Serum Biochemical Parameters, Rumen Fermentation, and Rumen Bacterial Communities Are Partly Driven by the Breed and Sex of Cattle When Fed High-Grain Diet
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals, Management, and Sampling
2.3. Chemical Analysis
2.4. DNA Extraction, PCR Amplification, and MiSeq Sequencing
2.5. Bioinformatic Analysis
2.6. Statistics Analysis
3. Results
3.1. Fattening Performance
3.2. Serum Biochemical Parameters
3.3. Rumen Fermentation
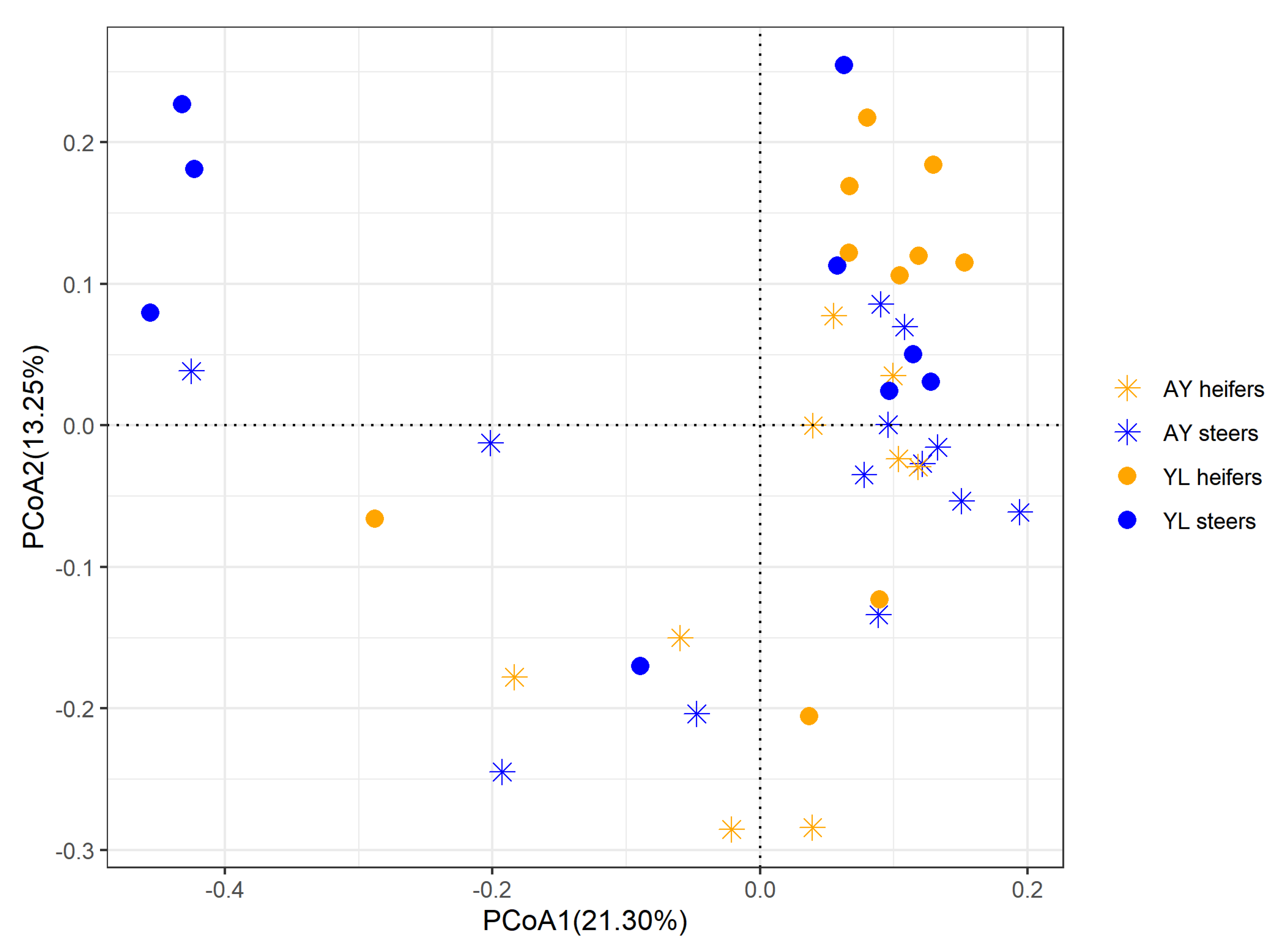
3.4. Bacterial Diversity and Composition
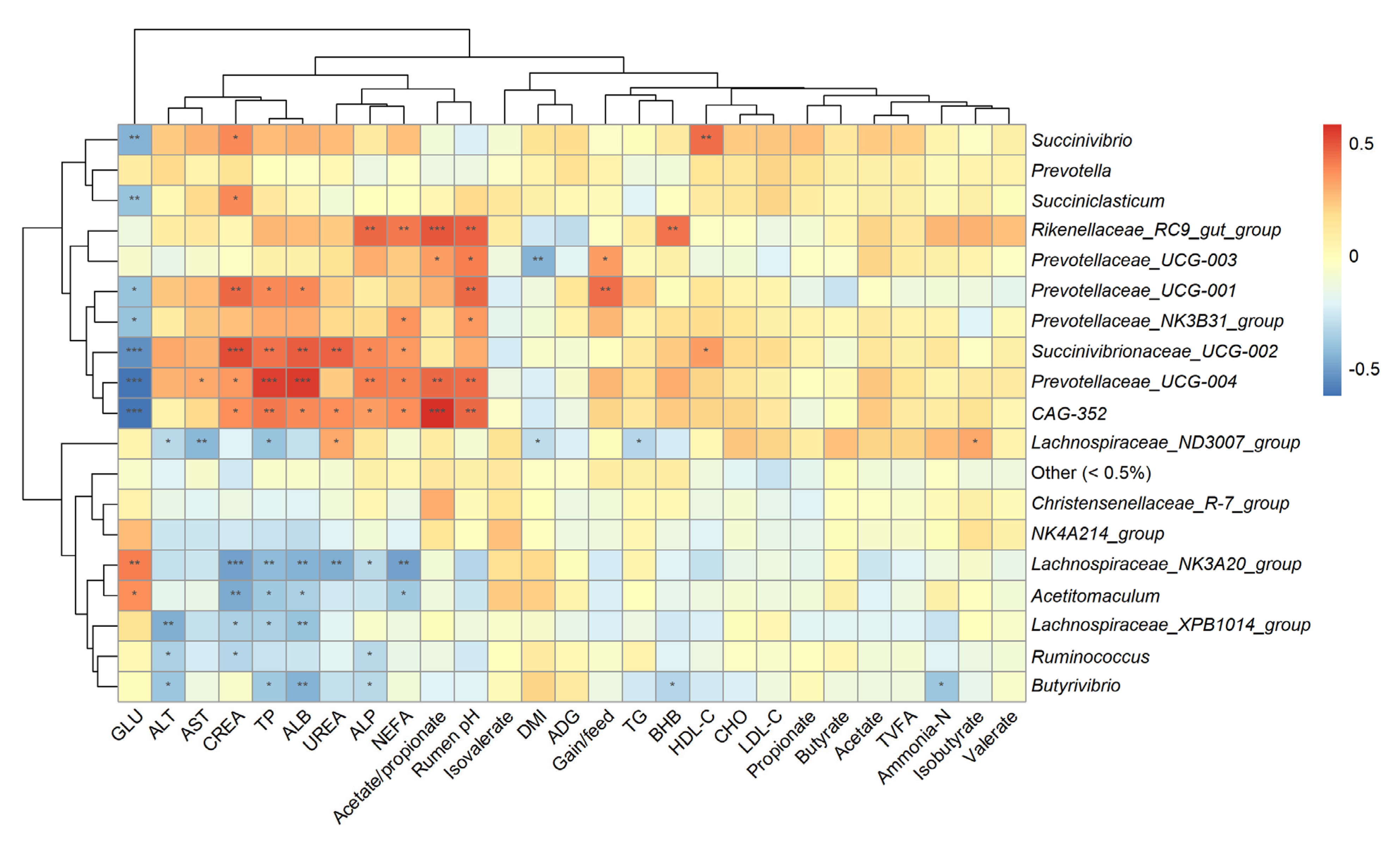
3.5. Correlation Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, W.; Gao, X.; Zhang, Y.; Zhao, Y.; Zhang, J.; Jia, Y.; Zhu, B.; Xu, L.; Zhang, L.; Gao, H.; et al. Genome-wide assessment of genetic diversity and population structure insights into admixture and introgression in Chinese indigenous cattle. BMC Genet. 2018, 19, 114. [Google Scholar] [CrossRef] [PubMed]
- Ling, X.; Zhang, W.-g.; Li, J.-y.; Zhu, D.-j.; Xu, X.-c.; Tian, Y.-z.; Xiong, X.; Guo, A.-z.; Cao, B.-h.; Niu, H.; et al. Genetic background analysis and breed evaluation of Yiling yellow cattle. J. Integr. Agric. 2017, 16, 2246–2256. [Google Scholar] [CrossRef]
- Qiu, X.; Qin, X.; Chen, L.; Qiu, Q.; Wang, H.; Aziz Ur Rahmanand, M.; Cao, B.; Su, H. Effects of age and rice straw inclusion levels in the diet of Yiling cull cows on growth performance, meat quality, and antioxidant status of tissues. Animals 2021, 11, 1732. [Google Scholar] [CrossRef] [PubMed]
- Newbold, C.J.; Ramos-Morales, E. Review: Ruminal microbiome and microbial metabolome: Effects of diet and ruminant host. Animal 2020, 14, s78–s86. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Niu, W.; Qiu, Q.; Xia, C.; Shao, T.; Wang, H.; Li, Q.; Yu, Z.; Gao, Z.; Rahman, M.A.U.; et al. Effect of calcium salt of long-chain fatty acids and alfalfa supplementation on performance of Holstein bulls. Oncotarget 2018, 9, 3029–3042. [Google Scholar] [CrossRef] [PubMed]
- Niu, W.; He, Y.; Xia, C.; Rahman, M.A.U.; Qiu, Q.; Shao, T.; Liang, Y.; Ji, L.; Wang, H.; Cao, B. Effects of replacing Leymus chinensis with whole-crop wheat hay on Holstein bull apparent digestibility, plasma parameters, rumen fermentation, and microbiota. Sci. Rep. 2017, 7, 2114. [Google Scholar] [CrossRef]
- Wang, H.; He, Y.; Li, H.; Wu, F.; Qiu, Q.; Niu, W.; Gao, Z.; Su, H.; Cao, B. Rumen fermentation, intramuscular fat fatty acid profiles and related rumen bacterial populations of Holstein bulls fed diets with different energy levels. Appl. Microbiol. Biotechnol. 2019, 103, 4931–4942. [Google Scholar] [CrossRef]
