Membrane Efflux Pumps of Pathogenic Vibrio Species: Role in Antimicrobial Resistance and Virulence
Abstract
1. Vibrio Species of Human Health Significance
1.1. Vibrio cholerae
1.2. Vibrio parahaemolyticus
1.3. Vibrio vulnificus
2. Efflux Pumps and Antibiotic Resistance
2.1. General Mechanisms of Antibiotic Efflux Pumps
2.2. Classification of Antimicrobial Efflux Pumps
2.3. Efflux Pumps of RND Family in Vibrio Species
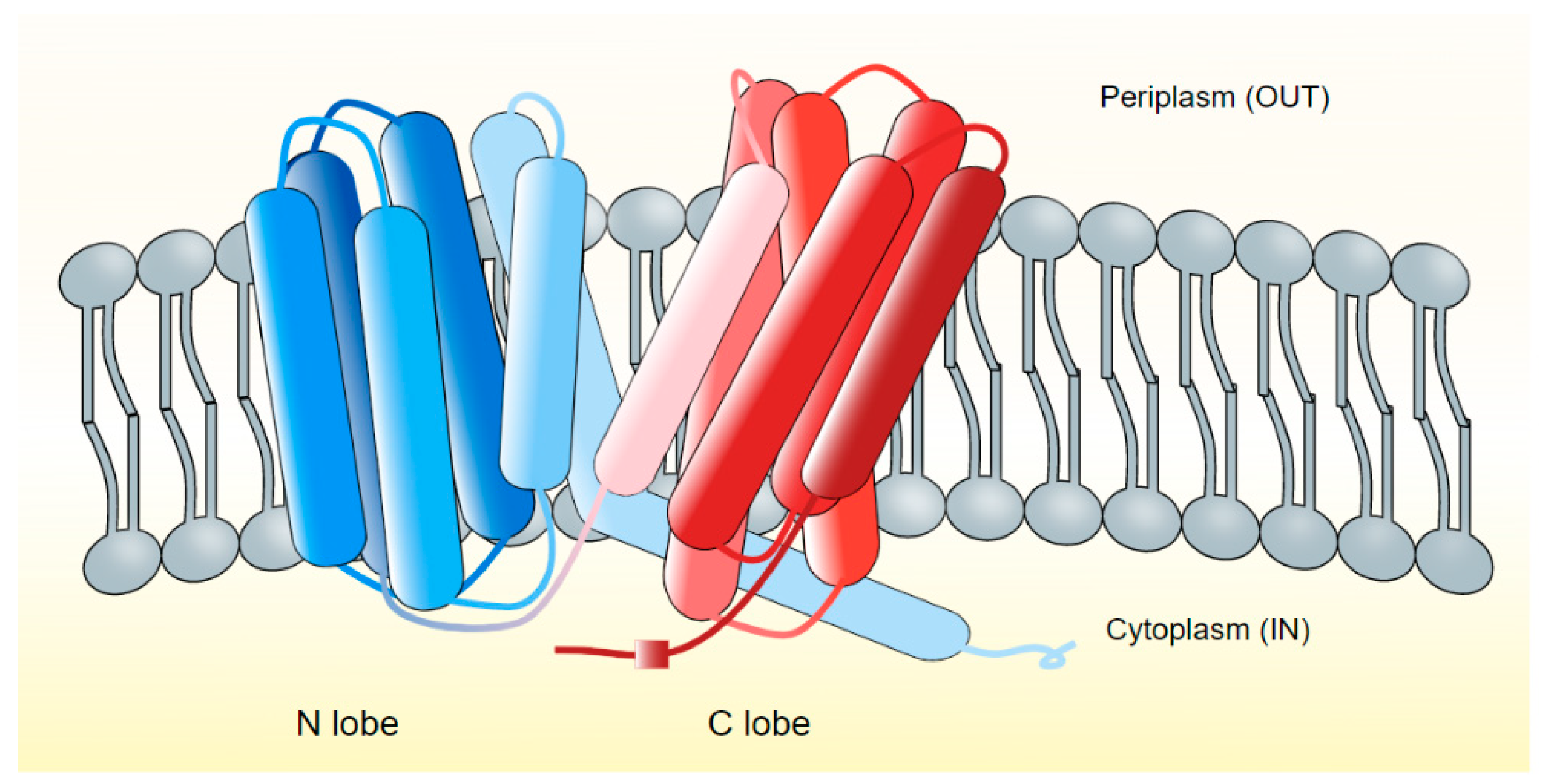
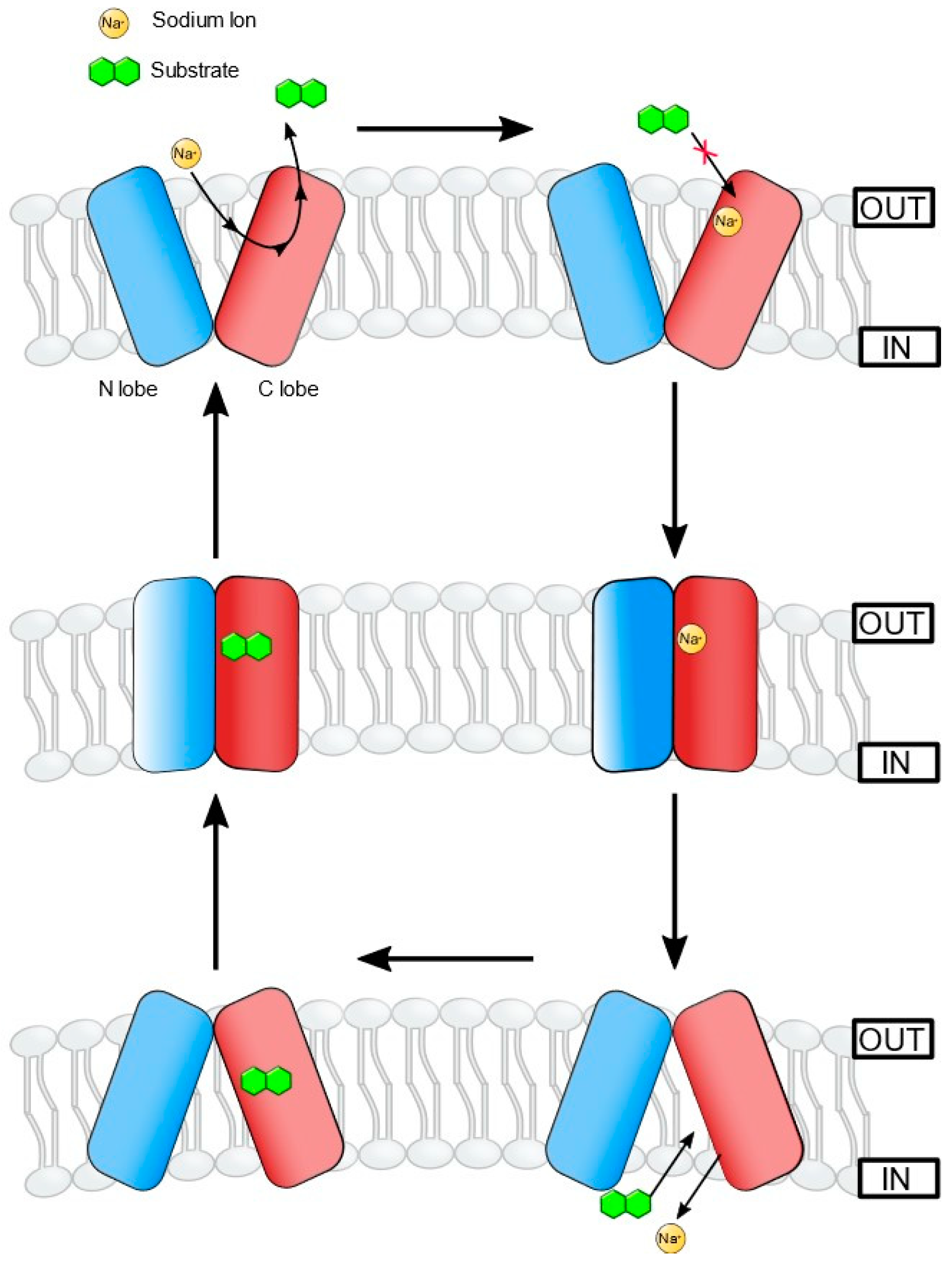
