Discrimination between the Two Closely Related Species of the Operational Group B. amyloliquefaciens Based on Whole-Cell Fatty Acid Profiling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacillus Strains and Growth Conditions
2.2. PCR Amplification of the gyrA and rpoB Genes
2.3. Phylogenetic Analysis
2.4. The Fatty Acid Methyl Ester (FAME) Analysis
2.5. Sample Pretreatment
2.6. Statistical Analysis
3. Results
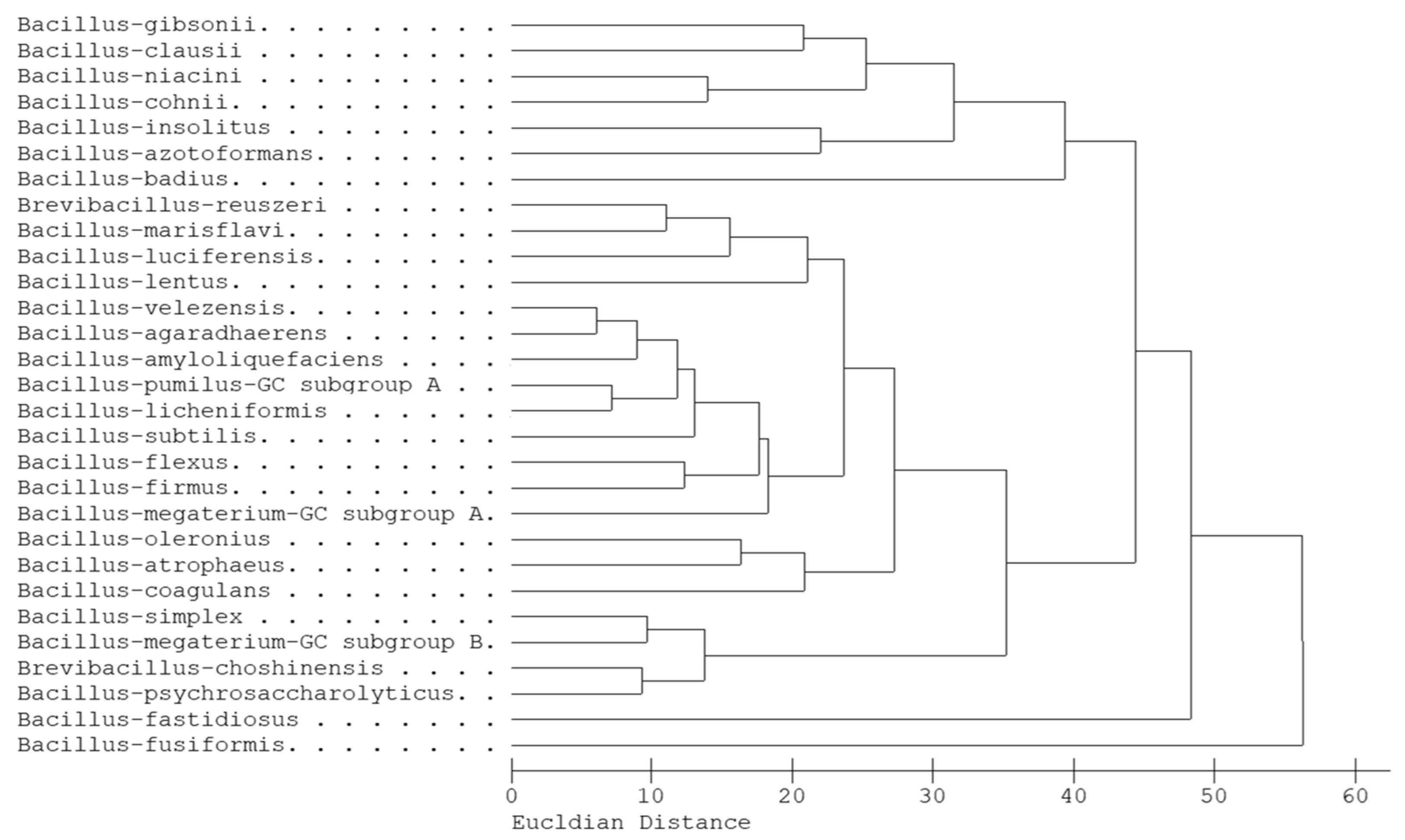
3.1. The Classification of B. velezensis and B. amyloliquefaciens Based on Molecular Markers
3.2. FAME Profiles of B. velezensis and B. amyloliquefaciens Strains
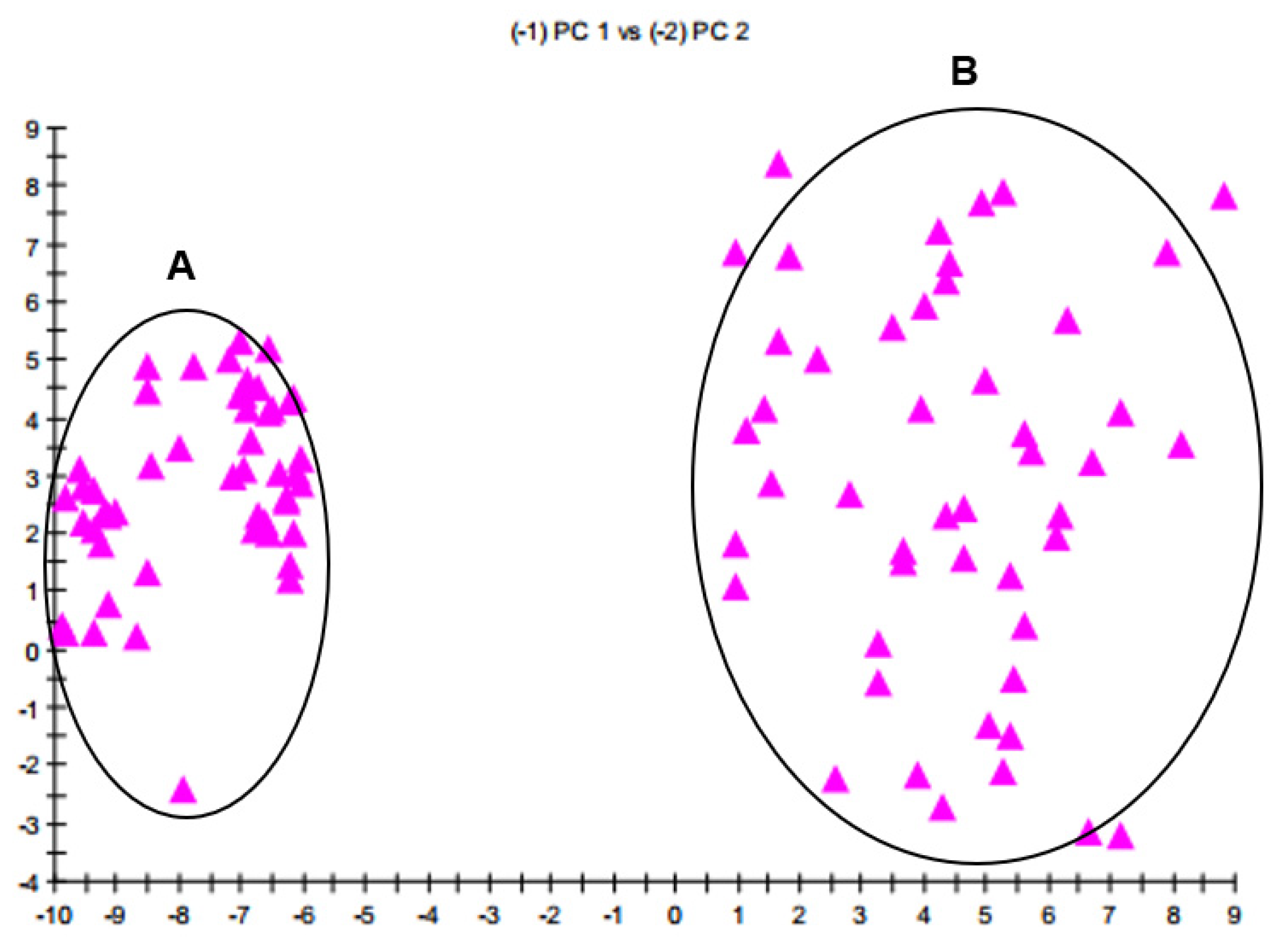
3.3. Differentiation of FAME Profiles between B. velezensis and B. amyloliquefaciens
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fan, B.; Blom, J.; Klenk, H.P.; Borriss, R. Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “Operational Group B. amyloliquefaciens” within the B. subtilis species complex. Front. Microbiol. 2017, 8, 22. [Google Scholar] [CrossRef] [Green Version]
- De Carvalho, C.C.C.R.; Caramujo, M.J. The various roles of fatty acids. Molecules 2018, 23, 2583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, J.; Nam, J.; Seo, M.-H. Complete genome sequence of Bacillus velezensis NST6 and comparison with the species belonging to operational group B. amyloliquefaciens. Genomics 2021, 113, 380–386. [Google Scholar] [CrossRef] [PubMed]
- da Costa, M.S.; Albuquerque, L.; Nobre, M.F.; Wait, R. The identification of fatty acids in bacteria. In Methods in Microbiology; Elsevier Ltd.: Amsterdam, The Netherlands, 2011; Volume 38, pp. 183–196. [Google Scholar]
- Diomandé, S.E.; Nguyen-The, C.; Guinebretière, M.H.; Broussolle, V.; Brillard, J. Role of fatty acids in Bacillus environmental adaptation. Front. Microbiol. 2015, 6, 813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rabbee, M.; Ali, M.S.; Choi, J.; Hwang, B.S.; Jeong, S.C.; Baek, K. Bacillus velezensis: A valuable member of bioactive molecules within plant microbiomes. Molecules 2019, 24, 1046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukumoto, J. Studies on the production of bacterial amylase. I. Isolation of bacteria secreting potent amylases and their distribution. J. Agric. Chem. Soc. Jpn. 1943, 19, 487–503. [Google Scholar] [CrossRef]
- Priest, F.G.; Goodfellow, M.; Shute, L.A.; Berkeley, W. Bacillus amyloliquefaciens sp. nov., nom. rev. Int. J. Syst. Bacteriol. 1987, 37, 69–71. [Google Scholar] [CrossRef]
