Abstract
The incidence of type 1 diabetes (T1D) has been dramatically increased in developed countries, and beyond the genetic impact, environmental factors, including diet, seem to play an important role in the onset and development of the disease. In this vein, five Lacticaseibacillus rhamnosus, isolated from traditional fermented Greek products, were screened for potential probiotic properties, aiming at maintaining gut homeostasis and antidiabetic capability to alleviate T1D symptoms. L. rhamnosus cell-free supernatants induced strong growth inhibitory activity against common food spoilage and foodborne pathogenic microorganisms, associated with several diseases, including T1D, and were also able to inhibit α-glucosidase activity (up to 44.87%), a promising property for alternatives to the antidiabetic drugs. In addition, survival rates up to 36.76% were recorded during the application of the static in vitro digestion model. The strains had no hemolytic activity and were sensitive to common antibiotics suggested by the European Food and Safety Association, apart from chloramphenicol. However, it is highly unlikely that the resistance has been acquired. In conclusion, our results suggest a great health-promoting potential of the newly isolated wild-type L. rhamnosus strains, but further confirmation of their efficiency in experimental animal models is considered an essential next research step.
1. Introduction
Today, an upsurge of interest in developing novel functional foods containing probiotic microorganisms is witnessed. According to the World Health Organization and Food and Agriculture Organization (WHO/FAO), probiotics are commonly defined as viable microorganisms (bacteria or yeasts) that, when administered in adequate amounts, confer a health benefit on the host [1]. Several studies available in the literature have clearly shown that probiotics are effective in reducing the risk of antibiotic-associated diarrhea and its duration in healthy children [2,3], reducing levels of coliforms in different parts of the intestine [4,5,6], modulating gut microbiota and preventing sensitization to foods [7,8], improving glucose intolerance and immune responses [9,10,11,12], etc.
To induce the health benefits, probiotics should be able to survive the acidic conditions of the upper gastrointestinal (GI) tract and proliferate and colonize in the gut [13,14], since they may influence interaction with the host and the other bacteria present, affect the local microbial composition, and/or stimulate the host’s immune system [12,15].
Type 1 diabetes (T1D) mellitus is characterized by the destruction of insulin-producing pancreatic beta cells, leading to insulin deficiency and increased glucose levels in blood and urine. Hyperglycemia in patients with T1D can be fatal if not treated with insulin, and some of its most recurrent clinical symptoms are weight loss, polyuria, polydipsia, and polyphagia [16]. In the last decades, the incidence of T1D has been dramatically increased in developed countries, and beyond the genetic impact, environmental factors, including diet, seem to play an important role in the onset and the development of the disease. The intestinal microbiome might affect the interaction between the GI tract and the immune system and result in altered immune responses, affecting the development of T1D [17]. Thus, the restoration of the normal microbiota composition, which could be accomplished with a probiotic-rich diet, constitutes a new target for the prevention and management of the disease.
At the same time, food spoilage and safety are considered among the most pressing public health issues, due to economic reasons and constant outbreaks of food-borne diseases. As a result, probiotics, such as lactic acid bacteria (LAB), have recently received increased interest in the food industry for maintaining human health and acting as a shield against food spoilage microorganisms and pathogens. The antimicrobial activity of LAB, which has been studied against a variety of spoilage microorganisms and several healthcare-associated pathogens [18,19,20], has been attributed to a wide spectrum of antimicrobial compounds, such as organic acids, bacteriocins, hydrogen peroxide, diacetyl, and others [21].
Indeed, foodborne pathogens have been associated with several diseases, including T1D. Due to the hyperglycemic environment, a dysfunction of the immune system is usually observed in T1D patients, resulting in the development of several serious either healthcare-associated or community-based infections. Thus, diabetic patients may have a predisposition to infections caused by enteric pathogenslike Escherichia coli, Salmonella Enteritidis, Listeria monocytogenes, Clostridium difficile, etc., due to gastrointestinal dysmotility syndromes associated with diabetes [22,23]. Furthermore, diabetic patients are susceptible to invasive fungal infections, such as pulmonary aspergillosis [24].
However, the health-promoting effects resulting from probiotics seem to be strain-specific rather than characteristic of a specific species. Therefore, the discovery and isolation of health-promoting bacteria are considered of utmost importance to any commercialization strategy of probiotic products.
In the present study, Lacticaseibacillus rhamnosus (previously classified as Lactobacillus rhamnosus) strains, isolated from traditional fermented Greek products, were screened in vitro for potential probiotic properties, aiming at maintaining gut homeostasis and antidiabetic capability to alleviate T1D symptoms. Data supporting significant growth inhibition activity of cell-free supernatants (CFSs) against common food spoilage microbes and food-borne pathogens, α-glucosidase inhibitory activity, and resistance to common antibiotics are presented.
2. Materials and Methods
2.1. Isolation of Lactic Acid Bacteria
Brine from naturally fermented Greek olives and olive fruits (Olea europaea var. rotunda) collected at the end of the fermentation process, as well as homemade cheese samples produced with no starter cultures, were collected for isolation of lactic acid bacteria. In brief, olives (10 g), cheese (10 g), and brine (1 mL) samples were macerated into de Man, Rogosa, and Sharpe (MRS) broth medium (Condalab, Madrid, Spain) and incubated at 37 °C for 24 h. Τhereafter, decimal dilutions were prepared in sterile quarter-strength Ringer’s solution, and 1 mL of each appropriate dilution was poured onto MRS agar (Condalab, Madrid, Spain) medium and incubated at 37 °C for 48 h. Colonies with typical characteristics were collected from the highest dilution and were picked for routinely streaking to obtain pure colonies. Each strain was maintained at −80 °C in MRS broth/glycerol (70:30).
2.2. Bacterial Identification
All isolates were tested for Gram staining, cell morphology, and catalase reaction, and only Gram-positive and catalase-negative strains were collected. Initial characterization was performed by the API50CH biochemical system (BioMèrieux, La Balme-les-Grottes, France), according to the manufacturer’s instructions and using the APIwebTM software. For molecular identification, the strains were inoculated into MRS broth and 1 mL of overnight culture was collected for genomic DNA extraction utilizing the Nucleospin tissue kit (Macherey-Nagel, Düren, Germany), according to the manufacturer’s instructions. Species-specific PCR reactions for the Lacticaseibacillus casei group were carried out in a volume of 20 μL with primers targeting mutL gene, i.e., CZfor (5′-CAGCGCTGGTGGAAGACTTG-3′), PC2a (5′-GGATTGGGTTTTGCGTGATGGTCGC-3′), RHfor (5′-GACTTCTCAACCAGCAGCGCAGA-3′), and CPRrev (5′-TGCATTTCCCCGCTTTCATGACT-3′), as described recently [25]. Amplification reactions were performed using an Eppendorf 5333 MasterCycler Thermal Cycler (Hamburg, Germany). PCR products were analyzed by electrophoresis (Bio-Rad Laboratories, Hercules CA, USA) in 1.5% agarose gel in 1% Tris-Borate-EDTA (TBE) and were visualized by UV transillumination (Bio-Rad Laboratories, Hercules, CA, USA).