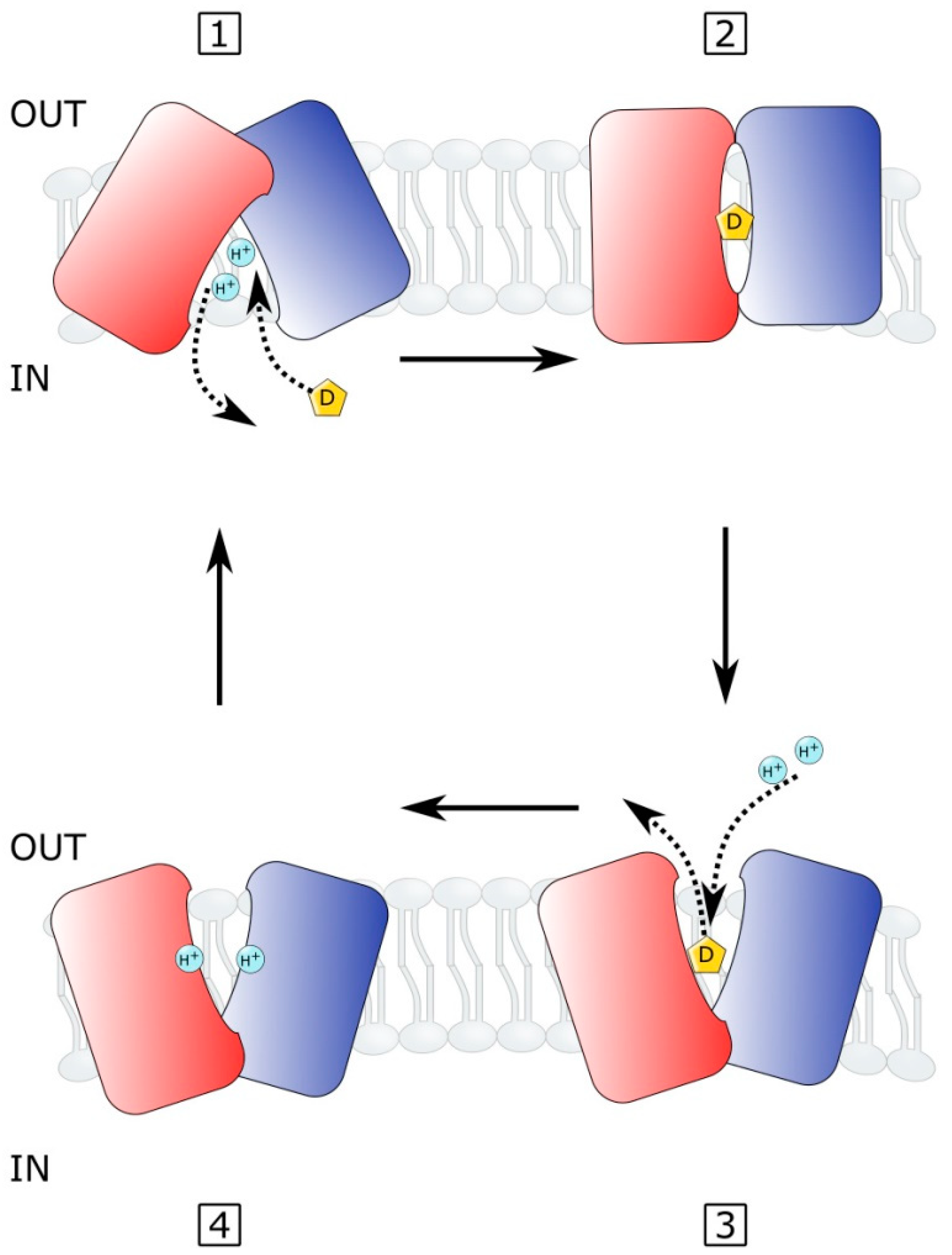
2.4. MATE Efflux Pumps in Vibrio Species
2.5. Efflux Pumps of MFS Family in Vibrio Species
2.6. Efflux Pumps of ABC Superfamily in Vibrio Species
3. Efflux Pumps and Virulence
Role in Quorum Sensing and Biofilm Formation
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Primers 2018, 4, 8. [Google Scholar] [CrossRef] [PubMed]
- CDC. Cholera—Vibrio cholerae infection|Cholera|CDC. Available online: https://www.cdc.gov/cholera/index.html (accessed on 10 October 2019).