- He, Y.; Yu, Z.; Qiu, Q.; Shao, T.; Niu, W.; Xia, C.; Wang, H.; Su, H.; Cao, B. Effects of dietary protein levels and calcium salts of long-chain fatty acids on nitrogen mobilization, rumen microbiota and plasma fatty acid composition in Holstein bulls. Anim. Feed Sci. Tech. 2018, 246, 1–10. [Google Scholar] [CrossRef]
- Qiu, Q.; Gao, C.; Aziz Ur Rahman, M.; Cao, B.; Su, H. Digestive ability, physiological characteristics, and rumen bacterial community of Holstein finishing steers in response to three nutrient density diets as fattening phases advanced. Microorganisms 2020, 8, 335. [Google Scholar] [CrossRef]
- Li, F.; Li, C.; Chen, Y.; Liu, J.; Zhang, C.; Irving, B.; Fitzsimmons, C.; Plastow, G.; Guan, L.L. Host genetics influence the rumen microbiota and heritable rumen microbial features associate with feed efficiency in cattle. Microbiome 2019, 7, 92. [Google Scholar] [CrossRef]
- Abbas, W.; Howard, J.T.; Paz, H.A.; Hales, K.E.; Wells, J.E.; Kuehn, L.A.; Erickson, G.E.; Spangler, M.L.; Fernando, S.C. Influence of host genetics in shaping the rumen bacterial community in beef cattle. Sci. Rep. 2020, 10, 15101. [Google Scholar] [CrossRef]
- Wallace, R.J.; Sasson, G.; Garnsworthy, P.C.; Tapio, I.; Gregson, E.; Bani, P.; Huhtanen, P.; Bayat, A.R.; Strozzi, F.; Biscarini, F.; et al. A heritable subset of the core rumen microbiome dictates dairy cow productivity and emissions. Sci. Adv. 2019, 5, eaav8391. [Google Scholar] [CrossRef] [PubMed]
- Sasson, G.; Kruger Ben-Shabat, S.; Seroussi, E.; Doron-Faigenboim, A.; Shterzer, N.; Yaacoby, S.; Berg Miller, M.E.; White, B.A.; Halperin, E.; Mizrahi, I. Heritable bovine rumen bacteria are phylogenetically related and correlated with the cow’s capacity to harvest energy from its feed. mBio 2017, 8, e00703-17. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wright, A.G.; Si, H.; Wang, X.; Qian, W.; Zhang, Z.; Li, G. Changes in the rumen microbiome and metabolites reveal the effect of host genetics on hybrid crosses. Environ. Microbiol. Rep. 2016, 8, 1016–1023. [Google Scholar] [CrossRef]
- Bainbridge, M.L.; Cersosimo, L.M.; Wright, A.D.; Kraft, J. Rumen bacterial communities shift across a lactation in Holstein, Jersey and Holstein x Jersey dairy cows and correlate to rumen function, bacterial fatty acid composition and production parameters. FEMS. Microbiol. Ecol. 2016, 92, fiw059. [Google Scholar] [CrossRef]
- Hernandez-Sanabria, E.; Goonewardene, L.A.; Wang, Z.; Zhou, M.; Moore, S.S.; Guan, L.L. Influence of sire breed on the interplay among rumen microbial populations inhabiting the rumen liquid of the progeny in beef cattle. PLoS ONE 2013, 8, e58461. [Google Scholar] [CrossRef]