2.3. Cell-Free Supernatant Preparation
Wild-type L. rhamnosus cultures were grown in MRS broth (pH 6.5) for 24 h at 37 °C, and then cell-free supernatants (CFSs) were obtained by centrifugation (8000× g for 20 min, 4 °C). CFSs were sterilized by filtration with a 0.22 μm filter (Merck, Darmstadt, Germany). L. rhamnosus GG (ATCC 53103) (Probiotical SpA, Novara, Italy) was used as a reference strain for comparison reasons.
2.4. Antimicrobial Activity of L. rhamnosus CFSs
2.4.1. Food Spoilage and Pathogenic Microbial Strains
Salmonella enterica subsp. enterica ser. Enteritidis FMCC B56 PT4 (kindly provided by the Laboratory of Microbiology and Biotechnology, Agricultural University of Athens, Athens, Greece), S. enterica subsp. enterica ser. Typhimurium B62 DSM 554, Escherichia coli ATCC 25922 (kindly provided by Dr. Nisiotou A., Athens Wine Institute, ELGO-DIMITRA, Athens, Greece), Listeria monocytogenes NCTC 10527 serotype 4b, Staphylococcus aureus ATCC 25923, and Clostridium difficile (kindly provided by the Laboratory of Clinical Microbiology, Sismanoglio General Hospital, Athens, Greece) were grown in brain heart infusion (BHI) broth (LAB M, Heywood, UK) at 37 °C for 24 h, except C. difficile, which was incubated at 37 °C for 48 h. Saccharomyces cerevisiae Uvaferm NEM (Lallemand, Montreal, QC, Canada) was grown in yeast extract peptone dextrose (YPD) broth (yeast extract 10 g/L, glucose 20 g/L, and peptone 20 g/L) at 28 °C for 24 h. Aspergillus niger 19111 (kindly provided by G.J.E. Nychas, Agricultural University of Athens, Athens, Greece) was grown on Malt agar (Condalab, Madrid, Spain) at 37 °C for 7 days.
2.4.2. Assessment of Growth Inhibition Activity of L. rhamnosus CFS
Growth inhibition activity against common food spoilage and pathogenic microorganisms was evaluated by the broth microdilution method, as described by Mayrhofer et al. [26], with slight modifications. Growth inhibition activity of various concentrations (3.12, 6.25, 12.5, 25, 50, 70, and 90%) of non-neutralized and neutralized (pH adjusted to 7.0 using 5 M NaOH) CFSs diluted to MRS broth was tested in a 96-well microplate. In each well, 100 μL of bacterial or yeast or fungus culture (105 cfu/mL or 105 spores/mL) and 100 μL of CFS were added to achieve a total volume of 200 μL [27], followed by incubation at 37 °C for 24 h, apart from C. difficile, which was incubated at 37 °C for 48 h, and S. cerevisiae Uvaferm NEM, which was incubated at 28 °C for 24 h. Wells with bacterial or yeast/fungal cultures with MRS broth containing no CFSs served as growth controls. The growth of spoilage microorganisms was monitored by measuring optical density (OD 620 nm) using VarioskanTM LUX Multimode Microplate Reader (Thermo Scientific, Vantaa, Finland). The percentage of growth inhibition was calculated based on the following equation:
where I (%) is the percentage of growth inhibition of the sample compared with the growth of the control, A620 (control) represents the changes in the optical density during growth of the control, and A620 (sample) represents the changes in the optical density of the sample [19].
2.5. Determination of α-Glucosidase Inhibition
α-Glucosidase inhibitory activity was assessed according to the method described by Zeng et al. [28], modified as follows. The reaction mixture containing 50 μL of CFS (pH adjusted to 7.4 with the addition of 5 M NaOH) or 10 mg/mL acarbose (reference inhibitor) (Sigma-Aldrich, St. Louis, MO, USA), 125 μL of phosphate-buffered saline (PBS) (0.80% NaCl, 0.02% KCl, 0.02% KH2PO4, and 0.22% Na2HPO4 (pH 7.4)), and 50 μL of 0.1 U/mL α-glucosidase from Saccharomyces cerevisiae (Type 1, ≥10 U/mg) (Sigma-Aldrich, St. Louis, MO, USA) was pre-incubated in a 96-well plate at 37 °C for 10 min. After pre-incubation, the reaction was initiated by adding 25 μL of 3 mM p-nitrophenyl α-D-glucopyranoside (pNPG) (Sigma-Aldrich, St. Louis, MO, USA), and incubation at 37 °C for 10 min was followed. Absorbance readings were recorded at 405 nm by VarioskanTM LUX Multimode Microplate Reader (Thermo Scientific, Vantaa, Finland), and α-glucosidase inhibition was calculated using the following equation:
where A405 (sample) is the absorbance with the sample and α-glucosidase, A405 (sample blank) is the absorbance with the sample without α-glucosidase, A405 (positive control) is the absorbance with α-glucosidase without the sample, and A405 (negative control) is the absorbance without α-glucosidase or the sample.
2.6. Static In Vitro Digestion Model
A static in vitro digestion model for probiotic strains screening was developed based on the previous study of Minekus et al. [29] with adaptations by Madureira et al. [30]. L. rhamnosus cells, grown in MRS broth, were harvested by centrifugation, washed twice with quarter-strength Ringer’s solution (Sigma-Aldrich, St. Louis, MO, USA), resuspended in simulated salivary fluid (SSF) [29] containing 75 U/mL α-amylase from Bacillus spp. (Type-IIA, ≥1500 U/mg) (Sigma-Aldrich, St. Louis, MO, USA) and 50 U/mL lysozyme from chicken egg white (≥20,000 U/mg) (Apollo Scientific, Cheshire, UK) and incubated at 37 °C for 2 min. Subsequently, cells were harvested once again by centrifugation, resuspended in simulated gastric fluid (SGF) [29] containing 2000 U/mL pepsin from porcine mucosa (≥3.200 U/mg) (Sigma-Aldrich, St. Louis, MO, USA) and incubated at 37 °C for 110 min. During incubation, pH in SGF was gradually reduced from 4.9 to 3.0 (10 min pH 4.5, 10 min pH 4.0, and 90 min pH 3.0), mimicking the transit from the oral cavity to stomach [30]. After the simulated gastric phase, cells were harvested by centrifugation again, resuspended in simulated intestinal fluid (SIF) [29] containing 100 U/mL pancreatin from porcine pancreas (8 × USP specifications) (Sigma-Aldrich, St. Louis, MO, USA) and incubated at 37 °C for 120 min. In all cases during the three simulated digestion phases, centrifugation conditions were 8000× g, 4 °C, 15 min, the harvested cells were resuspended in equal volumes, enzyme solutions were prepared fresh daily, and pH was adjusted with the addition of 0.5 M HCl or 1 M NaHCO3. Samples of 1 mL were collected right after the addition of SSF and after the end of each simulated digestion phase. Viable L. rhamnosus cell counts were determined by 10-fold serial dilution and pour plate method on MRS agar. MRS plates were incubated anaerobically at 37 °C for at least 72 h. The survival rates were calculated in accordance with the following equation [28]:
where α is the total count of viable cells after incubation for the specified digestion phase, and β is the total count of viable cells before the beginning of simulated digestion.