- Reidl, J.; Klose, K.E. Vibrio cholerae and cholera: Out of the water and into the host. FEMS Microbiol. Rev. 2002, 26, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Bonnin-Jusserand, M.; Copin, S.; Le Bris, C.; Brauge, T.; Gay, M.; Brisabois, A.; Grard, T.; Midelet-Bourdin, G. Vibrio species involved in seafood-borne outbreaks (Vibrio cholerae, V. parahaemolyticus and V. vulnificus): Review of microbiological versus recent molecular detection methods in seafood products. Crit. Rev. Food Sci. Nutr. 2019, 59, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.; Nelson, A.R.; Lopez, A.L.; Sack, D.A. Updated Global Burden of Cholera in Endemic Countries. PLoS Negl. Trop. Dis. 2015, 9, e0003832. [Google Scholar] [CrossRef]
- Colwell, R.R. Global climate and infectious disease: The cholera paradigm. Science 1996, 274, 2025–2031. [Google Scholar] [CrossRef] [PubMed]
- Sack, D.A.; Lyke, C.; McLaughlin, C.; Suwanvanichkij, V.; World Health Organization. Antimicrobial Resistance in Shigellosis, Cholera and Campylobacteriosis; World Health Organization: Geneva, Switzerland, 2001. [Google Scholar]
- Faruque, S.M.; Mekalanos, J.J. Phage-bacterial interactions in the evolution of toxigenic Vibrio cholerae. Virulence 2012, 3, 556–565. [Google Scholar] [CrossRef] [PubMed]
- Deepanjali, A.; Kumar, H.S.; Karunasagar, I. Seasonal variation in abundance of total and pathogenic Vibrio parahaemolyticus bacteria in oysters along the southwest coast of India. Appl. Environ. Microbiol. 2005, 71, 3575–3580. [Google Scholar] [CrossRef] [PubMed]
- Ghenem, L.; Elhadi, N.; Alzahrani, F.; Nishibuchi, M. Vibrio parahaemolyticus: A Review on Distribution, Pathogenesis, Virulence Determinants and Epidemiology. Saudi J. Med. Med. Sci. 2017, 5, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Nishibuchi, M.; Taniguchi, T.; Misawa, T.; Khaeomanee-Iam, V.; Honda, T.; Miwatani, T. Cloning and nucleotide sequence of the gene (trh) encoding the hemolysin related to the thermostable direct hemolysin of Vibrio parahaemolyticus. Infect. Immun. 1989, 57, 2691–2697. [Google Scholar] [CrossRef] [PubMed]
- Okuda, J.; Ishibashi, M.; Hayakawa, E.; Nishino, T.; Takeda, Y.; Mukhopadhyay, A.K.; Garg, S.; Bhattacharya, S.; Nair, G.B.; Nishibuchi, M. Emergence of a unique O3: K6 clone of Vibrio parahaemolyticus in Calcutta, India, and isolation of strains from the same clonal group from Southeast Asian travelers arriving in Japan. J. Clin. Microbiol. 1997, 35, 3150–3155. [Google Scholar] [CrossRef]
- Tran, L.; Nunan, L.; Redman, R.M.; Mohney, L.L.; Pantoja, C.R.; Fitzsimmons, K.; Lightner, D.V. Determination of the infectious nature of the agent of acute hepatopancreatic necrosis syndrome affecting penaeid shrimp. Dis. Aquat. Org. 2013, 105, 45–55. [Google Scholar] [CrossRef] [PubMed]
- Heng, S.-P.; Letchumanan, V.; Deng, C.-Y.; Ab Mutalib, N.-S.; Khan, T.M.; Chuah, L.-H.; Chan, K.-G.; Goh, B.-H.; Pusparajah, P.; Lee, L.-H. Vibrio vulnificus: An Environmental and Clinical Burden. Front. Microbiol. 2017, 8, 997. [Google Scholar] [CrossRef] [PubMed]
- Parvathi, A.; Kumar, H.S.; Karunasagar, I.; Karunasagar, I. Detection and enumeration of Vibrio vulnificus in oysters from two estuaries along the southwest coast of India, using molecular methods. Appl. Environ. Microbiol. 2004, 70, 6909–6913. [Google Scholar] [CrossRef]
- Huehn, S.; Eichhorn, C.; Urmersbach, S.; Breidenbach, J.; Bechlars, S.; Bier, N.; Alter, T.; Bartelt, E.; Frank, C.; Oberheitmann, B.; et al. Pathogenic vibrios in environmental, seafood and clinical sources in Germany. Int. J. Med. Microbiol. IJMM 2014, 304, 843–850. [Google Scholar] [CrossRef]
- Andersen, J.L.; He, G.X.; Kakarla, P.; Ranjana, K.C.; Kumar, S.; Lakra, W.S.; Mukherjee, M.M.; Ranaweera, I.; Shrestha, U.; Tran, T.; et al. Multidrug efflux pumps from Enterobacteriaceae, Vibrio cholerae and Staphylococcus aureus bacterial food pathogens. Int. J. Environ. Res. Public Health 2015, 12, 1487–1547. [Google Scholar] [CrossRef] [PubMed]
- Varela, M.F.; Stephen, J.; Lekshmi, M.; Ojha, M.; Wenzel, N.; Sanford, L.M.; Hernandez, A.J.; Parvathi, A.; Kumar, S.H. Bacterial Resistance to Antimicrobial Agents. Antibiotics 2021, 10, 593. [Google Scholar] [CrossRef]
- Lewis, K. Persister cells. Annu. Rev. Microbiol. 2010, 64, 357–372. [Google Scholar] [CrossRef]
- Lambert, P.A. Bacterial resistance to antibiotics: Modified target sites. Adv. Drug Deliv. Rev. 2005, 57, 1471–1485. [Google Scholar] [CrossRef]
- Koch, A.L. Bacterial Wall as Target for Attack. Clin. Microbiol. Rev. 2003, 16, 673–687. [Google Scholar] [CrossRef]
- Wilson, D.N.; Hauryliuk, V.; Atkinson, G.C.; O’Neill, A.J. Target protection as a key antibiotic resistance mechanism. Nat. Rev. Microbiol. 2020, 18, 637–648. [Google Scholar] [CrossRef]
- Walther-Rasmussen, J.; Hoiby, N. Cefotaximases (CTX-M-ases), an expanding family of extended-spectrum β-lactamases. Can. J. Microbiol. 2004, 50, 137–165. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 2003, 67, 593–656. [Google Scholar] [CrossRef] [PubMed]
- Li, X.Z.; Plesiat, P.; Nikaido, H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin. Microbiol. Rev. 2015, 28, 337–418. [Google Scholar] [CrossRef]
- Kumar, S.; Varela, M.F. Biochemistry of bacterial multidrug efflux pumps. Int. J. Mol. Sci. 2012, 13, 4484–4495. [Google Scholar] [CrossRef] [PubMed]
