaline pH Modulation
4.5. Other Roles of the SNF1 Complex in Yeast and Filamentous Fungi
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Popper, Z.A.; Michel, G.; Hervé, C.; Domozych, D.S.; Willats, W.G.T.; Tuohy, M.G.; Kloareg, B.; Stengel, D.B. Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants. Annu. Rev. Plant Biol. 2011, 62, 567–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polge, C.; Thomas, M. SNF1/AMPK/SnRK1 Kinases, Global Regulators at the Heart of Energy Control? Trends Plant Sci. 2007, 12, 20–28. [Google Scholar] [CrossRef] [PubMed]
- Ronne, H. Glucose Repression in Fungi. Trends Genet. 1995, 11, 12–17. [Google Scholar] [CrossRef]
- Rødkaer, S.V.; Faergeman, N.J. Glucose- and Nitrogen Sensing and Regulatory Mechanisms in Saccharomyces Cerevisiae. FEMS Yeast Res. 2014, 14, 683–696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, S.-P.; Carlson, M. Regulation of Snf1 Protein Kinase in Response to Environmental Stress. J. Biol. Chem. 2007, 282, 16838–16845. [Google Scholar] [CrossRef] [Green Version]
- Tonukari, N.J.; Scott-Craig, J.S.; Walton, J.D. Isolation of the Carbon Catabolite Repressor (CREA) Gene from the Plant-Pathogenic Fungus Cochliobolus Carbonum. DNA Seq. 2003, 14, 103–107. [Google Scholar] [CrossRef]
- Cho, Y.; Kim, K.-H.; La Rota, M.; Scott, D.; Santopietro, G.; Callihan, M.; Mitchell, T.K.; Lawrence, C.B. Identification of Novel Virulence Factors Associated with Signal Transduction Pathways in Alternaria Brassicicola. Mol. Microbiol. 2009, 72, 1316–1333. [Google Scholar] [CrossRef]
- Tonukari, N.J.; Scott-Craig, J.S.; Walton, J.D. The Cochliobolus Carbonum SNF1 Gene Is Required for Cell Wall-Degrading Enzyme Expression and Virulence on Maize. Plant Cell 2000, 12, 237. [Google Scholar] [CrossRef]
- Ospina-Giraldo, M.D.; Mullins, E.; Kang, S. Loss of Function of the Fusarium Oxysporum SNF1 Gene Reduces Virulence on Cabbage and Arabidopsis. Curr. Genet. 2003, 44, 49–57. [Google Scholar] [CrossRef]