- Ruiz-García, C.; Béjar, V.; Martínez-Checa, F.; Llamas, I.; Quesada, E. Bacillus velezensis sp. nov., a surfactant-producing bacterium isolated from the river Vélez in Málaga, southern Spain. Int. J. Syst. Evol. Microbiol. 2005, 55, 191–195. [Google Scholar] [CrossRef] [Green Version]
- Fan, B.; Wang, C.; Song, X.; Ding, X.; Wu, L.; Wu, H.; Gao, X.; Borriss, R. Bacillus velezensis FZB42 in 2018: The Gram-positive model strain for plant growth promotion and biocontrol. Front. Microbiol. 2018, 9, 2491. [Google Scholar] [CrossRef] [Green Version]
- Grady, E.N.; MacDonald, J.; Ho, M.T.; Weselowski, B.; McDowell, T.; Solomon, O.; Renaud, J.; Yuan, Z.C. Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6. BMC Microbiol. 2019, 19, 5. [Google Scholar] [CrossRef]
- Borriss, R.; Chen, X.-H.; Rückert, C.; Blom, J.; Becker, A.; Baumgarth, B.; Fan, B.; Pukall, R.; Schumann, P.; Spröer, C.; et al. Relationship of Bacillus amyloliquefaciens clades associated with strains DSM 7T and FZB42T: A proposal for Bacillus amyloliquefaciens subsp. amyloliquefaciens subsp. nov. and Bacillus amyloliquefaciens subsp. plantarum subsp. nov. based on complete genome sequence comparisons. Int. J. Syst. Evol. Microbiol. 2011, 61, 1786–1801. [Google Scholar] [CrossRef] [PubMed]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. Plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenom. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [CrossRef]
- Wang, L.T.; Lee, F.L.; Tai, C.J.; Kuo, H.P. Bacillus velezensis is a later heterotypic synonym of Bacillus amyloliquefaciens. Int. J. Syst. Evol. Microbiol. 2008, 58, 671–675. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Tian, Y.; Jia, M.; Lu, X.; Yue, L.; Zhao, X.; Jin, W.; Wang, Y.; Zhang, Y.; Xie, Z.; et al. Induction of salt tolerance in Arabidopsis thaliana by volatiles from Bacillus amyloliquefaciens FZB42 via the jasmonic acid signaling pathway. Front. Microbiol. 2020, 11, 562934. [Google Scholar] [CrossRef]
- Molina-Santiago, C.; Vela-Corcía, D.; Petras, D.; Díaz-Martínez, L.; Pérez-Lorente, A.I.; Sopeña-Torres, S.; Pearson, J.; Caraballo-Rodríguez, A.M.; Dorrestein, P.C.; de Vicente, A.; et al. Chemical interplay and complementary adaptative strategies toggle bacterial antagonism and co-existence. Cell Rep. 2021, 36, 109449. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Phylogenomic analysis shows that Bacillus amyloliquefaciens subsp. plantarum is a later heterotypic synonym of Bacillus methylotrophicus. Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 7, 2104–2109. [Google Scholar] [CrossRef]