- Floyd, J.T.; Kumar, S.; Mukherjee, M.M.; He, G.; Varela, M.F. A review of the molecular mechanisms of drug efflux in pathogenic bacteria: A structure-function perspective. Recent Res. Dev. Membr. Biol. 2013, 3, 15–66. [Google Scholar]
- Blair, J.M.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2015, 13, 42–51. [Google Scholar] [CrossRef]
- Levy, S.B. Active efflux, a common mechanism for biocide and antibiotic resistance. J. Appl. Microbiol. 2002, 92, 65S–71S. [Google Scholar] [CrossRef]
- Higgins, C.F. ABC transporters: Physiology, structure and mechanism--an overview. Res. Microbiol. 2001, 152, 205–210. [Google Scholar] [CrossRef]
- Parr, T.R., Jr.; Saier, M.H., Jr. The bacterial phosphotransferase system as a potential vehicle for the entry of novel antibiotics. Res. Microbiol. 1992, 143, 443–447. [Google Scholar] [CrossRef]
- Rambhatla, P.; Kumar, S.; Floyd, J.T.; Varela, M.F. Molecular cloning and characterization of mannitol-1-phosphate dehydrogenase from Vibrio cholerae. J. Microbiol. Biotechnol. 2011, 21, 914–920. [Google Scholar] [CrossRef]
- Kumar, S.; Smith, K.P.; Floyd, J.L.; Varela, M.F. Cloning and molecular analysis of a mannitol operon of phosphoenolpyruvate-dependent phosphotransferase (PTS) type from Vibrio cholerae O395. Arch. Microbiol. 2011, 193, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Poolman, B.; Konings, W.N. Secondary solute transport in bacteria. Biochim. Biophys. Acta 1993, 1183, 5–39. [Google Scholar] [CrossRef]
- West, I.C.; Mitchell, P. Proton/sodium ion antiport in Escherichia coli. Biochem. J. 1974, 144, 87–90. [Google Scholar] [CrossRef]
- Konings, W.N.; Poolman, B.; van Veen, H.W. Solute transport and energy transduction in bacteria. Antonie Van Leeuwenhoek 1994, 65, 369–380. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. RND transporters in the living world. Res. Microbiol. 2018, 169, 363–371. [Google Scholar] [CrossRef]
- Begum, A.; Rahman, M.M.; Ogawa, W.; Mizushima, T.; Kuroda, T.; Tsuchiya, T. Gene cloning and characterization of four MATE family multidrug efflux pumps from Vibrio cholerae non-O1. Microbiol. Immunol. 2005, 49, 949–957. [Google Scholar] [CrossRef]
- Huda, M.N.; Chen, J.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Gene cloning and characterization of VcrM, a Na+-coupled multidrug efflux pump, from Vibrio cholerae non-O1. Microbiol. Immunol. 2003, 47, 419–427. [Google Scholar] [CrossRef] [PubMed]
- Paulsen, I.T.; Skurray, R.A.; Tam, R.; Saier, M.H., Jr.; Turner, R.J.; Weiner, J.H.; Goldberg, E.B.; Grinius, L.L. The SMR family: A novel family of multidrug efflux proteins involved with the efflux of lipophilic drugs. Mol. Microbiol. 1996, 19, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Jack, D.L.; Yang, N.M.; Saier, M.H., Jr. The drug/metabolite transporter superfamily. Eur. J. Biochem. 2001, 268, 3620–3639. [Google Scholar] [CrossRef]
- Hassan, K.A.; Liu, Q.; Henderson, P.J.; Paulsen, I.T. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. mBio 2015, 6, e01982-14. [Google Scholar] [CrossRef]
- Rees, D.C.; Johnson, E.; Lewinson, O. ABC transporters: The power to change. Nat. Rev. Mol. Cell Biol. 2009, 10, 218–227. [Google Scholar] [CrossRef] [PubMed]
- Destoumieux-Garzón, D.; Duperthuy, M.; Vanhove, A.S.; Schmitt, P.; Wai, S.N. Resistance to Antimicrobial Peptides in Vibrios. Antibiotics 2014, 3, 540–563. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Structure and mechanism of RND-type multidrug efflux pumps. Adv Enzym. Relat. Areas Mol. Biol. 2011, 77, 1–60. [Google Scholar]
- Fernández, L.; Hancock, R.E.W. Adaptive and mutational resistance: Role of porins and efflux pumps in drug resistance. Clin. Microbiol. Rev. 2012, 25, 661–681. [Google Scholar] [CrossRef]
- Bavro, V.N.; Marshall, R.L.; Symmons, M.F. Architecture and roles of periplasmic adaptor proteins in tripartite efflux assemblies. Front. Microbiol. 2015, 6, 513. [Google Scholar]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef]
- Nikaido, H. Prevention of drug access to bacterial targets: Permeability barriers and active efflux. Science 1994, 264, 382–388. [Google Scholar] [CrossRef]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002, 419, 587–593. [Google Scholar] [CrossRef]
- Mikolosko, J.; Bobyk, K.; Zgurskaya, H.I.; Ghosh, P. Conformational flexibility in the multidrug efflux system protein AcrA. Structure 2006, 14, 577–587. [Google Scholar] [CrossRef]
- Akama, H.; Matsuura, T.; Kashiwagi, S.; Yoneyama, H.; Narita, S.; Tsukihara, T.; Nakagawa, A.; Nakae, T. Crystal structure of the membrane fusion protein, MexA, of the multidrug transporter in Pseudomonas aeruginosa. J. Biol. Chem. 2004, 279, 25939–25942. [Google Scholar] [CrossRef]
- Akama, H.; Kanemaki, M.; Yoshimura, M.; Tsukihara, T.; Kashiwagi, T.; Yoneyama, H.; Narita, S.; Nakagawa, A.; Nakae, T. Crystal structure of the drug discharge outer membrane protein, OprM, of Pseudomonas aeruginosa: Dual modes of membrane anchoring and occluded cavity end. J. Biol. Chem. 2004, 279, 52816–52819. [Google Scholar] [CrossRef] [PubMed]
- Higgins, M.K.; Bokma, E.; Koronakis, E.; Hughes, C.; Koronakis, V. Structure of the periplasmic component of a bacterial drug efflux pump. Proc. Natl. Acad. Sci. USA 2004, 101, 9994–9999. [Google Scholar] [CrossRef] [PubMed]
- Nikaido, H. Antibiotic resistance caused by gram-negative multidrug efflux pumps. Clin. Infect. Dis. 1998, 27 (Suppl. 1), S32–S41. [Google Scholar] [CrossRef]