- Yi, M.; Park, J.-H.; Ahn, J.-H.; Lee, Y.-H. MoSNF1 Regulates Sporulation and Pathogenicity in the Rice Blast Fungus Magnaporthe Oryzae. Fungal Genet. Biol. 2008, 45, 1172–1181. [Google Scholar] [CrossRef]
- Lee, S.-H.; Lee, J.; Lee, S.; Park, E.-H.; Kim, K.-W.; Kim, M.-D.; Yun, S.-H.; Lee, Y.-W. Gz SNF1 Is Required for Normal Sexual and Asexual Development in the Ascomycete Gibberella Zeae. Eukaryot. Cell 2009, 8, 116–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tzima, A.K.; Paplomatas, E.J.; Rauyaree, P.; Ospina-Giraldo, M.D.; Kang, S. VdSNF1, the Sucrose Nonfermenting Protein Kinase Gene of Verticillium Dahliae, Is Required for Virulence and Expression of Genes Involved in Cell-Wall Degradation. Mol. Plant-Microbe Interact. 2011, 24, 129–142. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Sun, X.; Xu, Q.; Zhu, C.; Li, Q.; Li, H. PdSNF1, a Sucrose Non-Fermenting Protein Kinase Gene, Is Required for Penicillium Digitatum Conidiation and Virulence. Appl. Microbiol. Biotechnol. 2013, 97, 5433–5445. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Zhang, H.; Strelkov, S.E.; Hwang, S.-F. The LmSNF1 Gene Is Required for Pathogenicity in the Canola Blackleg Pathogen Leptosphaeria Maculans. PLoS ONE 2014, 9, e92503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Islam, K.T.; Bond, J.P.; Fakhoury, A.M. FvSNF1, the Sucrose Non-Fermenting Protein Kinase Gene of Fusarium Virguliforme, Is Required for Cell-Wall-Degrading Enzymes Expression and Sudden Death Syndrome Development in Soybean. Curr. Genet. 2017, 63, 723–738. [Google Scholar] [CrossRef]
- Zhang, S.; Guo, Y.; Li, S.; Zhou, G.; Liu, J.; Xu, J.; Li, H. Functional Analysis of CfSnf1 in the Development and Pathogenicity of Anthracnose Fungus Colletotrichum Fructicola on Tea-Oil Tree. BMC Genet. 2019, 20, 94. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; Lv, W.; Zhang, Q.; Zhou, C. Coding the α-Subunit of SNF1 Kinase, Snf1 Is Required for the Conidiogenesis and Pathogenicity of the Alternaria Alternata Tangerine Pathotype. Fungal Biol. 2020, 124, 562–570. [Google Scholar] [CrossRef]