2.7. Safety Evaluation
Hemolytic activity of isolates was examined on blood agar plates containing 5% (w/v) sheep blood. Fresh overnight cultures were standardized to 108 cfu/mL and streaked on the plates. Colonies were investigated for surrounding zones (α and β hemolysis) after incubation at 30 °C for 48 h. Staphylococcus aureus MRSA, a clinical isolate kindly provided by the Laboratory of Clinical Microbiology, Sismanoglio General Hospital, Greece, was used as the positive control strain for β-hemolysis and Lacticaseibacillus rhamnosus GG as the negative control.
Furthermore, susceptibility to common antibiotics was also determined. The concentration ranges (mg/L) of the tested antibiotics were: ampicillin (0.032–16), clindamycin (0.032–16), erythromycin (0.016–8), gentamicin (0.5–256), streptomycin (0.5–256), tetracycline (0.125–64), kanamycin (2–1024), and chloramphenicol (0.125–64). Antibiotic susceptibility tests were performed using the broth microdilution method, according to the standard procedure of the International Organization for Standardization (ISO 10932/IDF 223:2010). Briefly, the inocula of the strains were prepared by suspending colonies in 0.85% NaCl solution and subsequently diluted in LAB susceptibility test medium (LSM) broth (HiMedia Laboratories, Mumbai, India). To each well, 100 μL of diluted inoculum and 100 μL of antibiotic solution were added, following incubation at 37 °C for 48 h. The minimum inhibitory concentration (MIC) was defined as the lowest concentration of antibiotics that inhibit the visible growth of the isolates, and the resulting values were compared with the microbiological cut-off values defined by the European Food Safety Authority’s (EFSA) recommendations [31].
2.8. Statistical Analysis
All treatments were carried out at least in duplicate. Statistical significance was determined by factorial analysis of variance (ANOVA), followed by Duncan’s multiple range test. Statistica version 10.0 was used to compute statistical significance at p < 0.05, coefficients, and ANOVA.
3. Results and Discussion
Probiotic bacteria should fulfill several requirements related to their ability in inhibiting the growth of food-spoilage microbes and pathogens, reducing α-glucosidase activity, reaching the lower GI tract intact, and being susceptible to antibiotics, among others.
3.1. L. rhamnosus Strains Isolation and Identification
Strains isolated from brine, fermented table olives, and cheese samples were examined for catalase reaction, Gram staining, and stable colony morphology after subculturing on MRS agar, and five strains were collected for further analysis. These strains were non-motile, Gram-positive, catalase-negative, rod-shaped and were preliminarily identified as Lacticaseibacillus rhamnosus using API50CH biochemical tests. Further molecular confirmation was based on species-specific primers for the Lacticaseibacillus casei group and utilizing Lacticaseibacillus rhamnosus GG, Lacticasebacillus casei ATCC 393, and Lacticaseibacillus paracasei DSM 20006 as reference strains. Targeting the mutL locus revealed genome diversity among the L. casei group and effectively differentiated the species of L. rhamnosus (801 bp PCR product), L. casei (666 bp PCR product), and L. paracasei (253 bp PCR product) [25]. PCR products of 800 bp confirmed the initial characterization of the new isolates as L. rhamnosus (Figure 1). Of note, the species L. rhamnosus is included in the Qualified Presumption of Safety (QPS), a list indicating its suitability to food industry applications [32].
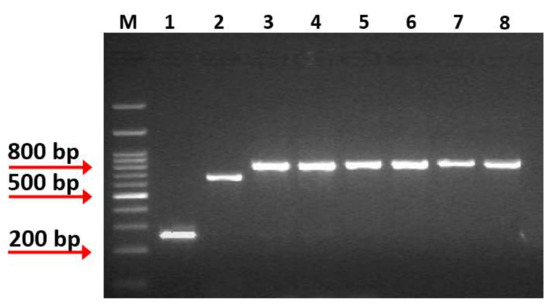
Figure 1.
Multiplex PCR amplification products obtained from species-specific mutL-targeting primers assay. Lane M: 1kb DNA Ladder (Takara, Shiga, Japan), Lane 1: PCR product from L. paracasei DSM 20006, Lane 2: PCR product from L. casei ATCC 393, Lane 3: PCR product from L. rhamnosus GG, Lane 4: PCR product from isolate OLXAL-1, Lane 5: PCR product from isolate OLXAL-2, Lane 6: PCR product from isolate OLXAL-3, Lane 7: PCR product from isolate OLXAL-4, Lane 8: PCR product from isolate CHTH-2.
3.2. Growth Inhibition Activity of L. rhamnosus CFSs against Food Spoilage and Pathogenic Microorganisms
In the present study, we investigated the antagonistic activity of five L. rhamnosus strains against common food spoilage and pathogenic microorganisms. Food spoilage and pathogenicity are mainly due to the growth and activity of several microbial species, including, among others, Escherichia coli, Salmonella spp., Listeria monocytogenes, Clostridium spp., Aspergillus spp., and Saccharomyces spp. [33,34,35]. Infections by enteric pathogens are also usual in diabetic patients. For example, it has been previously reported that insulin may stimulate E. coli growth and enhance its ability to form biofilms [36]. Considering healthcare-associated infections, C. difficile has been related to high-risk rates in T1D patients [37,38], as they usually need to be hospitalized for a long time, increasing the risk of nosocomial infections.