- Roehe, R.; Dewhurst, R.J.; Duthie, C.A.; Rooke, J.A.; McKain, N.; Ross, D.W.; Hyslop, J.J.; Waterhouse, A.; Freeman, T.C.; Watson, M.; et al. Bovine host genetic variation influences rumen microbial methane production with best selection criterion for low methane emitting and efficiently feed converting hosts based on metagenomic gene abundance. PLoS Genet. 2016, 12, e1005846. [Google Scholar] [CrossRef]
- Weatherburn, M.W. Phenol-hypochlorite reaction for determination of ammonia. Anal. Chem. 1967, 39, 971–974. [Google Scholar] [CrossRef]
- Berg, R.T.; Andersen, B.B.; Liboriussen, T. Growth of bovine tissues 1. Genetic influences on growth patterns of muscle, fat and bone in young bulls. Anim. Sci. 2010, 26, 245–258. [Google Scholar] [CrossRef]
- Walker, D.K.; Titgemeyer, E.C.; Baxa, T.J.; Chung, K.Y.; Johnson, D.E.; Laudert, S.B.; Johnson, B.J. Effects of ractopamine and sex on serum metabolites and skeletal muscle gene expression in finishing steers and heifers. J. Anim. Sci. 2010, 88, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Clare, M.; Richard, P.; Kate, K.; Sinead, W.; Mark, M.; David, K. Residual feed intake phenotype and gender affect the expression of key genes of the lipogenesis pathway in subcutaneous adipose tissue of beef cattle. J. Anim. Sci. Biot. 2018, 9, 68. [Google Scholar] [CrossRef]
- Byrne, C.J.; Fair, S.; English, A.M.; Urh, C.; Sauerwein, H.; Crowe, M.A.; Lonergan, P.; Kenny, D.A. Plane of nutrition before and after 6 months of age in Holstein-Friesian bulls: II. Effects on metabolic and reproductive endocrinology and identification of physiological markers of puberty and sexual maturation. J. Dairy Sci. 2018, 101, 3460–3475. [Google Scholar] [CrossRef]
- Kelly, A.K.; McGee, M.; Crews, D.H., Jr.; Fahey, A.G.; Wylie, A.R.; Kenny, D.A. Effect of divergence in residual feed intake on feeding behavior, blood metabolic variables, and body composition traits in growing beef heifers. J. Anim. Sci. 2010, 88, 109–123. [Google Scholar] [CrossRef]
- Li, M.; Gurram, B.; Lei, S.; Blum, N.T.; Huang, P.; Lin, J. Recent advances in fluorescence imaging of alkaline phosphatase. Chinese Chem. Lett. 2021, 32, 1316–1330. [Google Scholar] [CrossRef]
- Gade, T.P.; Motley, M.W.; Beattie, B.J.; Bhakta, R.; Boskey, A.L.; Koutcher, J.A.; Mayer-Kuckuk, P. Imaging of alkaline phosphatase activity in bone tissue. PLoS ONE 2011, 6, e22608. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Anderson, S.T.; Kidd, L.J.; Benvenutti, M.A.; Fletcher, M.T.; Dixon, R.M. New candidate markers of phosphorus status in beef breeder cows. Anim. Prod. Sci. 2017, 57, an17363. [Google Scholar] [CrossRef]