- Kunkle, D.E.; Bina, X.R.; Bina, J.E. The Vibrio cholerae VexGH RND Efflux System Maintains Cellular Homeostasis by Effluxing Vibriobactin. MBio 2017, 8, e00126-17. [Google Scholar] [CrossRef] [PubMed]
- Bina, J.E.; Provenzano, D.; Wang, C.; Bina, X.R.; Mekalanos, J.J. Characterization of the Vibrio cholerae vexAB and vexCD efflux systems. Arch. Microbiol. 2006, 186, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.L.; Bina, X.; Bina, J. Vibrio cholerae vexH Encodes a Multiple Drug Efflux Pump That Contributes to the Production of Cholera Toxin and the Toxin Co-Regulated Pilus. PLoS ONE 2012, 7, e38208. [Google Scholar] [CrossRef]
- Mukherjee, M.; Kakarla, P.; Kumar, S.; Gonzalez, E.; Floyd, J.T.; Inupakutika, M.; Devireddy, A.R.; Tirrell, S.R.; Bruns, M.; He, G.; et al. Comparative genome analysis of non-toxigenic non-O1 versus toxigenic O1 Vibrio cholerae. Genom. Discov. 2014, 2, 1–15. [Google Scholar] [CrossRef][Green Version]
- Rahman, M.M.; Matsuo, T.; Ogawa, W.; Koterasawa, M.; Kuroda, T.; Tsuchiya, T. Molecular cloning and characterization of all RND-type efflux transporters in Vibrio cholerae non-O1. Microbiol. Immunol. 2007, 51, 1061–1070. [Google Scholar] [CrossRef]
- Sistrunk, J.R.; Nickerson, K.P.; Chanin, R.B.; Rasko, D.A.; Faherty, C.S. Survival of the fittest: How bacterial pathogens utilize bile to enhance infection. Clin. Microbiol. Rev. 2016, 29, 819–836. [Google Scholar] [CrossRef]
- Provenzano, D.; Klose, K.E. Altered expression of the ToxR-regulated porins OmpU and OmpT diminishes Vibrio cholerae bile resistance, virulence factor expression, and intestinal colonization. Proc. Natl. Acad. Sci. USA 2000, 97, 10220–10224. [Google Scholar] [CrossRef]
- Wibbenmeyer, J.A.; Provenzano, D.; Landry, C.F.; Klose, K.E.; Delcour, A.H. Vibrio cholerae OmpU and OmpT porins are differentially affected by bile. Infect. Immun. 2002, 70, 121–126. [Google Scholar] [CrossRef] [PubMed]
- Bina, X.R.; Provenzano, D.; Nguyen, N.; Bina, J.E. Vibrio cholerae RND family efflux systems are required for antimicrobial resistance, optimal virulence factor production, and colonization of the infant mouse small intestine. Infect. Immun. 2008, 76, 3595–3605. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Huang, Y.; Xu, H.; Ying, S.; Pan, H.; Yu, W. Genomic and Phenotypic Characteristics for Vibrio vulnificus Infections. Infect. Drug Resist. 2021, 14, 3721–3726. [Google Scholar] [CrossRef]
- Lee, S.; Yeom, J.H.; Seo, S.; Lee, M.; Kim, S.; Bae, J.; Lee, K.; Hwang, J. Functional analysis of Vibrio vulnificus RND efflux pumps homologous to Vibrio cholerae VexAB and VexCD, and to Escherichia coli AcrAB. J. Microbiol. 2015, 53, 256–261. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Song, S.; Lee, K. Functional analysis of TolC homologs in Vibrio vulnificus. Curr. Microbiol. 2014, 68, 729–734. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Song, S.; Lee, M.; Hwang, S.; Kim, J.-S.; Ha, N.-C.; Lee, K. Interaction between the α-barrel tip of Vibrio vulnificus TolC homologs and AcrA implies the adapter bridging model. J. Microbiol. 2014, 52, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Misra, R.; Bavro, V.N. Assembly and transport mechanism of tripartite drug efflux systems. Biochim. Biophys. Acta 2009, 1794, 817–825. [Google Scholar] [CrossRef]
- Federici, L.; Du, D.; Walas, F.; Matsumura, H.; Fernandez-Recio, J.; McKeegan, K.S.; Borges-Walmsley, M.I.; Luisi, B.F.; Walmsley, A.R. The crystal structure of the outer membrane protein VceC from the bacterial pathogen Vibrio cholerae at 1.8 A resolution. J. Biol. Chem. 2005, 280, 15307–15314. [Google Scholar] [CrossRef]
- Kumar, S.; Ranjana, K.; Sanford, L.M.; Hernandez, A.J.; Kakarla, P.; Varela, M.F. Structural and functional roles of two evolutionarily conserved amino acid sequence motifs within solute transporters of the major facilitator superfamily. Trends Cell Mol. Biol. 2016, 11, 41–53. [Google Scholar]
- Yamanaka, H.; Tadokoro, S.; Miyano, M.; Takahashi, E.; Kobayashi, H.; Okamoto, K. Studies on the region involved in the transport activity of Escherichia coli TolC by chimeric protein analysis. Microb. Pathog. 2007, 42, 184–192. [Google Scholar] [CrossRef]
- Weng, Y.; Fields, E.G.; Bina, T.F.; Budnick, J.A.; Kunkle, D.E.; Bina, X.R.; Bina, J.E. Vibrio cholerae TolC Is Required for Expression of the ToxR Regulon. Infect. Immun. 2021, 89, e0024221. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Kataoka, A.; Shiota, S.; Mizushima, T.; Tsuchiya, T. NorM of Vibrio parahaemolyticus is an Na+-driven multidrug efflux pump. J. Bacteriol. 2000, 182, 6694–6697. [Google Scholar] [CrossRef] [PubMed]
- Morita, Y.; Kodama, K.; Shiota, S.; Mine, T.; Kataoka, A.; Mizushima, T.; Tsuchiya, T. NorM, a putative multidrug efflux protein, of Vibrio parahaemolyticus and its homolog in Escherichia coli. Antimicrob Agents Chemother 1998, 42, 1778–1782. [Google Scholar] [CrossRef] [PubMed]
- Brown, M.H.; Paulsen, I.T.; Skurray, R.A. The multidrug efflux protein NorM is a prototype of a new family of transporters. Mol. Microbiol. 1999, 31, 394–395. [Google Scholar] [CrossRef] [PubMed]
- Otsuka, M.; Yasuda, M.; Morita, Y.; Otsuka, C.; Tsuchiya, T.; Omote, H.; Moriyama, Y. Identification of essential amino acid residues of the NorM Na+/multidrug antiporter in Vibrio parahaemolyticus. J. Bacteriol. 2005, 187, 1552–1558. [Google Scholar] [CrossRef]
- Radchenko, M.; Symersky, J.; Nie, R.; Lu, M. Structural basis for the blockade of MATE multidrug efflux pumps. Nat. Commun. 2015, 6, 7995. [Google Scholar] [CrossRef]