The CFSs (pH 3.72–3.87) induced strong growth inhibitory activity at high concentrations (≥ 25%) against all bacterial pathogens tested, reaching values up to 90–99.9% (Pavlatou, C. Laboratory of Applied Microbiology and Biotechnology, Department of Molecular Biology & Genetics, Democritus University of Thrace, Alexandroupolis, Greece. Growth Inhibition Activity of L. rhamnosus CFSs against Food Spoilage and Pathogenic Microorganisms, 2022). Reducing the concentration of CFSs down to 12.5%, the majority of CFSs maintained strong inhibitory activity (88–98%) against S. Enteritidis, S. Typhimurium, E. coli, L. monocytogenes and S. aureus (Figure S1). Regarding C. difficile, a significant (p < 0.05) reduction in the growth inhibitory ability of all CFSs at 12.5% concentration was observed (38.1–56.8%) compared with higher concentrations (25 and 50%) (Figure S1). At concentrations below 6.25%, all CFSs exhibited significantly (p < 0.05) lower growth inhibitory activity in all pathogens examined (Figure S1), in accordance with Chen et al. [27] and Arena et al. [39]. The only exception noted concerned CFSs of L. rhamnosus OLXAL-1 and L. rhamnosus OLXAL-2 that strongly inhibited the growth of S. aureus (69.7% and 67.9%, respectively) even at 3.12% concentration. Similarly, Inturri et al. [40] reported very strong inhibitory activity of CFSs at concentrations under 12.5% against S. aureus.
Likewise, all CFSs exhibited strong growth inhibitory activity (88–100%) against S. cerevisiae and A. niger at 90% concentration. At lower concentrations (70%), a weaker (p < 0.05) growth inhibitory activity was observed (75–86% against S. cerevisiae and 65–79% against A. niger). Further decrease in CFSs concentration down to 50% resulted in even lower (p < 0.05) growth inhibition rates (55–63% against S. cerevisiae and 53–58% against A. niger) compared with higher concentrations (70% and 90%), while at CFSs concentrations below 25% a further growth inhibition (p < 0.05) was witnessed.
To investigate the nature of antimicrobial substances secreted by L. rhamnosus strains, the CFSs were submitted to neutralization and tested again as growth inhibitors. Neutralization of CFSs resulted in a drastic reduction (p < 0.05) in growth inhibitory activity against all bacterial pathogens tested (Figure 2), in accordance with Hor et al. [19] and Munoz et al. [41]. At 50% of neutralized CFSs concentration, the highest growth inhibitory activity was observed against L. monocytogenes and S. aureus (53.8% and 54.3%, respectively) (Figure 2). Lower concentrations (3.12, 6.25, 12.5, 25%) resulted in weaker (p < 0.05) antagonistic ability, reaching rates <36.2%.
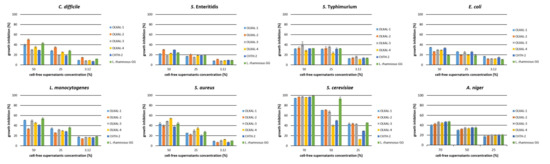
Figure 2.
Growth inhibition activity of neutralized cell-free supernatants (CFSs) of L. rhamnosus strains against food spoilage and pathogenic microorganisms. Values are expressed as a percentage of growth inhibition compared with the control.
Similar results were also observed for A. niger. In specific, at concentration 50% and 70% of neutralized CFSs, 30–35% and 40–47% inhibitory activity, respectively, was observed, and at lower concentrations, antifungal activity was reduced, in accordance with previous studies [42,43].
On the other hand, all neutralized CFSs at 90% concentration maintained strong growth inhibitory activity (97.3–99.4%) against S. cerevisiae Uvaferm NEM (Pavlatou, C. Laboratory of Applied Microbiology and Biotechnology, Department of Molecular Biology & Genetics, Democritus University of Thrace, Alexandroupolis, Greece. Growth Inhibition Activity of L. rhamnosus CFSs against Food Spoilage and Pathogenic Microorganisms, 2022), similar to untreated CFSs. Interestingly, at 70% CFSs concentration, the inhibitory activity of neutralized CFSs remained strong (p > 0.05), in contrast with untreated CFSs, where the antagonistic activity was reduced (p < 0.05), probably due to the increase in pH, as optimum pH range for growth is 4 to 4.54 [44]. Further 2-fold dilutions of neutralized CFSs resulted in lower inhibition rates, and the highest values were observed in L. rhamnosus GG, used as reference strain (p < 0.05).
The weaker growth inhibitory activity of neutralized CFSs against spoilage bacteria could be attributed to the organic acids resulting from cell metabolism. Production of organic acids leads to pH reduction, unsuitable for the growth of a wide spectrum of both Gram-positive and Gram-negative bacteria. More specifically, organic acids can pass through the cytoplasmic membrane of target microorganisms in their undissociated form, causing intracellular acidification and the collapse of the transmembrane proton motive force [39].
3.3. α-Glucosidase Inhibition
Insulin resistance is common among patients with T1D because of the constantly increasing doses of insulin, which in many cases results in weight gain. According to previous studies, concurrent administration of insulin and antidiabetic drugs, approved for the treatment of type 2 diabetes (T2D), may result in a reduction in insulin resistance and hence better weight management for individuals with T1D [45]. In particular, competitive inhibitors of α-glucosidase (miglitol and acarbose) have resulted in significant decreases in body mass index (BMI), total daily insulin dosages, hemoglobin A1c (HbA1c) and postprandial glucose levels [46,47,48]. However, in many cases, side effects related to the GI system, such as diarrhea and flatulence, have been reported [49,50,51]. In an attempt to avoid these adverse effects, oral supplementation of probiotic strains with α-glucosidase inhibitory activity is considered a promising alternative to the antidiabetic drugs [49,52,53]. In this vein, the α-glucosidase inhibitory activity of five wild-type L. rhamnosus strains was evaluated.
CFSs of all strains were able to inhibit α-glucosidase activity (Table 1). The inhibitory activities ranged from 35.43% to 44.87%, with the highest value recorded in CFS of L. rhamnosus OLXAL-1 strain (44.87%), significantly higher (p < 0.05) compared with L. rhamnosus GG (39.68%), which was used as a reference strain with well-documented antidiabetic properties [49]. According to previous literature, the α-glucosidase inhibitory activity of reference strain L. rhamnosus GG has been reported to vary between 13.5% and 37.9% [28,49,52,53], a result that leads to confusion concerning the comparability among different studies of the assay. This result disagreement probably stems from the several modifications implemented on each assay, such as the different origin of enzyme and varying concentrations of enzyme and substrate [54]. Nevertheless, in all the aforementioned studies, CFS of LAB strains exhibited α-glucosidase inhibition, which may be attributed to either secreted polysaccharides [52,55] or peptides produced from bacteria [28,56].

Table 1.
Basic characteristics of wild-type L. rhamnosus strains and α-glucosidase inhibition (%).
