Abstract
The discovery of heterotrophic nitrification-aerobic denitrification (HN-AD) microorganisms has opened a new window for wastewater treatment. The underlying mechanism of HN-AD, however, is not fully understood because of the phylogenetic diversity of HN-AD microbes. The isolation and characterization of new HN-AD microorganisms are encouraging for furthering the understanding of this process. In this study, we found an Alphaproteobacteria isolate W30 from a historically polluted river in China through an HN-AD microbes screening process, which we identified as Pannonibacter sp. A potential HN-AD pathway for W30 was proposed based on N conversion analyses and the successful amplification of the entire denitrification gene series. The isolate exhibited high efficiency of aerobic inorganic nitrogen transformation, which accounted for 97.11% of NH4+-N, 100% of NO3−-N, and 99.98% of NO2−-N removal with a maximum linear rate of 10.21 mg/L/h, 10.46 mg/L/h, and 10.77 mg/L/h, respectively. Assimilation rather than denitrification was the main mechanism for the environmental nitrogen depletion mediated by W30. The effect of environmental constraints on aerobic NO3−-N removal were characterized, following a membrane bioreactor effluent test under an oxic condition. Compared to known Alphaproteobacterial HN-AD microbes, we showed that Pannonibacter sp. W30 could deplete nitrogen with no NO2−-N or NO3−-N accumulation in the HN-AD process. Therefore, the application of Pannonibacter sp. W30 has the potential for developing a felicitous HN-AD technology to treat N-laden wastewater at the full-scale level.
1. Introduction
Traditional biological nitrogen removal technologies usually involve a two-step process with different types of microbes, autotrophic nitrifying bacteria, and denitrifiers (reducing NO3−-N or NO2−-N to gaseous nitrogen) [1]. Alternately, the heterotrophic nitrification-aerobic denitrification (HN-AD) process achieves simultaneous nitrification and denitrification in a system [2].
Heavy organic and inorganic nitrogen (N) pollution in water bodies is a burgeoning environmental issue worldwide [3,4,5,6]. Toggling between oxic and anoxic conditions, the traditional biological wastewater treatments that started from autotrophic nitrification (NH4+-N → NO2−-N → NO3−-N) compromise to low efficiency and incomplete N removal [1]. The development of heterotrophic nitrification-aerobic denitrification (HN-AD) processes, on the other hand, offers an alternate “one-pot” solution for wastewater treatment [2]. It has been well-recognized that HN-AD has several advantages over traditional methods in terms of carbon utilization, alkalinity demand, and space and energy requirements [7].
Fueled by organic carbons, microbes in HN-AD processes can simultaneously transform NH4+-N to NO3−-N and NO2−-N, and use O2, NO3−-N, and NO2−-N as the electron acceptors [8,9]. However, HN-AD encounters the same challenge found in traditional processes when tackling high N content wastewater. High N content in wastewater exerts at least two undesirable effects. On one hand, the removal of N from domestic wastewater with high N content can be limited by the fact that heterotrophic denitrifiers need sufficient carbon sources as electron donors. At the industry level, the removal of N from domestic wastewater with a low C/N ratio can also be limited by the fact that heterotrophic denitrifiers need sufficient carbon sources as electron donors [10]. Therefore, external carbon amendment is a common resort for treating this type of domestic wastewater in practice, leading to an extra carbon resource requirement. On the other hand, depending on functional genes and metabolism pathways, denitrification with high N content can sometimes accumulate nitrite, which inhibits the activity of denitrifying bacteria in wastewater treatments [11].
HN-AD microbes are ubiquitous and can prevail in activated sludge [12,13], aquaculture wastewater [14], marine sediments [15] and other different ecological environments. Microbes involved in HN-AD belong to various taxonomic groups, therefore their intra-group diversity is wide. Known HN-AD bacteria are included but not limited to Paracoccus denitrificans [16], Rhodococcus sp. [17], Pseudomonas sp. [18,19,20], Bacillus sp. [3,21], Alcaligenes sp. [22], and Halomonas sp. [23]. Among the known HN-AD microbes, only a few belong to the class of Alphaproteobacteria. Since most Alphaproteobacteria are oligotrophs that may adapt to nutrient variation in the environment, these microbes may apply novel mechanisms to tackle the low C/N ratio issue in HN-AD [24].