- Wang, X.-X.; He, P.-H.; Feng, M.-G.; Ying, S.-H. BbSNF1 Contributes to Cell Differentiation, Extracellular Acidification, and Virulence in Beauveria Bassiana, a Filamentous Entomopathogenic Fungus. Appl. Microbiol. Biotechnol. 2014, 98, 8657–8673. [Google Scholar] [CrossRef]
- Ming, Y.; Wei, Q.; Jin, K.; Xia, Y. MaSnf1, a Sucrose Non-Fermenting Protein Kinase Gene, Is Involved in Carbon Source Utilization, Stress Tolerance, and Virulence in Metarhizium Acridum. Appl. Microbiol. Biotechnol. 2014, 98, 10153–10164. [Google Scholar] [CrossRef]
- Wāng, Y.; Wang, R.; Wáng, Y.; Li, Y.; Yang, R.-H.; Gong, M.; Shang, J.-J.; Zhang, J.-S.; Mao, W.-J.; Zou, G.; et al. Diverse Function and Regulation of CmSnf1 in Entomopathogenic Fungus Cordyceps Militaris. Fungal Genet. Biol. 2020, 142, 103415. [Google Scholar] [CrossRef]
- Galarza, L.; Akagi, Y.; Takao, K.; Peralta, E.; Santos, E.; Kodama, M. Involvement of ThSNF1 in the Development and Virulence of Biocontrol Agent Trichoderma Harzianum. J. Gen. Plant Pathol. 2015, 81, 211–217. [Google Scholar] [CrossRef]
- Nadal, M.; Garcia-Pedrajas, M.D.; Gold, S.E. The Snf1 Gene of Ustilago Maydis Acts as a Dual Regulator of Cell Wall Degrading Enzymes. Phytopathology 2010, 100, 1364–1372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yan, P.; Lu, X.; Qiu, Y.; Liang, S.; Liu, G.; Li, S.; Mou, L.; Xie, N. Involvement of PaSNF1 in Fungal Development, Sterigmatocystin Biosynthesis, and Lignocellulosic Degradation in the Filamentous Fungus Podospora Anserina. Front. Microbiol. 2020, 11, 1038. [Google Scholar] [CrossRef] [PubMed]
- Casamayor, A.; Serrano, R.; Platara, M.; Casado, C.; Ruiz, A.; Ariño, J. The Role of the Snf1 Kinase in the Adaptive Response of Saccharomyces Cerevisiae to Alkaline PH Stress. Biochem. J. 2012, 444, 39–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, X.-Q.; Chen, G.-Q.; Liu, X.-H.; Dong, B.; Shi, H.-B.; Lu, J.