- Kusakizako, T.; Claxton, D.P.; Tanaka, Y.; Maturana, A.D.; Kuroda, T.; Ishitani, R.; McHaourab, H.S.; Nureki, O. Structural Basis of H+-Dependent Conformational Change in a Bacterial MATE Transporter. Structure 2019, 27, 293–301.e293. [Google Scholar] [CrossRef]
- Nie, L.; Grell, E.; Malviya, V.N.; Xie, H.; Wang, J.; Michel, H. Identification of the High-affinity Substrate-binding Site of the Multidrug and Toxic Compound Extrusion (MATE) Family Transporter from Pseudomonas stutzeri. J. Biol. Chem. 2016, 291, 15503–15514. [Google Scholar] [CrossRef]
- Jin, Y.; Nair, A.; van Veen, H.W. Multidrug transport protein NorM from Vibrio cholerae simultaneously couples to sodium- and proton-motive force. J. Biol. Chem. 2014, 289, 14624–14632. [Google Scholar] [CrossRef]
- Lu, M.; Radchenko, M.; Symersky, J.; Nie, R.; Guo, Y. Structural insights into H+-coupled multidrug extrusion by a MATE transporter. Nat. Struct. Mol. Biol. 2013, 20, 1310–1317. [Google Scholar] [CrossRef]
- He, X.; Szewczyk, P.; Karyakin, A.; Evin, M.; Hong, W.X.; Zhang, Q.; Chang, G. Structure of a cation-bound multidrug and toxic compound extrusion transporter. Nature 2010, 467, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Kusakizako, T.; Miyauchi, H.; Ishitani, R.; Nureki, O. Structural biology of the multidrug and toxic compound extrusion superfamily transporters. Biochim. Et Biophys. Acta (BBA)-Biomembr. 2020, 1862, 183154. [Google Scholar] [CrossRef] [PubMed]
- Vanni, S.; Campomanes, P.; Marcia, M.; Rothlisberger, U. Ion binding and internal hydration in the multidrug resistance secondary active transporter NorM investigated by molecular dynamics simulations. Biochemistry 2012, 51, 1281–1287. [Google Scholar] [CrossRef]
- Claxton, D.P.; Jagessar, K.L.; Steed, P.R.; Stein, R.A.; McHaourab, H.S. Sodium and proton coupling in the conformational cycle of a MATE antiporter from Vibrio cholerae. Proc. Natl. Acad. Sci. USA 2018, 115, E6182–E6190. [Google Scholar] [CrossRef] [PubMed]
- Huda, M.N.; Morita, Y.; Kuroda, T.; Mizushima, T.; Tsuchiya, T. Na+-driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a non-halophilic bacterium. FEMS Microbiol. Lett. 2001, 203, 235–239. [Google Scholar] [CrossRef]
- Heidelberg, J.F.; Eisen, J.A.; Nelson, W.C.; Clayton, R.A.; Gwinn, M.L.; Dodson, R.J.; Haft, D.H.; Hickey, E.K.; Peterson, J.D.; Umayam, L.; et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature 2000, 406, 477–483. [Google Scholar] [CrossRef] [PubMed]
- Zakrzewska, S.; Mehdipour, A.R.; Malviya, V.N.; Nonaka, T.; Koepke, J.; Muenke, C.; Hausner, W.; Hummer, G.; Safarian, S.; Michel, H. Inward-facing conformation of a multidrug resistance MATE family transporter. Proc. Natl. Acad. Sci. USA 2019, 116, 12275–12284. [Google Scholar] [CrossRef]
- Mousa, J.J.; Newsome, R.C.; Yang, Y.; Jobin, C.; Bruner, S.D. ClbM is a versatile, cation-promiscuous MATE transporter found in the colibactin biosynthetic gene cluster. Biochem. Biophys. Res. Commun. 2017, 482, 1233–1239. [Google Scholar] [CrossRef]
- Lu, M.; Symersky, J.; Radchenko, M.; Koide, A.; Guo, Y.; Nie, R.; Koide, S. Structures of a Na+-coupled, substrate-bound MATE multidrug transporter. Proc. Natl. Acad. Sci. USA 2013, 110, 2099–2104. [Google Scholar] [CrossRef]
- Lu, M. Structures of multidrug and toxic compound extrusion transporters and their mechanistic implications. Channels 2016, 10, 88–100. [Google Scholar] [CrossRef]
- Omote, H.; Hiasa, M.; Matsumoto, T.; Otsuka, M.; Moriyama, Y. The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol. Sci. 2006, 27, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Yan, N. Structural biology of the major facilitator superfamily transporters. Annu. Rev. Biophys. 2015, 44, 257–283. [Google Scholar] [CrossRef] [PubMed]
- Castellano, S.; Claxton, D.P.; Ficici, E.; Kusakizako, T.; Stix, R.; Zhou, W.; Nureki, O.; McHaourab, H.S.; Faraldo-Gómez, J.D. Conserved binding site in the N-lobe of prokaryotic MATE transporters suggests a role for Na+ in ion-coupled drug efflux. J. Biol. Chem. 2021, 296, 100262. [Google Scholar] [CrossRef] [PubMed]
- Claxton, D.P.; Jagessar, K.L.; McHaourab, H.S. Principles of Alternating Access in Multidrug and Toxin Extrusion (MATE) Transporters. J. Mol. Biol. 2021, 433, 166959. [Google Scholar] [CrossRef] [PubMed]
- Ficici, E.; Zhou, W.; Castellano, S.; Faraldo-Gómez, J.D. Broadly conserved Na+-binding site in the N-lobe of prokaryotic multidrug MATE transporters. Proc. Natl. Acad. Sci. USA 2018, 115, E6172–E6181. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Hipolito, C.J.; Maturana, A.D.; Ito, K.; Kuroda, T.; Higuchi, T.; Katoh, T.; Kato, H.E.; Hattori, M.; Kumazaki, K. Structural basis for the drug extrusion mechanism by a MATE multidrug transporter. Nature 2013, 496, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Burse, A.; Weingart, H.; Ullrich, M.S. NorM, an Erwinia amylovora multidrug efflux pump involved in in vitro competition with other epiphytic bacteria. Appl. Environ. Microbiol. 2004, 70, 693–703. [Google Scholar] [CrossRef]
- Colmer, J.A.; Fralick, J.A.; Hamood, A.N. Isolation and characterization of a putative multidrug resistance pump from Vibrio cholerae. Mol. Microbiol. 1998, 27, 63–72. [Google Scholar] [CrossRef]