- Kunkel, H.O.; Stokes, D.K.; Anthony, W.B.; Futrell, M.F. Serum alkaline phosphatase activity in European and Brahman breeds of cattle and their crossbred types. J. Anim. Sci. 1953, 12, 765–770. [Google Scholar] [CrossRef]
- Cole, N.A.; Brown, M.A.; Phillips, W.A. Genetic x environment interactions on blood constituents of Angus, Brahman, and reciprocal-cross cows and calves grazing common bermudagrass or endophyte-infected tall fescue. J. Anim. Sci. 2001, 79, 1151–1161. [Google Scholar] [CrossRef]
- He, Y.; Wang, H.; Yu, Z.; Niu, W.; Qiu, Q.; Su, H.; Cao, B. Effects of the gender differences in cattle rumen fermentation on anaerobic fermentation of wheat straw. J. Clean. Prod. 2018, 205, 845–853. [Google Scholar] [CrossRef]
- Chen, Z.J. Genomic and epigenetic insights into the molecular bases of heterosis. Nat. Rev. Genet. 2013, 14, 471–482. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Wang, Z.; Hu, R.; Wang, X.; Li, F.; Zhang, X.; Zou, H.; Peng, Q.; Xue, B.; Wang, L. Comparative study of the bacterial communities throughout the gastrointestinal tract in two beef cattle breeds. Appl. Microbiol. Biotechnol. 2021, 105, 313–325. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Z.; Hu, R.; Peng, Q.; Xue, B.; Wang, L. Comparison of carcass characteristics and meat quality between Simmental crossbred cattle, cattle-yaks and Xuanhan yellow cattle. J. Sci. Food. Agric. 2021, 101, 3927–3932. [Google Scholar] [CrossRef]
- Yurkovetskiy, L.; Burrows, M.; Khan, A.A.; Graham, L.; Volchkov, P.; Becker, L.; Antonopoulos, D.; Umesaki, Y.; Chervonsky, A.V. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013, 39, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Org, E.; Mehrabian, M.; Parks, B.W.; Shipkova, P.; Liu, X.; Drake, T.A.; Lusis, A.J. Sex differences and hormonal effects on gut microbiota composition in mice. Gut. Microbes 2016, 7, 313–322. [Google Scholar] [CrossRef]
- Davenport, E.R.; Cusanovich, D.A.; Michelini, K.; Barreiro, L.B.; Ober, C.; Gilad, Y. Genome-wide association studies of the human gut microbiota. PLoS ONE 2015, 10, e0140301. [Google Scholar] [CrossRef] [PubMed]
- Danielsson, R.; Dicksved, J.; Sun, L.; Gonda, H.; Muller, B.; Schnurer, A.; Bertilsson, J. Methane production in dairy cows correlates with rumen methanogenic and bacterial community structure. Front. Microbiol. 2017, 8, 226. [Google Scholar] [CrossRef] [PubMed]
- Joch, M.; Mrazek, J.; Skrivanova, E.; Cermak, L.; Marounek, M. Effects of pure plant secondary metabolites on methane production, rumen fermentation and rumen bacteria populations in vitro. J. Anim. Physiol. Anim. Nutr. 2018, 102, 869–881. [Google Scholar] [CrossRef]