- Liu, G.; Kong, Y.; Fan, Y.; Geng, C.; Peng, D.; Sun, M. Data on genome analysis of Bacillus velezensis LS69. Data Br. 2017, 13, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Reva, O.N.; Dixelius, C.; Meijer, J.; Priest, F.G. Taxonomic characterization and plant colonizing abilities of some bacteria related to Bacillus amyloliquefaciens and Bacillus subtilis. FEMS Microbiol. Ecol. 2004, 48, 249–259. [Google Scholar] [CrossRef]
- Reva, O.N.; Swanevelder, D.Z.H.; Mwita, L.A.; Mwakilili, A.D.; Muzondiwa, D.; Joubert, M.; Chan, W.Y.; Lutz, S.; Ahrens, C.H.; Avdeeva, L.V.; et al. Genetic, epigenetic and phenotypic diversity of four Bacillus velezensis strains used for plant protection or as probiotics. Front. Microbiol. 2019, 10, 2610. [Google Scholar] [CrossRef] [Green Version]
- MIDI Inc. The Sherlock Chromatographic Analysis System Operating Manual, 6th ed.; MIDI, Inc.: Newark, DE, USA, 2018. [Google Scholar]
- Vágvölgyi, C.; Sajben-Nagy, E.; Bóka, B.; Vörös, M.; Berki, A.; Palágyi, A.; Krisch, J.; Škrbić, B.; Đurišić-Mladenović, N.; Manczinger, L. Isolation and characterization of antagonistic Bacillus strains capable to degrade ethylenethiourea. Curr. Microbiol. 2013, 66, 243–250. [Google Scholar] [CrossRef]
- Stefanic, P.; Mandic-Mulec, I. Social interactions and distribution of Bacillus subtilis pherotypes at microscale. J. Bacteriol. 2009, 191, 1756–1764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Roberts, M.S.; Nakamura, L.K.; Cohan, F.M. Bacillus mojavensis sp. nov., distinguishable from Bacillus subtilis by sexual isolation, divergence in DNA sequence, and differences in fatty acid composition. Int. J. Syst. Bacteriol. 1994, 44, 256–264. [Google Scholar] [CrossRef] [Green Version]
- Slabbinck, B.; De Baets, B.; Dawyndt, P.; De Vos, P. Genus-wide Bacillus species identification through proper artificial neural network experiments on fatty acid profiles. Antonie Van Leeuwenhoek 2008, 94, 187–198. [Google Scholar] [CrossRef]
- Truong, T.V.; Nackos, A.N.; Williams, J.R.; Vanderwerken, D.N.; Kimball, J.A.; Murray, J.A.; Hawkes, J.E.; Harvey, D.J.; Tolley, H.D.; Robison, R.A.; et al. Differentiation of Bacillus endospore species from fatty acid methyl ester biomarkers. Anal. Methods 2010, 2, 638–644. [Google Scholar] [CrossRef]