- Varela, M.F.; Sansom, C.E.; Griffith, J.K. Mutational analysis and molecular modelling of an amino acid sequence motif conserved in antiporters but not symporters in a transporter superfamily. Mol. Membr. Biol. 1995, 12, 313–319. [Google Scholar] [CrossRef]
- Woolley, R.C.; Vediyappan, G.; Anderson, M.; Lackey, M.; Ramasubramanian, B.; Jiangping, B.; Borisova, T.; Colmer, J.A.; Hamood, A.N.; McVay, C.S.; et al. Characterization of the Vibrio cholerae vceCAB multiple-drug resistance efflux operon in Escherichia coli. J. Bacteriol. 2005, 187, 5500–5503. [Google Scholar] [CrossRef]
- Alatoom, A.A.; Aburto, R.; Hamood, A.N.; Colmer-Hamood, J.A. VceR negatively regulates the vceCAB MDR efflux operon and positively regulates its own synthesis in Vibrio cholerae 569B. Can. J. Microbiol. 2007, 53, 888–900. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Mosley, L.; Fralick, J.A. Evidence that the C-terminus of OprM is involved in the assembly of the VceAB-OprM efflux pump. FEBS Lett. 2010, 584, 1493–1497. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.P.; Kumar, S.; Varela, M.F. Identification, cloning, and functional characterization of EmrD-3, a putative multidrug efflux pump of the major facilitator superfamily from Vibrio cholerae O395. Arch. Microbiol. 2009, 191, 903–911. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Lindquist, I.E.; Sundararajan, A.; Rajanna, C.; Floyd, J.T.; Smith, K.P.; Andersen, J.L.; He, G.; Ayers, R.M.; Johnson, J.A.; et al. Genome Sequence of Non-O1 Vibrio cholerae PS15. Genome Announc. 2013, 1, e00227-12. [Google Scholar] [CrossRef] [PubMed]
- Kakarla, P.; Kc, R.; Shrestha, U.; Ranaweera, I.; Mukherjee, M.; Willmon, T.M.; Hernandez, A.J.; Barr, S.R.; Varela, M.F. Functional Roles of Highly Conserved Amino Acid Sequence Motifs A and C in Solute Transporters of the Major Facilitator Superfamily. In Drug Resistance in Bacteria, Fungi, Malaria, and Cancer; Arora, G., Sajid, A., Kalia, V., Eds.; Springer: Cham, Switzerland, 2017. [Google Scholar] [CrossRef]
- Tanabe, T.; Nakao, H.; Kuroda, T.; Tsuchiya, T.; Yamamoto, S. Involvement of the Vibrio parahaemolyticus pvsC gene in export of the siderophore vibrioferrin. Microbiol. Immunol. 2006, 50, 871–876. [Google Scholar] [PubMed]
- Tanabe, T.; Funahashi, T.; Nakao, H.; Miyoshi, S.; Shinoda, S.; Yamamoto, S. Identification and characterization of genes required for biosynthesis and transport of the siderophore vibrioferrin in Vibrio parahaemolyticus. J. Bacteriol. 2003, 185, 6938–6949. [Google Scholar] [CrossRef]
- Amin, S.A.; Green, D.H.; Kupper, F.C.; Carrano, C.J. Vibrioferrin, an unusual marine siderophore: Iron binding, photochemistry, and biological implications. Inorg. Chem. 2009, 48, 11451–11458. [Google Scholar] [CrossRef]
- Chen, S.; Wang, H.; Katzianer, D.S.; Zhong, Z.; Zhu, J. LysR family activator-regulated major facilitator superfamily transporters are involved in Vibrio cholerae antimicrobial compound resistance and intestinal colonisation. Int. J. Antimicrob. Agents 2013, 41, 188–192. [Google Scholar] [CrossRef]
- Liu, H.; Xie, J. Comparative genomics of Mycobacterium tuberculosis drug efflux pumps and their transcriptional regulators. Crit. Rev.TM Eukaryot. Gene Expr. 2014, 24, 163–180. [Google Scholar] [CrossRef]
- Bruns, M.M.; Kakarla, P.; Floyd, J.T.; Mukherjee, M.M.; Ponce, R.C.; Garcia, J.A.; Ranaweera, I.; Sanford, L.M.; Hernandez, A.J.; Willmon, T.M.; et al. Modulation of the multidrug efflux pump EmrD-3 from Vibrio cholerae by Allium sativum extract and the bioactive agent allyl sulfide plus synergistic enhancement of antimicrobial susceptibility by A. sativum extract. Arch. Microbiol. 2017, 199, 1103–1112. [Google Scholar] [CrossRef]
- Varela, M.F.; Kumar, S. Strategies for discovery of new molecular targets for anti-infective drugs. Curr. Opin. Pharmacol. 2019, 48, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; He, G.; Kakarla, P.; Shrestha, U.; Ranjana, K.C.; Ranaweera, I.; Mark Willmon, T.; Barr, S.R.; Hernandez, A.J.; Varela, M.F. Bacterial multidrug efflux pumps of the major facilitator superfamily as targets for modulation. Infect. Disord.-Drug Targets (Former. Curr. Drug Targets-Infect. Disord.) 2016, 16, 28–43. [Google Scholar]
- Varela, M.F.; Griffith, J.K. Nucleotide and deduced protein sequences of the class D tetracycline resistance determinant: Relationship to other antimicrobial transport proteins. Antimicrob. Agents Chemother. 1993, 37, 1253–1258. [Google Scholar] [CrossRef] [PubMed]
- Griffith, J.K.; Baker, M.E.; Rouch, D.A.; Page, M.G.; Skurray, R.A.; Paulsen, I.T.; Chater, K.F.; Baldwin, S.A.; Henderson, P.J. Membrane transport proteins: Implications of sequence comparisons. Curr. Opin. Cell Biol. 1992, 4, 684–695. [Google Scholar] [CrossRef]
- Kim, J.; Cater, R.J.; Choy, B.C.; Mancia, F. Structural Insights into Transporter-Mediated Drug Resistance in Infectious Diseases. J. Mol. Biol. 2021, 433, 167005. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; van Veen, H.W.; Murakami, S.; Pos, K.M.; Luisi, B.F. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr. Opin. Struct. Biol. 2015, 33, 76–91. [Google Scholar] [CrossRef]
- Law, C.J.; Maloney, P.C.; Wang, D.N. Ins and outs of major facilitator superfamily antiporters. Annu. Rev. Microbiol. 2008, 62, 289–305. [Google Scholar] [CrossRef]
- Krämer, R. Functional principles of solute transport systems: Concepts and perspectives. Biochim. Biophys. Acta 1994, 1185, 1–34. [Google Scholar] [CrossRef]