- Pope, P.B.; Smith, W.; Denman, S.E.; Tringe, S.G.; Barry, K.; Hugenholtz, P.; McSweeney, C.S.; McHardy, A.C.; Morrison, M. Isolation of succinivibrionaceae implicated in low methane emissions from tammar wallabies. Science 2011, 333, 646–648. [Google Scholar] [CrossRef]
- Auffret, M.D.; Stewart, R.D.; Dewhurst, R.J.; Duthie, C.A.; Watson, M.; Roehe, R. Identification of microbial genetic capacities and potential mechanisms within the rumen microbiome explaining differences in beef cattle feed efficiency. Front. Microbiol. 2020, 11, 1229. [Google Scholar] [CrossRef]
- Zhang, Y.K.; Zhang, X.X.; Li, F.D.; Li, C.; Li, G.Z.; Zhang, D.Y.; Song, Q.Z.; Li, X.L.; Zhao, Y.; Wang, W.M. Characterization of the rumen microbiota and its relationship with residual feed intake in sheep. Animal 2021, 15, 100161. [Google Scholar] [CrossRef]
- Zened, A.; Combes, S.; Cauquil, L.; Mariette, J.; Klopp, C.; Bouchez, O.; Troegeler-Meynadier, A.; Enjalbert, F. Microbial ecology of the rumen evaluated by 454 GS FLX pyrosequencing is affected by starch and oil supplementation of diets. FEMS Microbiol. Ecol. 2013, 83, 504–514. [Google Scholar] [CrossRef]
- Huang, C.; Ge, F.; Yao, X.; Guo, X.; Bao, P.; Ma, X.; Wu, X.; Chu, M.; Yan, P.; Liang, C. Microbiome and metabolomics reveal the effects of different feeding systems on the growth and ruminal development of Yaks. Front. Microbiol. 2021, 12, 682989. [Google Scholar] [CrossRef]
- Dai, X.; Tian, Y.; Li, J.; Luo, Y.; Liu, D.; Zheng, H.; Wang, J.; Dong, Z.; Hu, S.; Huang, L. Metatranscriptomic analyses of plant cell wall polysaccharide degradation by microorganisms in the cow rumen. Appl. Environ. Microbiol. 2015, 81, 1375–1386. [Google Scholar] [CrossRef] [PubMed]
- Walker, N.D.; McEwan, N.R.; Wallace, R.J. Cloning and functional expression of dipeptidyl peptidase IV from the ruminal bacterium Prevotella albensis M384(T). Microbiology 2003, 149, 2227–2234. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.Y.; Sun, H.Z.; Wu, X.H.; Liu, J.X.; Guan, L.L. Multi-omics reveals that the rumen microbiome and its metabolome together with the host metabolome contribute to individualized dairy cow performance. Microbiome 2020, 8, 64. [Google Scholar] [CrossRef] [PubMed]
- Paz, H.A.; Hales, K.E.; Wells, J.E.; Kuehn, L.A.; Freetly, H.C.; Berry, E.D.; Flythe, M.D.; Spangler, M.L.; Fernando, S.C. Rumen bacterial community structure impacts feed efficiency in beef cattle. J. Anim. Sci. 2018, 96, 1045–1058. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Zhang, Y.; Yu, Z.; Xu, Q.; Zheng, N.; Zhao, S.; Huang, G.; Wang, J. Ruminal microbiota-host interaction and its effect on nutrient metabolism. Anim. Nutr. 2021, 7, 49–55. [Google Scholar] [CrossRef]