- Perez, K.J.; dos Viana, J.S.; Lopes, F.C.; Pereira, J.Q.; dos Santos, D.M.; Oliveira, J.S.; Velho, R.V.; Crispim, S.M.; Nicoli, J.R.; Brandelli, A.; et al. Bacillus spp. isolated from puba as a source of biosurfactants and antimicrobial lipopeptides. Front. Microbiol. 2017, 8, 61. [Google Scholar] [CrossRef] [Green Version]


| Characteristics a | B. velezensisb | B. amyloliquefaciensb | References |
|---|---|---|---|
| Pigmentation | Creamy white | Creamy white | [9] |
| Oxidase | + | + | |
| Acid in API system from: | |||
| - Glycogen | + | nd | |
| - Lactose | + | + | |
| - Melibiose | − | + | |
| - Methyl α-ᴅ-glycoside | + | + | |
| - ᴅ-Raffinose | + | + | |
| - ᴅ-Turanose | − | + | |
| Hydrolysis of | |||
| - Tween 20 | − | + | |
| - Tween 80 | − | nd | |
| - DNA | − | − | |
| Arginine dihydrolase | − | − | |
| ONPG | + | − | |
| Non-ribosomally synthesized secondary metabolites | [1,10,12] | ||
| - Surfactin | + | + | |
| - Macrolactin | + | − | |
| - Bacillaene | + | + | |
| - Fengycin | + | − | |
| - Difficidin | + | − | |
| - Bacillibactin | + | + | |
| - Bacilysin | + | + | |
| Ribosomally synthesized antimicrobial compounds | |||
| - Sublancin | − | − | |
| - Subtilosin | − | − | |
| - Amylocyclicin | + | + | |
| - Plantazolicin | + | − | |
| Plant colonization | + | − | [12] |
| AZCL-HE cellulose liquefaction | + | − | |
| Growth in lactose minimal medium | + | + | |
| Amylase AmyA | − | + | |
| Amylase AmyE | + | − | |
| Cellulase BglC | + | − | |
| Xylanase XynA | + | − | |
| 16S rRNA gene sequence similarity (%) | 99.7% | [14] | |
| rpoB gene sequence similarity (%) | >98% | [1] | |
| gyrB gene sequence similarity (%) | >95.5% | [14] | |
| DNA relatedness value (%) | 74–84% | [14] | |
| Strains a | Genbank Accession Number | Origin | |
|---|---|---|---|
| gyrA | rpoB | ||
| B. velezensis | |||
| SZMC 24980 | OK256097 | OK256115 | soil sample from pepper field, Totovo selo, Serbia |
| SZMC 24981 | OK256098 | OK256116 | soil sample from pepper field, Totovo selo, Serbia |
| SZMC 24982 | OK256099 | OK256117 | soil sample from pepper field, Totovo selo, Serbia |
| SZMC 24983 | OK256100 | OK256118 | soil sample from pepper field, Totovo selo, Serbia |
| SZMC 24984 | OK256101 | OK256119 | soil sample from pepper field, Cantavir, Serbia |