-P.; Lin, F. Crosstalk between SNF1 Pathway and the Peroxisome-Mediated Lipid Metabolism in Magnaporthe Oryzae. PLoS ONE 2014, 9, e103124. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Li, Y.; Wang, H.; Wei, D.; Akhberdi, O.; Liu, Y.; Xiang, B.; Hao, X.; Zhu, X. The AMP-Activated Protein Kinase Homolog Snf1 Concerts Carbon Utilization, Conidia Production and the Biosynthesis of Secondary Metabolites in the Taxol-Producer Pestalotiopsis Microspora. Genes 2018, 9, 59. [Google Scholar] [CrossRef] [Green Version]
- Amselem, J.; Cuomo, C.A.; van Kan, J.A.L.; Viaud, M.; Benito, E.P.; Couloux, A.; Coutinho, P.M.; de Vries, R.P.; Dyer, P.S.; Fillinger, S.; et al. Genomic Analysis of the Necrotrophic Fungal Pathogens Sclerotinia Sclerotiorum and Botrytis Cinerea. PLoS Genet. 2011, 7, e1002230. [Google Scholar] [CrossRef] [Green Version]
- Kelloniemi, J.; Trouvelot, S.; Héloir, M.-C.; Simon, A.; Dalmais, B.; Frettinger, P.; Cimerman, A.; Fermaud, M.; Roudet, J.; Baulande, S.; et al. Analysis of the Molecular Dialogue Between Gray Mold (Botrytis Cinerea) and Grapevine (Vitis Vinifera) Reveals a Clear Shift in Defense Mechanisms During Berry Ripening. Mol. Plant-Microbe Interact. 2015, 28, 1167–1180. [Google Scholar] [CrossRef] [Green Version]
- Choquer, M.; Fournier, E.; Kunz, C.; Levis, C.; Pradier, J.-M.; Simon, A.; Viaud, M. Botrytis Cinerea Virulence Factors: New Insights into a Necrotrophic and Polyphageous Pathogen. FEMS Microbiol. Lett. 2007, 277, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Fillinger, S.; Elad, Y. (Eds.) Botrytis–The Fungus, the Pathogen and Its Management in Agricultural Systems; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-23370-3. [Google Scholar]
- Have, A.T.; Mulder, W.; Visser, J.; van Kan, J.A.L. The Endopolygalacturonase Gene Bcpg1 Is Required for Full Virulence of Botrytis cinerea. Mol. Plant-Microbe Interact. 1998, 11, 1009–1016. [Google Scholar] [CrossRef] [Green Version]