- Malmuthuge, N.; Guan, L.L. Understanding host-microbial interactions in rumen: Searching the best opportunity for microbiota manipulation. J. Anim. Sci. Biotechnol. 2017, 8, 8. [Google Scholar] [CrossRef]
| Item 3 | AY 1 | YL 2 | Pooled SEM | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| Heifers | Steers | Heifers | Steers | Breed | Sex | B × S | ||
| Initial BW (kg) | 445 | 489 | 274 | 327 | 10.2 | <0.001 | <0.001 | 0.665 |
| Final BW (kg) | 564a | 569a | 331c | 393b | 13.8 | <0.001 | 0.020 | 0.044 |
| DMI (kg/d) | 7.99 | 8.07 | 4.04 | 5.51 | 0.42 | <0.001 | 0.073 | 0.109 |
| ADG (g/d) | 989a | 663b | 469c | 546bc | 53.1 | <0.001 | 0.025 | 0.001 |
| G/F (g/kg) | 127 | 83 | 122 | 112 | 13.6 | 0.384 | 0.053 | 0.232 |
| Item 3 | AY 1 | YL 2 | Pooled SEM | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| Heifers | Steers | Heifers | Steers | Breed | Sex | B × S | ||
| GLU (mmol/L) | 2.55 | 3.05 | 2.70 | 3.33 | 0.284 | 0.435 | 0.047 | 0.857 |
| TG (mmol/L) | 0.29 | 0.28 | 0.31 | 0.27 | 0.023 | 0.810 | 0.256 | 0.448 |
| CHO (mmol/L) | 4.08 | 4.02 | 4.09 | 3.93 | 0.266 | 0.882 | 0.685 | 0.837 |
| NEFA (mmol/L) | 0.13 | 0.19 | 0.16 | 0.20 | 0.025 | 0.401 | 0.073 | 0.711 |
| BHB (mmol/L) | 0.13 | 0.18 | 0.14 | 0.20 | 0.019 | 0.422 | 0.007 | 0.538 |
| HDL-C (mmol/L) | 0.29 | 0.28 | 0.31 | 0.27 | 0.089 | 0.105 | 0.596 | 0.289 |
| LDL-C (mmol/L) | 0.93 | 0.96 | 1.00 | 0.92 | 0.069 | 0.880 | 0.745 | 0.423 |
| CREA (mmol/L) | 0.10 | 0.11 | 0.10 | 0.11 | 0.007 | 0.777 | 0.329 | 0.742 |
| UREA (mmol/L) | 4.46 | 4.62 | 4.87 | 4.86 | 0.234 | 0.169 | 0.756 | 0.727 |
| AST (U/L) | 74.8 | 70.4 | 56.9 | 65.3 | 4.90 | 0.025 | 0.685 | 0.198 |
| ALT (U/L) | 23.6 | 24.6 | 21.3 | 24.6 | 1.57 | 0.455 | 0.174 | 0.479 |
| ALP (U/L) | 76.6 | 102.2 | 122.0 | 156.7 | 12.6 | <0.001 | 0.023 | 0.722 |
| TP (U/L) | 56.0 | 59.9 | 59.1 | 60.2 | 2.86 | 0.559 | 0.381 | 0.621 |
| ALB (U/L) | 32.4 | 33.4 | 33.2 | 33.6 | 1.20 | 0.645 | 0.554 | 0.793 |
| Item 3 | AY 1 | YL 2 | Pooled SEM | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| Heifers | Steers | Heifers | Steers | Breed | Sex | B × S | ||
| pH value | 6.50 | 6.65 | 6.73 | 6.68 | 0.062 | 0.046 | 0.419 | 0.128 |
| Ammonia-N (mg/dL) | 3.95b | 3.72b | 3.07b | 5.76a | 0.554 | 0.303 | 0.032 | 0.012 |
| VFA (mmol/d) | ||||||||
| TVFA | 119.8 | 120.7 | 105.0 | 121.0 | 8.01 | 0.374 | 0.302 | 0.350 |
| Acetate | 75.4 | 75.0 | 67.5 | 77.4 | 5.30 | 0.606 | 0.377 | 0.342 |
| Propionate | 25.3 | 26.4 | 21.8 | 23.3 | 1.82 | 0.081 | 0.491 | 0.908 |