| SZMC 24985 | OK256102 | OK256120 | soil sample from pepper field, Cantavir, Serbia |
| SZMC 24986 | OK256103 | OK256121 | soil sample from tomato field, Cantavir, Serbia |
| SZMC 24995 | OK256104 | OK256122 | soil sample from tomato field, Cantavir, Serbia |
| SZMC 25020 | OK256105 | OK256123 | soil sample from tomato field, Cenej, Serbia |
| SZMC 25646 | OK256106 | OK256124 | pea rhizosphere, Madaras, Hungary |
| SZMC 25647 | OK256107 | OK256125 | pea rhizosphere, Madaras, Hungary |
| SZMC 25610 | OK256108 | OK256126 | maize rhizosphere, Vaszar, Hungary |
| SZMC 6046 | OK256109 | OK256127 | tomato rhizosphere, Hungary |
| SZMC 16093B | OK256110 | OK256128 | tomato rhizosphere, Hungary |
| SZMC 6387J | OK256111 | OK256129 | tomato rhizosphere, Hungary |
| DSM 23117T (=BGSC 10A6 = FZB42 = LMG 26770 = SZMC 27497) | OK256112 | OK256130 | plant pathogen-infested soil of a sugar beet field, Brandenburg, Germany |
| B. amyloliquefaciens | |||
| DSM 7T (=ATCC 23350 = SZMC 6027) | OK256113 | OK256131 | soil and industrial amylase fermentations, Japan |
| DSM 1061T (=IAM 1523 = SZMC 6222) | OK256114 | OK256132 | unknown origin |
| B. subtilis | |||
| ATCC 6633T | CP039755.1 | CP039755.1 | Japan |
| Feature/FA | B. velezensis | B. amyloliquefaciens |
|---|---|---|
| 12:0 | 0.48 ± 0.23 | 0.54 ± 0.17 |
| 13:0 iso | 0.89 ± 0.22 | 0.50 ± 0.19 |
| 14:0 iso | 1.18 ± 0.58 | 1.44 ± 0.11 |
| 14:0 | 2.87 ± 0.70 | 0.61 ± 0.14 |
| 15:0 iso | 30.39 ± 2.53 | 27.84 ± 1.65 |
| 15:0 anteiso | 32.13 ± 2.33 | 31.92 ± 1.98 |
| 16:0 iso | 1.70 ± 0.77 | 3.51 ± 0.19 |
| 16:1 ω11c | 1.65 ± 0.42 | 1.09 ± 0.28 |
| 16:0 | 12.53 ± 1.82 | 4.57 ± 0.55 |
| 17:1 iso ω10c | 0.85 ± 0.47 | 1.07 ± 0.37 |
| 17:0 iso | 8.52 ± 0.96 | 15.92 ± 1.96 |
| 17:0 anteiso | 5.50 ± 0.85 | 8.99 ± 0.73 |
| 18:0 | 0.60 ± 0.14 | 0.59 ± 0.23 |
| Feature | This Study | Ruiz-García et al. [9] | Wang et al. [14] | Borriss et al. [12] | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| B. v.a | B. a.b | B. v. | B. a. | B. v. | B. v. | B. a. | B. a. | B. v. | B. a. | |
| CR-502T | DSM 7T | BCRC 17467T | BCRC 14193 | BCRC 11601T | BCRC 17038 | DSM 23117T | DSM 7T | |||
| 12:0 | 0.48 | 0.54 | - | - | - | - | - | - | - | - |
| 13:0 iso | 0.89 | 0.50 | 0.87 | - | - | - | - | - | 0.31 | 0.38 |
| 14:0 iso | 1.18 | 1.44 | 1.08 | 2.46 | - | 1.3 | 1.5 | 1.7 | 0.43 | 0.99 |
| 14:0 | 2.87 | 0.61 | 2.96 | - | 3.8 | 3.1 | - | - | 1.21 | 0.36 |