3.4. L. rhamnosus Cell Survival during In Vitro Digestion
Tolerance to gastric acidity, bile salts, and digestive enzymes consists an essential prerequisite for in vitro characterization of probiotics. However, in many cases, tolerance of probiotic strains to the harsh conditions of the GI tract is evaluated by separated static in vitro assays, which fail to simulate the successive stress conditions that occur in vivo [57,58]. Thus, the implementation of an in vitro digestion assay that incorporates oral, gastric, and intestinal phases consecutively can be a more efficient way to study the survival of presumptive probiotic strains.
According to our results (Table 2), incubation in the simulated oral phase did not affect the initial viable cell counts of all L. rhamnosus strains tested, while incubation in the simulated gastric and intestinal phase led to a significant (p < 0.05) reduction. The survival rate in the simulated gastric phase ranged from 73.26% to 77.12%, indicating high tolerance to the acidic conditions for all strains. Simulated intestinal phase resulted in survival rates from 0 to 36.76%, while the reference strain L. rhamnosus GG showed zero survival rate, in accordance with Zeng et al. [28]. Overall, strains L. rhamnosus OLXAL-1, L. rhamnosus OLXAL-2, L. rhamnosus OLXAL-3, and L. rhamnosus OLXAL-4 demonstrated a survival rate >33.07% after simulated digestion, while L. rhamnosus CHTH-2 showed a significantly (p < 0.05) lower survival rate (17.49%).

Table 2.
Tolerance of LAB strains to simulated digestion phases.
3.5. Hemolytic Activity and Susceptibility to Antibiotics
Probiotic strains intended for use in the food industry should meet safety requirements apart from potential health-promoting effects.
Since the hemolytic activity of LAB cultures has been reported in previous studies [49,59], the type of hemolysis of isolated L. rhamnosus strains was investigated. According to our results, no clearing zone on blood agar was observed in any of the wild-type L. rhamnosus cultures (Figure S2). Hence, all strains were considered as γ-hemolytic.
Antibiotics resistance (AR) in the Lactobacillus genus is under continuous review, and many studies have investigated the resistant phenotype linked with genetic alterations [60,61]. The acquired resistance of LAB and the possible horizontal transfer of the resistance to the gut pathogens or human commensal bacteria are of major concern, and within this context, microbiological cut-off values intend to distinguish susceptible from resistant strains [31].
In our study, MIC values of the five L. rhamnosus isolates for common antibiotics were determined, and according to the results, all strains were susceptible to all tested antibiotics, apart from chloramphenicol (Table 3). Despite the phenotypical resistance to chloramphenicol, the resulting MIC value (8 mg/L) was just higher than the recommended cut-off value for L. rhamnosus species (4 mg/L) [31]. Thus, it is highly unlikely that the resistance was acquired, as microbial growth would be expected even after exposure to a much higher concentration [62]. Similar results were reported for L. rhamnosus strains of human and food origin, although for some antibiotics a wide range of MIC values has been presented [63,64]. For example, the MIC values for clindamycin and erythromycin ranged from 0.032 to 8 and 0.016 to 32 mg/L, respectively, and for chloramphenicol from 0.5 to 8 mg/L [63], in accordance with the present study. L. rhamnosus L-455 strain displayed high resistance to erythromycin, streptomycin, and clindamycin, despite the absence of resistance genes [63]. Another study determined the MIC values of L. rhamnosus strains [64] and presented comparable outcomes with our study, as well. Briefly, MIC values of ampicillin ranged from 0.5 to 8 mg/L by agar dilution and broth microdilution methods, and thus, a new cut-off value of 8 mg/L was proposed. Some strains were characterized as highly resistant to clindamycin, erythromycin, streptomycin, and tetracycline because the corresponding MIC values were higher (i.e., ≥256 mg/L of streptomycin compared with 32 mg/L, which is the cut-off value) than the breakpoints suggested by EFSA. A microarray method suggested the presence of resistance genes; however, it was not confirmed by PCR reactions [64].

Table 3.
MICs values (mg/L) determined for new wild-type L. rhamnosus strains in LAB susceptibility test medium (LSM) broth by the microdilution method [31].
In line with all mentioned above, a recent phylogenetic analysis revealed that within the L. casei-manihotivorans group, over 80% of the studied strains displayed chloramphenicol resistance, which was associated with the cat gene coding for chloramphenicol acetyltransferase [60].
Undoubtedly, antibiotic resistance is an issue under continued discussion that may be affected by many factors, such as the strain’s origin, applied methods, etc. Hence, future research is needed to gain insight into the safe use of probiotics or starter cultures in food production.
4. Conclusions
In summary, the results presented in the present work suggest the great potential of the newly isolated wild-type L. rhamnosus strains in maintaining gut homeostasis and as antidiabetic agents to alleviate T1D symptoms. However, further confirmation of their efficiency in experimental animal models is considered an essential next research step.
Supplementary Materials
The following are available online at: https://www.mdpi.com/article/10.3390/microorganisms10020272/s1, Figure S1: Growth inhibition activity of cell-free supernatants (CFSs) of L. rhamnosus strains against food spoilage and pathogenic microorganisms, Figure S2: Hemolytic activity of (a) L. rhamnosus OLXAL-1, (b) L. rhamnosus GG, and (c,d) S. aureus MRSA.
Author Contributions
Conceptualization, Y.K.; methodology, G.N., C.P., V.S. and D.D.; validation, G.N., V.S. and Y.K.; formal analysis, G.N. and V.S.; investigation, G.N., C.P., V.S. and D.D.; resources, Y.K.; data curation, G.N., C.P., V.S., D.D. and Y.K.; writing—original draft preparation, G.N., C.P. and V.S.; writing—review and editing, Y.K.; visualization, D.D., V.S. and Y.K.; supervision, Y.K.; project administration, Y.K.; funding acquisition, Y.K. All authors have read and agreed to the published version of the manuscript.
Funding
The research work was supported by the Hellenic Foundation for Research and Innovation (H.F.R.I.) under the “First Call for H.F.R.I. Research Projects to support Faculty members and Researchers and the procurement of high-cost research equipment grant” (Project Number: HFRI-FM17-2496, acronym: iFUNcultures).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to restrictions of the funding authorities.
Acknowledgments
We would like to thank Maria Tokamani for technical support on species-specific PCR and Evangelos Nennes for his artwork on the graphical abstract.
Conflicts of Interest
The authors declare no conflict of interest.
References
- World Health Organization (WHO); Food and Agriculture Organization of the United Nations (FAO). Guidelines for the evaluation of probiotics in food. In Report of a Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food; World Health Organization (WHO): London, ON, Canada, 2002; pp. 1–11. Available online: https://www.who.int/foodsafety/fs_management/en/probiotic_guidelines.pdf (accessed on 10 December 2021).