In this study, we screened a historically eutrophic domestic river in Beijing, China, for isolating aerobic nitrogen removal bacteria. Isolate W30 was identified as Pannonibacter sp., which belongs to Alphaproteobacteria. Its HN-AD capability was investigated using NH4+-N, NO3−-N, and NO2−-N as sole nitrogen sources, respectively. Based on N transformation assay and functional genes amplification, we suggest that isolate W30 is capable of carrying out nitrification and denitrification pathways. Since aerobic denitrification is the critical step for HN-AD, the species-specific N removal constraints were investigated. These factors were C/N ratio, carbon resource, dissolved oxygen, initial pH, temperature, and inoculation volume. Subsequently, the feasibility of isolate W30 in treating high N content wastewater at the full-scale level was tested using the effluent of a pilot membrane bioreactor (MBR). Our results suggest that (1) Pannonibacter sp. W30 is able to conduct HN-AD; (2) oxygen, however, is still a limiting factor affecting aerobic denitrification mediated by W30; (3) Pannonibacter sp. W30 adapts to a broad range of C/N ratios in wastewaters.
2. Materials and Methods
2.1. Media
Enrichment medium (EM): The EM was composed of 0.5 g/L (NH4)2SO4, 0.36 g/L KNO3, 4.0 g/L sodium citrate, and 0.05% (ratio of volume) of the trace element solution which was composed of 6.5 g/L K2HPO4·3H2O, 2.5 g/L MgSO4·7H2O, 2.5 g/L NaCl, 0.05 g/L FeSO4·7H2O, and 0.04 g/L MnSO4·H2O. The final pH of the EM was adjusted to 7.0.
Denitrifying medium (DM): The DM was composed of 0.36 g/L KNO3, 10.55 g/L Na2HPO4·12H2O, 1.5 g/L KH2PO4, 0.1 g/L MgSO4·7H2O, 4.0 g/L sodium citrate, and 0.2% (volume ratio) of trace element solution which included 50.0 g/L EDTA-Na2, 2.2 g/L ZnSO4, 5.5 g/L CaCl2, 5.06 g/L MnCl2·4H2O, 5.0 g/L FeSO4·7H2O, 1.57 g/L CuSO4·5H2O, and 1.61 g/L CoCl2·6H2O. The final pH of the DM was adjusted to 7.0.
Screen medium (GN): The GN for the denitrifying bacteria was composed of 1.0 g/L KNO3, 8.5 g/L sodium citrate, 1.0 g/L L-asparagine, 1.0 g/L KH2PO4, 1.0 g/L MgSO4·7H2O, 0.2 g/L CaCl2·6H2O, 0.05 g/L FeCl3·6H2O, and 0.1% (volume ratio) of the 1% (ratio of weight/volume) alcoholic dissolved bromothymol blue (BTB). The final pH of the DM was adjusted to 7.0. The solid GN was made by adding 2% of agar powder to the liquid GN.
Luria-Bertani broth medium (LB): The LB broth liquid medium consisted of 10 g/L tryptone, 10 g/L yeast extract and 5 g/L NaCl, and 2% (g/L) of agar powder was added to LB broth liquid medium when solid plate medium was needed.
HNM medium consisted of the following components (per liter): 2.87 g of C6H5Na3O7, 0.24 g of (NH4)2SO4, 50 mL of HNM trace elements solution. The HNM trace elements solution contained (per liter): 5 g of K2HPO4 or 6.5 g of K2HPO4·3H2O, 2.5 g of MgSO4·7H2O and NaCl, 0.05 g of FeSO4·7H2O and MnSO4·4H2O [25].
DM medium consisted of the following components (per liter): 2.87 g of C6H5Na3O7, 0.36 g of KNO3 (DM1) or 0.25 g of NaNO2 (DM2), 0.2 g of MgSO4·7H2O, 1.5 g of KH2PO4, 10.55 g of Na2HPO4·12H2O or 5.3 g of Na2HPO4·2H2O, 2 mL of DM trace elements solution. The DM trace elements solution contained (per liter): 50 g of EDTA-Na2, 2.2 g of ZnSO4·7H2O, 5.5 g of CaCl2, 5.06 g of MnCl2·4H2O, 5.0 g of FeSO4, 1.57 g of CuSO4·5H2O, and 1.6 g of CoCl2·6H2O.
All media were sterilized for 30 min at 0.11–0.15 MPa and 121 °C.
2.2. Isolation of Potential HN-AD Microorganisms
The method for isolating HN-AD microbes has been described previously [26]. Briefly, an aliquot of water sample from Liangshui River (39.90° N 116.22° E) in Beijing, China was inoculated into 250 mL enrichment medium and incubated at 30 °C with shaking (150 rpm). A total of 25 mL cell culture was replaced with an equal volume of fresh enrichment medium every 48 h for 30 d and a following replacement with 25 mL of fresh denitrifying medium every 24 h for 14 d. Then, 1 mL of the cultivated medium was diluted (10−1) with sterilized distilled H2O and spread on solid screen medium with the pH indicator bromothymol blue at 30 °C. After cell colonies formed, the blue ones primitively confirmed as aerobic denitrifiers were picked up and incubated individually in liquid screen medium at 30 °C with shaking (150 rpm). Luria-Bertani (LB) broth medium was used to preserve the isolates at 4 °C.