| 15:0 iso | 30.39 | 27.84 | 29.86 | 30.50 | 30.4 | 24.0 | 26.3 | 23.7 | 31.00 | 40.29 |
| 15:0 anteiso | 32.13 | 31.92 | 32.70 | 36.48 | 27.6 | 28.7 | 32.3 | 33.8 | 31.73 | 28.32 |
| 16:0 iso | 1.70 | 3.51 | 1.31 | 4.52 | 1.0 | 2.2 | 3.8 | 4.3 | 1.01 | 2.13 |
| 16:1 ω5c | - | - | - | 2.14 | - | - | - | - | - | - |
| 16:1 ω7c | - | - | - | - | - | - | - | - | 0.19 | 0.42 |
| 16:1 ω9c | - | - | - | 0.62 | - | - | - | - | - | - |
| 16:1 ω11c | 1.65 | 1.09 | 4.42 | - | 3.5 | 2.7 | 1.7 | - | 2.59 | 1.23 |
| 16:0 | 12.53 | 4.57 | 13.41 | 4.52 | 18.3 | 19.0 | 5.8 | 7.0 | 7.60 | 3.02 |
| 17:1 iso ω7c | - | - | - | 1.67 | - | - | - | - | - | - |
| 17:1 iso ω10c | 0.85 | 1.07 | 1.44 | - | 1.3 | 1.3 | 1.7 | - | 2.70 | 2.59 |
| 17:0 iso | 8.52 | 15.92 | 7.67 | 9.01 | 7.8 | 10.3 | 16.3 | 17.6 | 12.11 | 13.14 |
| 17:0 anteiso | 5.50 | 8.99 | 4.27 | 7.06 | 3.4 | 5.4 | 9.0 | 10.0 | - | - |
| 17:0 | 0.17 | 0.22 | - | - | - | - | - | - | 7.70 | 6.46 |
| 18:0 | 0.60 | 0.59 | - | - | - | 1.1 | - | - | - | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huynh, T.; Vörös, M.; Kedves, O.; Turbat, A.; Sipos, G.; Leitgeb, B.; Kredics, L.; Vágvölgyi, C.; Szekeres, A. Discrimination between the Two Closely Related Species of the Operational Group B. amyloliquefaciens Based on Whole-Cell Fatty Acid Profiling. Microorganisms 2022, 10, 418. https://doi.org/10.3390/microorganisms10020418
Huynh T, Vörös M, Kedves O, Turbat A, Sipos G, Leitgeb B, Kredics L, Vágvölgyi C, Szekeres A. Discrimination between the Two Closely Related Species of the Operational Group B. amyloliquefaciens Based on Whole-Cell Fatty Acid Profiling. Microorganisms. 2022; 10(2):418. https://doi.org/10.3390/microorganisms10020418
Chicago/Turabian StyleHuynh, Thu, Mónika Vörös, Orsolya Kedves, Adiyadolgor Turbat, György Sipos, Balázs Leitgeb, László Kredics, Csaba Vágvölgyi, and András Szekeres. 2022. "Discrimination between the Two Closely Related Species of the Operational Group B. amyloliquefaciens Based on Whole-Cell Fatty Acid Profiling" Microorganisms 10, no. 2: 418. https://doi.org/10.3390/microorganisms10020418
APA StyleHuynh, T., Vörös, M., Kedves, O., Turbat, A., Sipos, G., Leitgeb, B., Kredics, L., Vágvölgyi, C., & Szekeres, A. (2022). Discrimination between the Two Closely Related Species of the Operational Group B. amyloliquefaciens Based on Whole-Cell Fatty Acid Profiling. Microorganisms, 10(2), 418. https://doi.org/10.3390/microorganisms10020418