- Zakordonets, L.; Tolstanova, G.; Yankovskiy, D.; Dyment, H.; Kramarev, S. Different regimes of multiprobiotic for prevention of immediate and delayed side effects of antibiotic therapy in children. Res. J. Pharm. Biol. Chem. Sci. 2016, 7, 2194–2201. [Google Scholar]
- Guo, Q.; Goldenberg, J.Z.; Humphrey, C.; El Dib, R.; Johnston, B.C. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst. Rev. 2019, 4, CD004827. [Google Scholar] [CrossRef] [PubMed]
- Savino, F.; Cordisco, L.; Tarasco, V.; Locatelli, E.; Di Gioia, D.; Oggero, R.; Matteuzzi, D. Antagonistic effect of Lactobacillus strains against gas-producing coliforms isolated from colicky infants. BMC Microbiol. 2011, 11, 157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaikham, P.; Apichartsrangkoon, A. Effects of encapsulated Lactobacillus acidophilus along with pasteurized longan juice on the colon microbiota residing in a dynamic simulator of the human intestinal microbial ecosystem. Appl. Microbiol. Biotechnol. 2014, 98, 485–495. [Google Scholar] [CrossRef] [PubMed]
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Wu, W.; Qin, H. Probiotics modify human intestinal mucosa-associated microbiota in patients with colorectal cancer. Mol. Med. Rep. 2015, 12, 6119–6127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azad, M.A.K.; Sarker, M.; Li, T.; Yin, J. Probiotic species in the modulation of gut microbiota: An overview. Biomed Res. Int. 2018, 2018, 9478630. [Google Scholar] [CrossRef] [Green Version]
- Zhang, G.Q.; Hu, H.J.; Liu, C.Y.; Zhang, Q.; Shakya, S.; Li, Z.Y. Probiotics for prevention of atopy and food hypersensitivity in early childhood A PRISMA-compliant systematic review and meta-analysis of randomized controlled trials. Medicine 2016, 95, e2562. [Google Scholar] [CrossRef]
- Delzenne, N.M.; Neyrinck, A.M.; Bäckhed, F.; Cani, P.D. Targeting gut microbiota in obesity: Effects of prebiotics and probiotics. Nat. Rev. Endocrinol. 2011, 7, 639–646. [Google Scholar] [CrossRef]
- Stenman, L.K.; Waget, A.; Garret, C.; Klopp, P.; Burcelin, R.; Lahtinen, S. Potential probiotic Bifidobacterium animalis ssp. lactis 420 prevents weight gain and glucose intolerance in diet-induced obese mice. Benef. Microbes 2014, 5, 437–445. [Google Scholar] [CrossRef] [Green Version]
- van Baarlen, P.; Wells, J.M.; Kleerebezem, M. Regulation of intestinal homeostasis and immunity with probiotic lactobacilli. Trends Immunol. 2013, 34, 208–215. [Google Scholar] [CrossRef]
- Fang, Z.; Lu, W.; Zhao, J.; Zhang, H.; Qian, L.; Wang, Q.; Chen, W. Probiotics modulate the gut microbiota composition and immune responses in patients with atopic dermatitis: A pilot study. Eur. J. Nutr. 2020, 59, 2119–2130. [Google Scholar] [CrossRef] [PubMed]
- Santarmaki, V.; Kourkoutas, Y.; Zoumpopoulou, G.; Mavrogonatou, E.; Kiourtzidis, M.; Chorianopoulos, N.; Tassou, C.; Tsakalidou, E.; Simopoulos, C.; Ypsilantis, P. Survival, intestinal mucosa adhesion, and immunomodulatory potential of Lactobacillus plantarum strains. Curr. Microbiol. 2017, 74, 1061–1067. [Google Scholar] [CrossRef] [PubMed]
- Argyri, A.A.; Zoumpopoulou, G.; Karatzas, K.A.G.; Tsakalidou, E.; Nychas, G.J.E.; Panagou, E.Z.; Tassou, C.C. Selection of potential probiotic lactic acid bacteria from fermented olives by in vitro tests. Food Microbiol. 2013, 33, 282–291. [Google Scholar] [CrossRef] [PubMed]
- Duysburgh, C.; Van den Abbeele, P.; Morera, M.; Marzorati, M. Lacticaseibacillus rhamnosus GG and Saccharomyces cerevisiae boulardii supplementation exert protective effects on human gut microbiome following antibiotic administration in vitro. Benef. Microbes 2021, 12, 365–379. [Google Scholar] [CrossRef]
- Abdelazez, A.; Abdelmotaal, H.; Evivie, S.E.; Melak, S.; Jia, F.F.; Khoso, M.H.; Zhu, Z.T.; Zhang, L.J.; Sami, R.; Meng, X.C. Screening potential probiotic characteristics of Lactobacillus brevis strains in vitro and intervention effect on type I diabetes in vivo. Biomed Res. Int. 2018, 2018, 7356173. [Google Scholar] [CrossRef] [Green Version]
- Needell, J.C.; Zipris, D. The role of the intestinal microbiome in type 1 diabetes pathogenesis. Curr. Diab. Rep. 2016, 16, 89. [Google Scholar] [CrossRef]
- Shokryazdan, P.; Sieo, C.C.; Kalavathy, R.; Liang, J.B.; Alitheen, N.B.; Jahromi, M.F.; Ho, Y.W. Probiotic Potential of Lactobacillus Strains with antimicrobial activity against some human pathogenic strains. Biomed. Res. Int. 2014, 2014, 927268. [Google Scholar] [CrossRef] [Green Version]
- Hor, Y.Y.; Liong, M.T. Use of extracellular extracts of lactic acid bacteria and bifidobacteria for the inhibition of dermatological pathogen Staphylococcus aureus. Dermatol. Sin. 2014, 32, 141–147. [Google Scholar] [CrossRef] [Green Version]
- Ratsep, M.; Naaber, P.; Kõljalg, S.; Smidt, I.; Shkut, E.; Sepp, E. Effect of Lactobacillus plantarum strains on clinical isolates of Clostridium difficile in vitro. J. Probiotics Health 2014, 2, 1000119. [Google Scholar] [CrossRef] [Green Version]
- Şanlıbaba, P.; Güçer, Y. Antimicrobial activity of lactic acid bacteria. J. Int. Sci. Publ. 2015, 3, 451–457. [Google Scholar]
- Calvet, H.M.; Yoshikawa, T.T. Infections in diabetes. Infect. Dis. Clin. N. Am. 2001, 15, 407–421. [Google Scholar] [CrossRef]
- Casqueiro, J.; Casqueiro, J.; Alves, C. Infections in patients with diabetes mellitus: A review of pathogenesis. Indian J. Endocrinol. Metab. 2012, 16, 27–36. [Google Scholar] [CrossRef]