| Isobutyrate | 1.05 | 1.23 | 0.94 | 1.70 | 0.203 | 0.385 | 0.027 | 0.156 |
| Butyrate | 14.4 | 14.1 | 11.7 | 14.7 | 1.00 | 0.292 | 0.188 | 0.120 |
| Isovalerate | 2.39 | 2.53 | 1.90 | 2.52 | 0.202 | 0.222 | 0.070 | 0.250 |
| Valerate | 1.34 | 1.41 | 1.17 | 1.51 | 0.090 | 0.680 | 0.033 | 0.141 |
| Acetate/propionate | 2.96 | 2.90 | 3.14 | 3.37 | 0.145 | 0.030 | 0.555 | 0.334 |
| Item | AY 1 | YL 2 | Pooled SEM | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| Heifers | Steers | Heifers | Steers | Breed | Sex | B × S | ||
| Observed OTUs | 1727 | 1719 | 1943 | 1753 | 83.6 | 0.138 | 0.240 | 0.279 |
| Good’s coverage (%) | 98.3 | 98.3 | 98.2 | 98.4 | 0.002 | 0.991 | 0.477 | 0.381 |
| Chao1 | 2374 | 2378 | 2677 | 2387 | 121 | 0.206 | 0.245 | 0.232 |
| Shannon | 7.80 | 7.86 | 8.30 | 7.92 | 0.167 | 0.104 | 0.333 | 0.186 |
| Simpson | 0.977 | 0.985 | 0.987 | 0.981 | 0.006 | 0.527 | 0.848 | 0.204 |
| PD whole tree | 137 | 139 | 149 | 142 | 4.53 | 0.102 | 0.611 | 0.358 |
| Item | AY 1 | YL 2 | Pooled SEM | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| Heifers | Steers | Heifers | Steers | Breed | Sex | B × S | ||
| Bacteroidetes | 63.6 | 60.1 | 64.6 | 56.0 | 3.57 | 0.677 | 0.100 | 0.482 |
| Firmicutes | 30.5 | 29.8 | 29.3 | 38.6 | 3.74 | 0.320 | 0.260 | 0.187 |
| Proteobacteria | 2.91b | 6.69a | 3.24b | 2.09b | 1.11 | 0.063 | 0.246 | 0.033 |
| .Actinobacteria | 0.956 | 0.888 | 0.369 | 0.592 | 0.345 | 0.209 | 0.824 | 0.676 |
| Patescibacteria | 0.600 | 0.637 | 0.463 | 0.692 | 0.129 | 0.754 | 0.309 | 0.463 |
| Fibrobacteres | 0.287 | 0.467 | 0.524 | 0.302 | 0.120 | 0.766 | 0.864 | 0.103 |
| Spirochaetes | 0.339 | 0.360 | 0.461 | 0.321 | 0.070 | 0.558 | 0.396 | 0.255 |
| Verrucomicrobia | 0.226 | 0.300 | 0.371 | 0.401 | 0.0470 | 0.013 | 0.278 | 0.641 |
| Desulfobacteria | 0.199 | 0.226 | 0.268 | 0.328 | 0.0273 | 0.003 | 0.118 | 0.553 |
| Cyanobacteria | 0.173 | 0.299 | 0.201 | 0.255 | 0.0598 | 0.890 | 0.140 | 0.555 |
| Others | 0.203 | 0.271 | 0.208 | 0.389 | 0.0453 | 0.178 | 0.009 | 0.218 |
| Item | AY 1 | YL 2 | Pooled SEM | p Value | ||||
|---|---|---|---|---|---|---|---|---|
| Heifers | Steers | Heifers | Steers | Breed | Sex | B × S | ||
| Prevotella | 46.4 | 43.5 | 42.4 | 36.5 | 4.13 | 0.189 | 0.296 | 0.721 |
| Christensenellaceae R-7 group | 3.41 | 3.87 | 3.72 | 6.98 | 1.05 | 0.112 | 0.086 | 0.190 |
| Ruminococcus | 4.10 | 3.19 | 3.48 | 3.67 | 0.634 | 0.910 | 0.574 | 0.392 |
| Succiniclasticum | 3.67 | 3.39 | 3.59 | 2.85 | 0.569 | 0.586 | 0.376 | 0.688 |
| NK4A214 group | 2.21 | 2.58 | 2.59 | 4.63 | 0.671 | 0.080 | 0.082 | 0.222 |
| Rikenellaceae RC9 gut group | 1.98 | 2.72 | 2.75 | 4.36 | 0.421 | 0.007 | 0.008 | 0.304 |