- Wubben, J.P.; Have, A.T.; van Kan, J.A.L.; Visser, J. Regulation of Endopolygalacturonase Gene Expression in Botrytis Cinerea by Galacturonic Acid, Ambient PH and Carbon Catabolite Repression. Curr. Genet. 2000, 37, 152–157. [Google Scholar] [CrossRef] [PubMed]
- Have, A.T.; Breuil, W.O.; Wubben, J.P.; Visser, J.; van Kan, J.A.L. Botrytis Cinerea Endopolygalacturonase Genes Are Differentially Expressed in Various Plant Tissues. Fungal Genet. Biol. 2001, 33, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Poinssot, B.; Vandelle, E.; Bentéjac, M.; Adrian, M.; Levis, C.; Brygoo, Y.; Garin, J.; Sicilia, F.; Coutos-Thévenot, P.; Pugin, A. The Endopolygalacturonase 1 from Botrytis Cinerea Activates Grapevine Defense Reactions Unrelated to Its Enzymatic Activity. Mol. Plant-Microbe Interact. 2003, 16, 553–564. [Google Scholar] [CrossRef] [Green Version]
- Valette-Collet, O.; Cimerman, A.; Reignault, P.; Levis, C.; Boccara, M. Disruption of Botrytis Cinerea Pectin Methylesterase Gene Bcpme1 Reduces Virulence on Several Host Plants. Mol. Plant Microbe Interact. 2003, 16, 360–367. [Google Scholar] [CrossRef] [Green Version]
- Kars, I.; van Kan, J.A.L. Extracellular Enzymes and Metabolites Involved in Pathogenesis of Botrytis. In Botrytis: Biology, Pathology and Control; Elad, Y., Williamson, B., Tudzynski, P., Delen, N., Eds.; Springer: Dordrecht, The Netherlands, 2007; pp. 99–118. ISBN 978-1-4020-2624-9. [Google Scholar]
- Brito, N.; Espino, J.J.; González, C. The Endo-β-1,4-Xylanase Xyn11A Is Required for Virulence in Botrytis Cinerea. Mol. Plant-Microbe Interact. 2006, 19, 25–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noda, J.; Brito, N.; González, C. The Botrytis Cinerea Xylanase Xyn11A Contributes to Virulence with Its Necrotizing Activity, Not with Its Catalytic Activity. BMC Plant Biol. 2010, 10, 38. [Google Scholar] [CrossRef] [Green Version]
- García, N.; González, M.A.; González, C.; Brito, N. Simultaneous Silencing of Xylanase Genes in Botrytis Cinerea. Front. Plant Sci. 2017, 8, 2174. [Google Scholar] [CrossRef] [PubMed]