- Udeani, T.K.; Asogwa, V.N.; Ezenwaka, U. Assessment of systemic fungal infections among diabetic patients in Enugu, Nigeria. J. Infect Dis. Epidemiol. 2018, 4, 051. [Google Scholar] [CrossRef]
- Bottari, B.; Felis, G.E.; Salvetti, E.; Castioni, A.; Campedelli, I.; Torriani, S.; Bernini, V.; Gatti, M. Effective identification of Lactobacillus casei group species: Genome-based selection of the gene mutL as the target of a novel multiplex PCR assay. Microbiology 2017, 163, 950–960. [Google Scholar] [CrossRef]
- Mayrhofer, S.; Domig, K.J.; Mair, C.; Zitz, U.; Huys, G.; Kneifel, W. Comparison of broth microdilution, Etest, and agar disk diffusion methods for antimicrobial susceptibility testing of Lactobacillus acidophilus group members. Appl. Environ. Microbiol. 2008, 74, 3745–3748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Lai, C.; Huang, H.; Su, Y.; Chiu, Y.; Toh, H.; Chiang, S.; Chuang, Y.; Lu, Y.; Tang, H. Antimicrobial ability and mechanism analysis of Lactobacillus species against carbapenemase-producing Enterobacteriaceae. J. Microbiol. Immunol. Infect. 2021, 54, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Luo, J.; Zuo, F.; Zhang, Y.; Ma, H.; Chen, S. Screening for potential novel probiotic Lactobacillus strains based on high dipeptidyl peptidase IV and α-glucosidase inhibitory activity. J. Funct. Foods 2016, 20, 486–495. [Google Scholar] [CrossRef]
- Minekus, M.; Alminger, M.; Alvito, P.; Ballance, S.; Bohn, T.; Bourlieu, C.; Carrière, F.; Boutrou, R.; Corredig, M.; Dupont, D.; et al. A standardised static in vitro digestion method suitable for food-an international consensus. Food Funct. 2014, 5, 1113–1124. [Google Scholar] [CrossRef] [Green Version]
- Madureira, A.R.; Amorim, M.; Gomes, A.M.; Pintado, M.E.; Malcata, F.X. Protective effect of whey cheese matrix on probiotic strains exposed to simulated gastrointestinal conditions. Food Res. Int. 2011, 44, 465–470. [Google Scholar] [CrossRef]
- EFSA Panel on Additives and Products or Substances Used in Animal Feed (FEEDAP). Guidance on the Assessment of Bacterial Susceptibility to Antimicrobials of Human and Veterinary Importance. EFSA J. 2012, 10, 2740. [Google Scholar] [CrossRef]
- Koutsoumanis, K.; Allende, A.; Alvarez-Ordóñez, A.; Bolton, D.; Bover-Cid, S.; Chemaly, M.; Davies, R.; De Cesare, A.; Hilbert, F.; Lindqvist, R.; et al. Scientific Opinion on the update of the list of QPS-recommended biological agents intentionally added to food or feed as notified to EFSA (2017–2019). EFSA J. 2020, 18, 5966. [Google Scholar] [CrossRef] [Green Version]
- Mitropoulou, G.; Oreopoulou, A.; Papavassilopoulou, E.; Vamvakias, M.; Panas, P.; Fragias, S.; Kourkoutas, Y. Origanum vulgare ssp. hirtum essential oil as a natural intrinsic hurdle against common spoilage and pathogenic microbes of concern in tomato juice. Appl. Microbiol. 2021, 1, 1–10. [Google Scholar] [CrossRef]
- Balali, G.I.; Yar, D.D.; Afua Dela, V.G.; Adjei-Kusi, P. Microbial contamination, an increasing threat to the consumption of fresh fruits and vegetables in today’s world. Int. J. Microbiol. 2020, 2020, 3029295. [Google Scholar] [CrossRef] [PubMed]
- Dinev, T.; Beev, G.; Tzanova, M.; Denev, S.; Dermendzhieva, D. Antimicrobial activity of Lactobacillus plantarum against pathogenic and food spoilage microorganisms: A review. Bulg. J. Vet. Med. 2018, 21, 253–268. [Google Scholar] [CrossRef]
- Madacki-Todorović, K.; Eminović, I.; Mehmedinović, N.I.; Ibrišimović, M. Insulin acts as stim-ulatory agent in diabetes-related Escherichia coli pathogenesis. Int. J. Diabetes Clin. Res. 2018, 5, 098. [Google Scholar] [CrossRef]
- Ambalam, P.; Kondepudi, K.K.; Balusupati, P.; Nilsson, I.; Wadström, T.; Ljungh, A. Prebiotic preferences of human lactobacilli strains in co-culture with bifidobacteria and antimicrobial activity against Clostridium difficile. J. Appl. Microbiol. 2015, 119, 1672–1682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, H.Q.; Jiang, Z.D. Clostridium difficile infection in diabetes. Diabetes Res. Clin. Pract. 2014, 105, 285–294. [Google Scholar] [CrossRef] [PubMed]
- Arena, M.P.; Silvain, A.; Normanno, G.; Grieco, F.; Drider, D.; Spano, G.; Fiocco, D. Use of Lactobacillus plantarum strains as a bio-control strategy against food-borne pathogenic microorganisms. Front. Microbiol. 2016, 7, 464. [Google Scholar] [CrossRef] [Green Version]
- Inturri, R.; Trovato, L.; Li, G.; Oliveri, S.; Blandino, G. In vitro inhibitory activity of Bifidobacterium longum BB536 and Lactobacillus rhamnosus HN001 alone or in combination against bacterial and Candida reference strains and clinical isolates. Heliyon 2019, 5, e02891. [Google Scholar] [CrossRef] [Green Version]
- Muñoz-Quezada, S.; Bermudez-Brito, M.; Chenoll, E.; Genovćs, S.; Gomez-Llorente, C.; Plaza-Diaz, J.; Matencio, E.; Gil, A.; Romero, F.; Ramon, D.; et al. Competitive inhibition of three novel bacteria isolated from faeces of breast milk-fed infants against selected enteropathogens. Br. J. Nutr. 2013, 109, S63–S69. [Google Scholar] [CrossRef] [Green Version]
- Taheur, F.B.; Mansour, C.; Chaieb, K. Inhibitory effect of kefir on Aspergillus growth and mycotoxin production. Euro-Mediterr. J. Environ. Integr. 2020, 5, 5. [Google Scholar] [CrossRef]