- Yamato, I. Ordered binding model as a general mechanistic mechanism for secondary active transport systems. FEBS Lett. 1992, 298, 1–5. [Google Scholar] [CrossRef]
- Yaffe, D.; Radestock, S.; Shuster, Y.; Forrest, L.R.; Schuldiner, S. Identification of molecular hinge points mediating alternating access in the vesicular monoamine transporter VMAT2. Proc. Natl. Acad. Sci. USA 2013, 110, E1332–E1341. [Google Scholar] [CrossRef]
- Heng, J.; Zhao, Y.; Liu, M.; Liu, Y.; Fan, J.; Wang, X.; Zhang, X.C. Substrate-bound structure of the E. coli multidrug resistance transporter MdfA. Cell Res. 2015, 25, 1060–1073. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Lekshmi, M.; Parvathi, A.; Ojha, M.; Wenzel, N.; Varela, M.F. Functional and Structural Roles of the Major Facilitator Superfamily Bacterial Multidrug Efflux Pumps. Microorganisms 2020, 8, 266. [Google Scholar] [CrossRef] [PubMed]
- Ranaweera, I.; Shrestha, U.; Ranjana, K.C.; Kakarla, P.; Willmon, T.M.; Hernandez, A.J.; Mukherjee, M.M.; Barr, S.R.; Varela, M.F. Structural comparison of bacterial multidrug efflux pumps of the major facilitator superfamily. Trends Cell Mol. Biol. 2015, 10, 131–140. [Google Scholar] [PubMed]
- Nagarathinam, K.; Jaenecke, F.; Nakada-Nakura, Y.; Hotta, Y.; Liu, K.; Iwata, S.; Stubbs, M.T.; Nomura, N.; Tanabe, M. The multidrug-resistance transporter MdfA from Escherichia coli: Crystallization and X-ray diffraction analysis. Acta Crystallogr. F Struct. Biol. Commun. 2017, 73, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Padyana, S.; Dipin, K.; Kumar, S.; Nayak, B. Antimicrobial compounds of plant origin as efflux pump inhibitors: New avenues for controlling multidrug resistant pathogens. J. Antimicrob. Agents 2018, 4, 1–6. [Google Scholar]
- Moussatova, A.; Kandt, C.; O’Mara, M.L.; Tieleman, D.P. ATP-binding cassette transporters in Escherichia coli. Biochim. Biophys. Acta 2008, 1778, 1757–1771. [Google Scholar] [CrossRef] [PubMed]
- Higgins, C.F. ABC transporters: From microorganisms to man. Annu. Rev. Cell Biol. 1992, 8, 67–113. [Google Scholar] [CrossRef]
- Lu, W.J.; Lin, H.J.; Janganan, T.K.; Li, C.Y.; Chin, W.C.; Bavro, V.N.; Lin, H.V. ATP-Binding Cassette Transporter VcaM from Vibrio cholerae is Dependent on the Outer Membrane Factor Family for Its Function. Int. J. Mol. Sci. 2018, 19, 1000. [Google Scholar] [CrossRef]
- Cerda-Maira, F.A.; Ringelberg, C.S.; Taylor, R.K. The bile response repressor BreR regulates expression of the Vibrio cholerae breAB efflux system operon. J. Bacteriol. 2008, 190, 7441–7452. [Google Scholar] [CrossRef]
- Alvarez-Ortega, C.; Olivares, J.; Martinez, J.L. RND multidrug efflux pumps: What are they good for? Front. Microbiol. 2013, 4, 7. [Google Scholar] [CrossRef]
- Bina, X.R.; Philippart, J.A.; Bina, J.E. Effect of the efflux inhibitors 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-beta-naphthylamide on antimicrobial susceptibility and virulence factor production in Vibrio cholerae. J. Antimicrob. Chemother. 2009, 63, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.; Lee, N.Y.; Kim, J.; Lee, M.A.; Kim, K.S.; Lee, K.H.; Park, S.J. Functional characterization of EpsC, a component of the type II secretion system, in the pathogenicity of Vibrio vulnificus. Infect. Immun. 2011, 79, 4068–4080. [Google Scholar] [CrossRef] [PubMed]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial Biofilm Eradication Agents: A Current Review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef] [PubMed]
- Stoodley, P.; Sauer, K.; Davies, D.G.; Costerton, J.W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002, 56, 187–209. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, S.K.; Chowdhury, I.; Singh, R. Understanding the mechanism of bacterial biofilms resistance to antimicrobial agents. Open Microbiol. J. 2017, 11, 53. [Google Scholar] [CrossRef]
- Szemerédi, N.; Kincses, A.; Rehorova, K.; Hoang, L.; Salardón-Jiménez, N.; Sevilla-Hernández, C.; Viktorová, J.; Domínguez-Álvarez, E.; Spengler, G. Ketone-and Cyano-Selenoesters to Overcome Efflux Pump, Quorum-Sensing, and Biofilm-Mediated Resistance. Antibiotics 2020, 9, 896. [Google Scholar] [CrossRef] [PubMed]
- Alcalde-Rico, M.; Hernando-Amado, S.; Blanco, P.; Martínez, J.L. Multidrug Efflux Pumps at the Crossroad between Antibiotic Resistance and Bacterial Virulence. Front. Microbiol. 2016, 7, 1483. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stephen, J.; Lekshmi, M.; Ammini, P.; Kumar, S.H.; Varela, M.F. Membrane Efflux Pumps of Pathogenic Vibrio Species: Role in Antimicrobial Resistance and Virulence. Microorganisms 2022, 10, 382. https://doi.org/10.3390/microorganisms10020382
Stephen J, Lekshmi M, Ammini P, Kumar SH, Varela MF. Membrane Efflux Pumps of Pathogenic Vibrio Species: Role in Antimicrobial Resistance and Virulence. Microorganisms. 2022; 10(2):382. https://doi.org/10.3390/microorganisms10020382
Chicago/Turabian StyleStephen, Jerusha, Manjusha Lekshmi, Parvathi Ammini, Sanath H. Kumar, and Manuel F. Varela. 2022. "Membrane Efflux Pumps of Pathogenic Vibrio Species: Role in Antimicrobial Resistance and Virulence" Microorganisms 10, no. 2: 382. https://doi.org/10.3390/microorganisms10020382
APA StyleStephen, J., Lekshmi, M., Ammini, P., Kumar, S. H., & Varela, M. F. (2022). Membrane Efflux Pumps of Pathogenic Vibrio Species: Role in Antimicrobial Resistance and Virulence. Microorganisms, 10(2), 382. https://doi.org/10.3390/microorganisms10020382