- Choquer, M.; Rascle, C.; Gonçalves, I.R.; Vallée, A.; Ribot, C.; Loisel, E.; Smilevski, P.; Ferria, J.; Savadogo, M.; Souibgui, E.; et al. The Infection Cushion of Botrytis Cinerea: A Fungal ‘Weapon’ of Plant-biomass Destruction. Environ. Microbiol. 2021, 23, 2293–2314. [Google Scholar] [CrossRef] [PubMed]
- De Vallée, A.; Bally, P.; Bruel, C.; Chandat, L.; Choquer, M.; Dieryckx, C.; Dupuy, J.W.; Kaiser, S.; Latorse, M.-P.; Loisel, E.; et al. A Similar Secretome Disturbance as a Hallmark of Non-Pathogenic Botrytis Cinerea ATMT-Mutants? Front. Microbiol. 2019, 10, 2829. [Google Scholar] [CrossRef] [PubMed]
- Souibgui, E.; Bruel, C.; Choquer, M.; de Vallée, A.; Dieryckx, C.; Dupuy, J.W.; Latorse, M.-P.; Rascle, C.; Poussereau, N. Clathrin Is Important for Virulence Factors Delivery in the Necrotrophic Fungus Botrytis Cinerea. Front. Plant Sci. 2021, 12, 668937. [Google Scholar] [CrossRef]
- Billon-Grand, G.; Rascle, C.; Droux, M.; Rollins, J.A.; Poussereau, N. PH Modulation Differs during Sunflower Cotyledon Colonization by the Two Closely Related Necrotrophic Fungi Botrytis Cinerea and Sclerotinia Sclerotiorum: Botrytis and Sclerotinia Differ in PH Modulation. Mol. Plant Pathol. 2012, 13, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Catlett, N.L.; Lee, B.-N.; Yoder, O.C.; Turgeon, B.G. Split-Marker Recombination for Efficient Targeted Deletion of Fungal Genes. Fungal Genet. Rep. 2003, 50, 9–11. [Google Scholar] [CrossRef] [Green Version]
- Patel, R.M.; Heneghan, M.N.; van Kan, J.A.L.; Bailey, A.M.; Foster, G.D. The POT and PLOB Vector Systems: Improving Ease of Transgene Expression in Botrytis Cinerea. J. Gen. Appl. Microbiol. 2008, 54, 367–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.-H.; Hamari, Z.; Han, K.-H.; Seo, J.-A.; Reyes-Domínguez, Y.; Scazzocchio, C. Double-Joint PCR: A PCR-Based Molecular Tool for Gene Manipulations in Filamentous Fungi. Fungal Genet. Biol. 2004, 41, 973–981. [Google Scholar] [CrossRef] [PubMed]