- Poornachandra Rao, K.; Deepthi, B.V.; Rakesh, S.; Ganesh, T.; Achar, P.; Sreenivasa, M.Y. Antiaflatoxigenic Potential of Cell-Free Supernatant from Lactobacillus plantarum MYS44 Against Aspergillus parasiticus. Probiotics Antimicrob. Proteins 2019, 11, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Salari, R.; Salari, R. Investigation of the best Saccharomyces cerevisiae growth condition. Electron. Physician 2017, 9, 3592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degeeter, M.; Williamson, B. Alternative agents in type 1 diabetes in addition to insulin therapy. J. Pharm. Pract. 2016, 29, 144–159. [Google Scholar] [CrossRef] [PubMed]
- Nagai, E.; Katsuno, T.; Miyagawa, J.I.; Konishi, K.; Miuchi, M.; Ochi, F.; Kusunoki, Y.; Tokuda, M.; Murai, K.; Hamaguchi, T.; et al. Effects of miglitol in combination with intensive insulin therapy on blood glucose control with special reference to incretin responses in type 1 diabetes mellitus. Endocr. J. 2011, 58, 869–877. [Google Scholar] [CrossRef] [Green Version]
- Hollander, P.; Pi-Sunyer, X.; Coniff, R.F. Acarbose in the treatment of type I. Diabetes Care 1997, 20, 248–253. [Google Scholar] [CrossRef]
- Sels, J.P.J.E.; Verdonk, H.E.R.; Wolffenbuttel, B.H.R. Effects of acarbose (Glucobay®) in persons with type 1 diabetes: A multicentre study. Diabetes Res. Clin. Pract. 1998, 41, 139–145. [Google Scholar] [CrossRef]
- Koh, W.Y.; Utra, U.; Ahmad, R.; Rather, I.A.; Park, Y.H. Evaluation of probiotic potential and anti-hyperglycemic properties of a novel Lactobacillus strain isolated from water kefir grains. Food Sci. Biotechnol. 2018, 27, 1369–1376. [Google Scholar] [CrossRef]
- Lee, S.H.; Park, M.H.; Han, J.S.; Jeong, Y.; Kim, M.; Jeon, Y.J. Bioactive compounds extracted from Gamtae (Ecklonia cava) by using enzymatic hydrolysis, a potent α-glucosidase and α-amylase inhibitor, alleviates postprandial hyperglycemia in diabetic mice. Food Sci. Biotechnol. 2012, 21, 1149–1155. [Google Scholar] [CrossRef]
- Serra-Barcellona, C.; Habib, N.C.; Honoré, S.M.; Sánchez, S.S.; Genta, S.B. Enhydrin regulates postprandial hyperglycemia in diabetic rats by inhibition of α-glucosidase activity. Plant Foods Hum. Nutr. 2017, 72, 156–160. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, Q.; Dang, H.; Liu, X.; Tian, F.; Zhao, J.; Chen, Y.; Zhang, H.; Chen, W. Screening for potential new probiotic based on probiotic properties and α-glucosidase inhibitory activity. Food Control 2014, 35, 65–72. [Google Scholar] [CrossRef]
- Muganga, L.; Liu, X.; Tian, F.; Zhao, J.; Zhang, H.; Chen, W. Screening for lactic acid bacteria based on antihyperglycaemic and probiotic potential and application in synbiotic set yoghurt. J. Funct. Foods 2015, 16, 125–136. [Google Scholar] [CrossRef]
- Promyos, N.; Temviriyanukul, P.; Suttisansanee, U. Evaluation of α-glucosidase inhibitory assay using different sub-classes of flavonoids. Curr. Appl. Sci. Technol. 2017, 17, 172–180. [Google Scholar]
- Ramchandran, L.; Shah, N.P. Effect of exopolysaccharides and inulin on the proteolytic, angiotensin-I-converting enzyme- and α-glucosidase-inhibitory activities as well as on textural and rheological properties of low-fat yogurt during refrigerated storage. Dairy Sci. Technol. 2009, 89, 583–600. [Google Scholar] [CrossRef]
- Yu, Z.; Yin, Y.; Zhao, W.; Yu, Y.; Liu, B.; Liu, J.; Chen, F. Novel peptides derived from egg white protein inhibiting alpha-glucosidase. Food Chem. 2011, 129, 1376–1382. [Google Scholar] [CrossRef]
- Byakika, S.; Mukisa, I.M.; Byaruhanga, Y.B.; Muyanja, C. A review of criteria and methods for evaluating the probiotic potential of microorganisms. Food Rev. Int. 2019, 35, 427–466. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Zoumpopoulou, G.; Foligné, B.; Alexandraki, V.; Kazou, M.; Pot, B.; Tsakalidou, E. Discovering probiotic microorganisms: In vitro, in vivo, genetic and omics approaches. Front. Microbiol. 2015, 6, 58. [Google Scholar] [CrossRef] [Green Version]
- Chao, S.H.; Tomii, Y.; Watanabe, K.; Tsai, Y.C. Diversity of lactic acid bacteria in fermented brines used to make stinky tofu. Int. J. Food Microbiol. 2008, 123, 134–141. [Google Scholar] [CrossRef]
- Campedelli, I.; Mathur, H.; Salvetti, E.; Rea, M.C.; Torriani, S.; Ross, R.P.; Hill, C.; O'Toole, P.W. Genus-Wide assessment of antibiotic resistance in Lactobacillus spp. Appl. Environ. Microbiol. 2019, 85, e01738-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rozman, V.; Lorbeg, P.M.; Accetto, T.; Matijašić, B.B. Characterization of antimicrobial resistance in lactobacilli and bifidobacteria used as probiotics or starter cultures based on integration of phenotypic and in silico data. Int. J. Food Microbiol. 2020, 314, 108388. [Google Scholar] [CrossRef]
- Lüdin, P.; Roetschi, A.; Wüthrich, D.; Bruggmann, R.; Berthoud, H.; Shani, N. Update on tetracycline susceptibility of Pediococcus acidilactici based on strains isolated from Swiss cheese and whey. J. Food Prot. 2018, 81, 1582–1589. [Google Scholar] [CrossRef] [PubMed]
- Klare, I.; Konstabel, C.; Werner, G.; Huys, G.; Vankerckhoven, V.; Kahlmeter, G.; Hildebrandt, B.; Müller-Bertling, S.; Witte, W.; Goossens, H. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J. Antimicrob. Chemother. 2007, 59, 900–912. [Google Scholar] [CrossRef] [PubMed]
- Korhonen, J.M.; Van Hoek, A.H.A.M.; Saarela, M.; Huys, G.; Tosi, L.; Mayrhofer, S.; Von Wright, A. Antimicrobial susceptibility of Lactobacillus rhamnosus. Benef. Microbes 2010, 1, 75–80. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).