| Succinivibrionaceae UCG-002 | 1.46b | 4.51a | 2.33ab | 1.27b | 0.885 | 0.191 | 0.269 | 0.026 |
| Prevotellaceae UCG-003 | 1.85 | 1.93 | 3.01 | 2.36 | 0.219 | 0.001 | 0.206 | 0.105 |
| Prevotellaceae UCG-001 | 1.83b | 1.76b | 2.59a | 1.32b | 0.249 | 0.532 | 0.011 | 0.021 |
| Lachnospiraceae NK3A20 group | 1.04 | 1.38 | 0.92 | 2.38 | 0.533 | 0.416 | 0.100 | 0.299 |
| Lachnospiraceae ND3007 group | 0.607 | 0.773 | 1.018 | 0.848 | 0.289 | 0.408 | 0.995 | 0.566 |
| Lachnospiraceae XPB1014 group | 0.575 | 0.722 | 0.636 | 1.287 | 0.246 | 0.212 | 0.114 | 0.313 |
| Prevotellaceae NK3B31 group | 0.591 | 0.626 | 0.947 | 0.606 | 0.151 | 0.272 | 0.318 | 0.221 |
| Prevotellaceae UCG-004 | 0.533 | 0.542 | 1.020 | 0.474 | 0.200 | 0.304 | 0.188 | 0.174 |
| Acetitomaculum | 0.502 | 0.530 | 0.367 | 1.034 | 0.211 | 0.388 | 0.108 | 0.139 |
| Butyrivibrio | 0.639 | 0.699 | 0.458 | 0.544 | 0.124 | 0.182 | 0.560 | 0.917 |
| CAG-352 | 0.504 | 0.543 | 0.840 | 0.399 | 0.127 | 0.453 | 0.123 | 0.067 |
| Succinivibrio | 0.717 | 0.886 | 0.210 | 0.142 | 0.215 | 0.006 | 0.817 | 0.584 |
| Others (<0.5%) | 27.3 | 25.8 | 27.2 | 28.3 | 1.78 | 0.517 | 0.924 | 0.451 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qiu, X.; Qin, X.; Chen, L.; Chen, Z.; Hao, R.; Zhang, S.; Yang, S.; Wang, L.; Cui, Y.; Li, Y.; et al. Serum Biochemical Parameters, Rumen Fermentation, and Rumen Bacterial Communities Are Partly Driven by the Breed and Sex of Cattle When Fed High-Grain Diet. Microorganisms 2022, 10, 323. https://doi.org/10.3390/microorganisms10020323
Qiu X, Qin X, Chen L, Chen Z, Hao R, Zhang S, Yang S, Wang L, Cui Y, Li Y, et al. Serum Biochemical Parameters, Rumen Fermentation, and Rumen Bacterial Communities Are Partly Driven by the Breed and Sex of Cattle When Fed High-Grain Diet. Microorganisms. 2022; 10(2):323. https://doi.org/10.3390/microorganisms10020323
Chicago/Turabian StyleQiu, Xinjun, Xiaoli Qin, Liming Chen, Zhiming Chen, Rikang Hao, Siyu Zhang, Shunran Yang, Lina Wang, Yafang Cui, Yingqi Li, and et al. 2022. "Serum Biochemical Parameters, Rumen Fermentation, and Rumen Bacterial Communities Are Partly Driven by the Breed and Sex of Cattle When Fed High-Grain Diet" Microorganisms 10, no. 2: 323. https://doi.org/10.3390/microorganisms10020323
APA StyleQiu, X., Qin, X., Chen, L., Chen, Z., Hao, R., Zhang, S., Yang, S., Wang, L., Cui, Y., Li, Y., Ma, Y., Cao, B., & Su, H. (2022). Serum Biochemical Parameters, Rumen Fermentation, and Rumen Bacterial Communities Are Partly Driven by the Breed and Sex of Cattle When Fed High-Grain Diet. Microorganisms, 10(2), 323. https://doi.org/10.3390/microorganisms10020323