- Lalève, A.; Gamet, S.; Walker, A.-S.; Debieu, D.; Toquin, V.; Fillinger, S. Site-Directed Mutagenesis of the P225, N230 and H272 Residues of Succinate Dehydrogenase Subunit B from Botrytis Cinerea Highlights Different Roles in Enzyme Activity and Inhibitor Binding: Roles of Key SdhB Residues in SDH Activity and Inhibition. Environ. Microbiol. 2014, 16, 2253–2266. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Lever, M. A New Reaction for Colorimetric Determination of Carbohydrates. Anal. Biochem. 1972, 47, 273–279. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient Requirements of Suspension Cultures of Soybean Root Cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Reid, K.E.; Olsson, N.; Schlosser, J.; Peng, F.; Lund, S.T. An Optimized Grapevine RNA Isolation Procedure and Statistical Determination of Reference Genes for Real-Time RT-PCR during Berry Development. BMC Plant Biol. 2006, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- He, P.-H.; Wang, X.-X.; Chu, X.-L.; Feng, M.-G.; Ying, S.-H. RNA Sequencing Analysis Identifies the Metabolic and Developmental Genes Regulated by BbSNF1 during Conidiation of the Entomopathogenic Fungus Beauveria Bassiana. Curr. Genet. 2015, 61, 143–152. [Google Scholar] [CrossRef]
- Serra-Cardona, A.; Canadell, D.; Arino, J. Coordinate Responses to Alkaline PH Stress in Budding Yeast. Microb. Cell 2015, 2, 182–196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rascle, C.; Dieryckx, C.; Dupuy, J.W.; Muszkieta, L.; Souibgui, E.; Droux, M.; Bruel, C.; Girard, V.; Poussereau, N. The PH Regulator PacC: A Host-Dependent Virulence Factor in Botrytis Cinerea: The PH Regulator PacC. Environ. Microbiol. Rep. 2018, 10, 555–568. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Yang, X.; Lin, X.; Jiang, H.-Y.; Hu, X.-P.; Liu, S.-X. Effect of Overexpression of SNF1 on the Transcriptional and Metabolic Landscape of Baker’s Yeast under Freezing Stress. Microb. Cell Factories 2021, 20, 10. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.C. Beta-Subunits of Snf1 Kinase Are Required for Kinase Function and Substrate Definition. EMBO J. 2000, 19, 4936–4943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Son, H.; Park, A.R.; Lee, S.-H.; Choi, G.J.; Kim, J.-C.; Lee, Y.-W. Functional Characterization of Sucrose Non-Fermenting 1 Protein Kinase Complex Genes in the Ascomycete Fusarium Graminearum. Curr. Genet. 2014, 60, 35–47. [Google Scholar] [CrossRef]

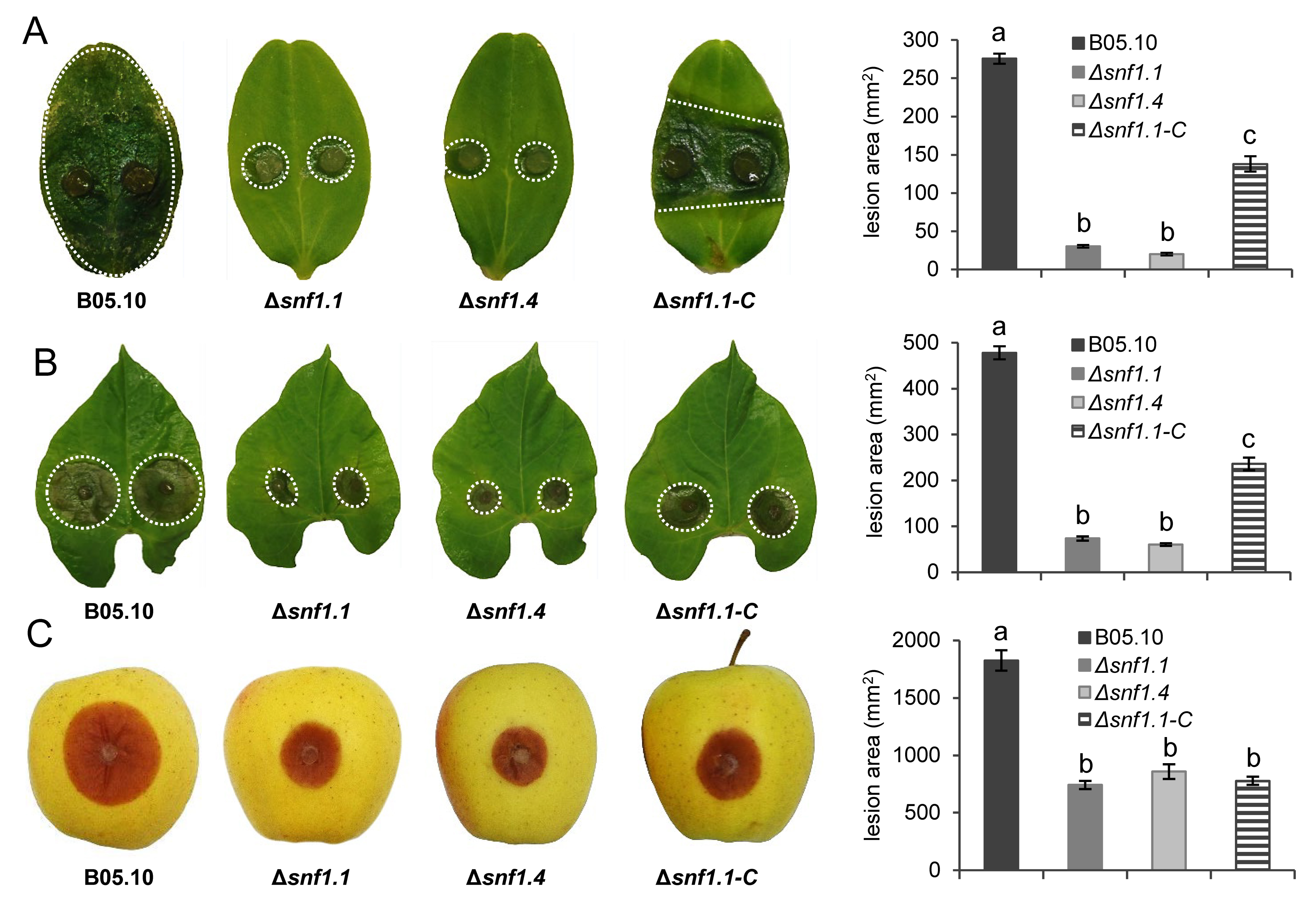
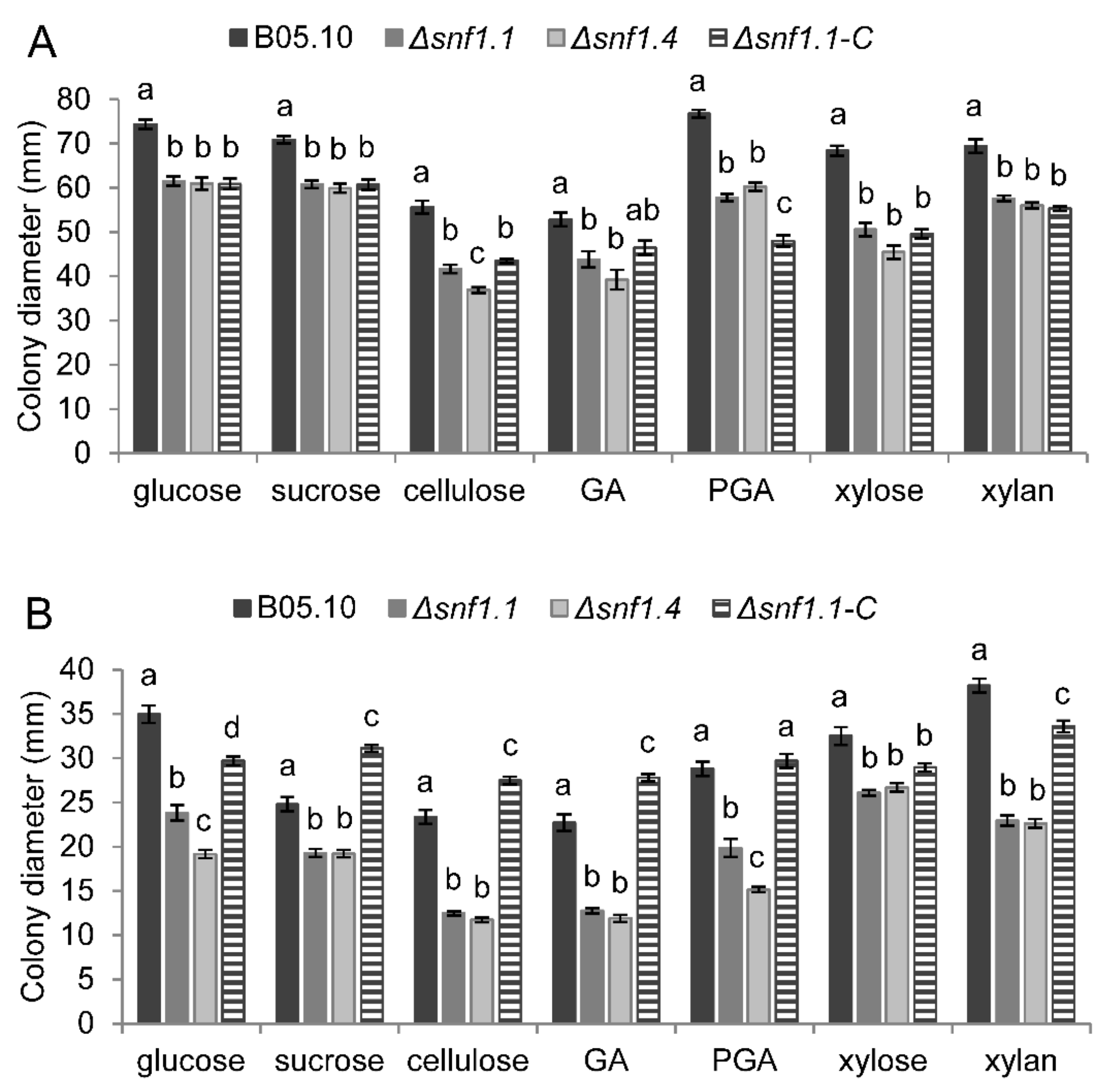
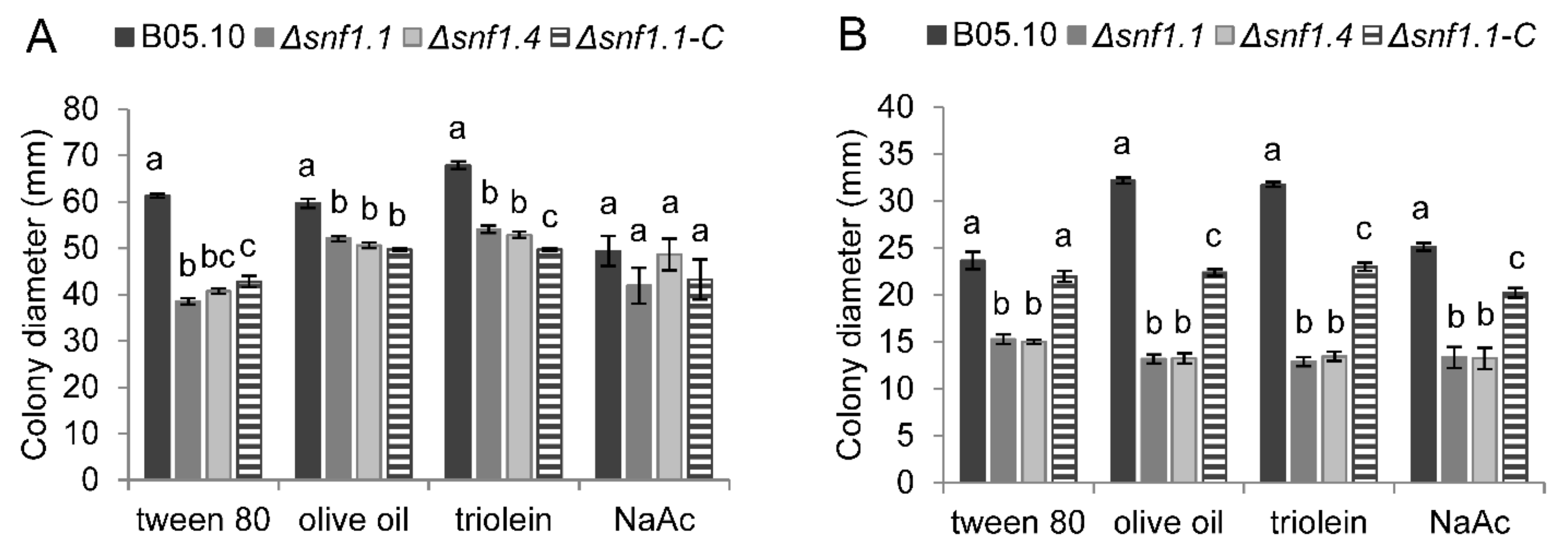
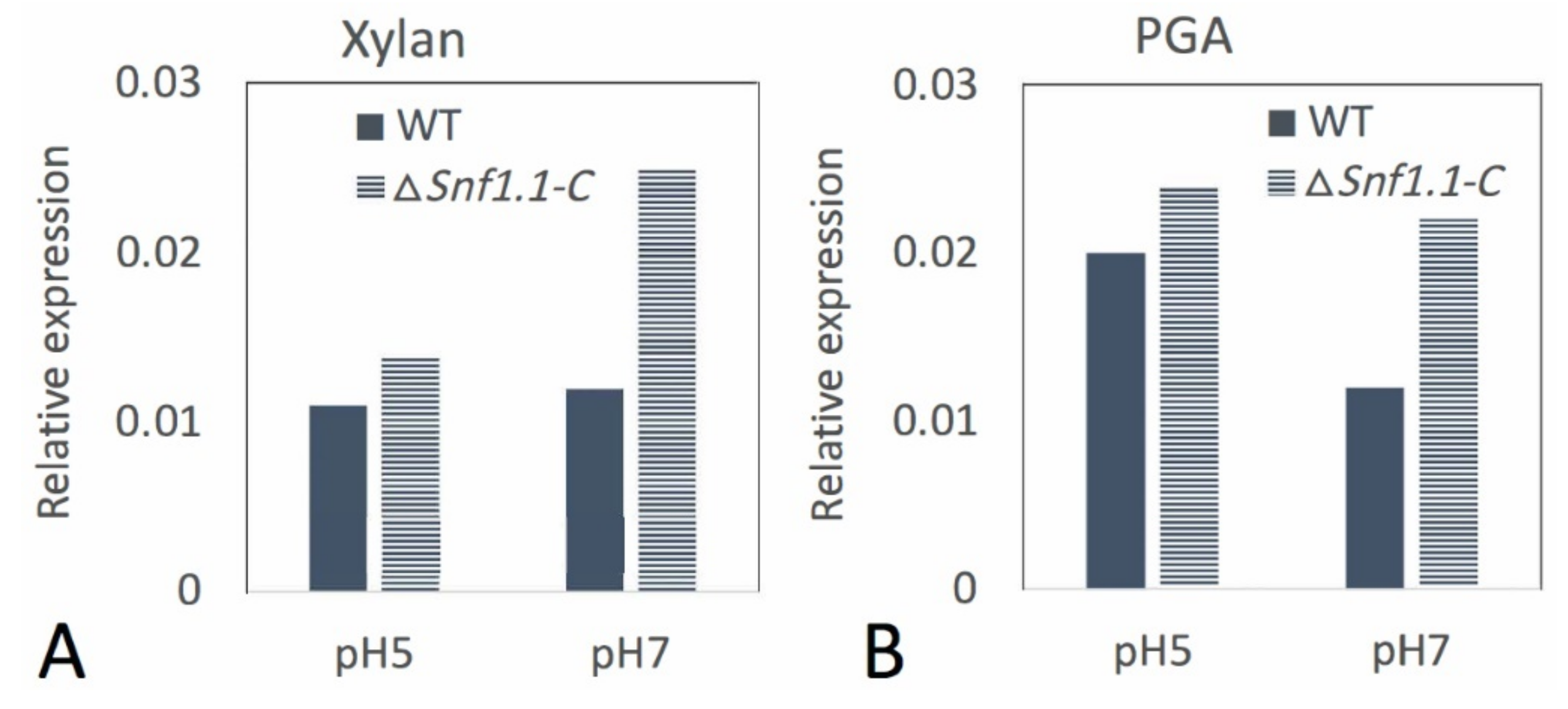
| Strain | 1 Dpi | 2 Dpi | 3 Dpi |
|---|---|---|---|
| Wild type | 4.95 | 5.58 | 6.78 |
| Δsnf1.1 | 4.85 | 7.03 | 7.53 |
| Δsnf1.4 | 4.64 | 6.56 | 7.39 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lengyel, S.; Rascle, C.; Poussereau, N.; Bruel, C.; Sella, L.; Choquer, M.; Favaron, F. Snf1 Kinase Differentially Regulates Botrytis cinerea Pathogenicity according to the Plant Host. Microorganisms 2022, 10, 444. https://doi.org/10.3390/microorganisms10020444
Lengyel S, Rascle C, Poussereau N, Bruel C, Sella L, Choquer M, Favaron F. Snf1 Kinase Differentially Regulates Botrytis cinerea Pathogenicity according to the Plant Host. Microorganisms. 2022; 10(2):444. https://doi.org/10.3390/microorganisms10020444
Chicago/Turabian StyleLengyel, Szabina, Christine Rascle, Nathalie Poussereau, Christophe Bruel, Luca Sella, Mathias Choquer, and Francesco Favaron. 2022. "Snf1 Kinase Differentially Regulates Botrytis cinerea Pathogenicity according to the Plant Host" Microorganisms 10, no. 2: 444. https://doi.org/10.3390/microorganisms10020444
APA StyleLengyel, S., Rascle, C., Poussereau, N., Bruel, C., Sella, L., Choquer, M., & Favaron, F. (2022). Snf1 Kinase Differentially Regulates Botrytis cinerea Pathogenicity according to the Plant Host. Microorganisms, 10(2), 444. https://doi.org/10.3390/microorganisms10020444