2.3. Identification of Isolate W30
Isolate W30 was purified through the plate streaking method on LB solid medium at 30 °C. Cells were fixed with 2.5% glutaraldehyde (pH 6.8) at 4 °C overnight and dehydrated for 20 min each in an ascending ethanol dehydration series (30, 50, 70, 85, 90, and 100%). Cells were dried through a CO2 critical point dryer (HCP-2, Hitachi, Tokyo, Japan) and coated with gold (E-1010, Hitachi, Tokyo, Japan). Cell morphology was observed using a scanning electron microscope (SEM, JSM-5800, Hitachi, Tokyo, Japan) at an accelerating voltage of 15 kV. The 16S rRNA gene of W30 was amplified using the primer pair 27F/1492R [27] and sequenced by RuiBiotech company (Beijing, China). The obtained sequence (1353 bp) was aligned using an in-house software CExpress by the company. The phylogenetic tree of W30 was constructed by MEGA 7.0 [28] based on the neighbor-joining method with the partial sequenced 16S rRNA gene of W30 and that of known HN-AD strains after the phylogeny of W30 was determined through the Basic Local Alignment Search Tool program (BLAST) at https://blast.ncbi.nlm.nih.gov/Blast.cgi/ (accessed on 1 November 2021). The information for denitrification genes was collected from Kyoto Encyclopedia of Genes and Genomes (KEGG) database at https://www.genome.jp/kegg/ (accessed on 1 November 2021).
2.4. Characterization of the Aerobic N Removal Capacity and the Constraints
2.4.1. Amplification and Identification of HN-AD Related Genes
HN-AD related genes including napA, narZ, narH, nirK, nirS, norB and nosZ which were supposed in the genome of W30 were amplified with corresponding primer pairs (Table S1). The PCR mixture (30 μL) was composed of 15 μL 2×EasyTaq PCR SuperMix (RuiBiotech, Beijing), 1 μL of each primer (10 μM), 2 μL DNA template, and 11 μL ddH2O. The PCR was carried out as follows: pre-denaturation at 94 °C for 5 min, 35 cycles of denaturation at 94 °C for 30 sec, annealing for 30 sec at 61 °C for narZ, at 59 °C for narH, at 60 °C for napA, at 55 °C for nirK, at 53 °C for nirS, at 57 °C for norB, at 56 °C for nosZ. The elongation was at 72 °C for 1 min with a final extension at 72 °C for 7 min. The PCR products were analyzed and purified by 1% agarose gel electrophoresis and sequenced by RuiBiotech company (Beijing, China).
2.4.2. Nitrogen Removal Capacity
A heterotrophic nitrification medium (HNM) and two aerobic denitrification media (DM1 and DM2) were prepared as basic media for characterizing W30′s N removal capacity under oxic conditions [8,29,30]. Ammonium sulfate was the sole N species in HNM, whereas potassium nitrate and sodium nitrite were the N sources in DM1 and DM2, respectively. The initial inorganic N concentration was set at 50 mg/L with a C/N ratio of 16:1 by using 800 mg/L carbon from sodium citrate. The detailed components of HNM, DM1, and DM2 are described in the supplementary information. The initial pH of all media was set at 7.0. An aliquot (5 mL) of W30 cell suspension at the exponential phase was inoculated into 500 mL HNM, 500 mL DM1, and 500 mL DM2 in a 1 L Erlenmeyer flask, respectively. The initial OD600 value of each mixture was determined by a spectrometer (VIS-7220N, Beijing Beifen-Ruili Analytical Instrument, Beijing, China). The initial OD600 values were 0.148 ± 0.005 for HNM, 0.151 ± 0.007 for DM1, and 0.153 ± 0.004 for DM2. The flasks were incubated at 30 °C for 24 h with shaking at 150 rpm to provide 6.1 mg/L dissolved oxygen in the system. OD600, NH4+-N, NO3−-N, NO2−-N, dissolved oxygen (DO), temperature, and Chemical Oxygen Demand (COD) were assayed every 3 h. Total nitrogen (TN) and dissolved total nitrogen (DTN) were tested at 0 h and 24 h, respectively. Except for OD600 and TN, the rest of the parameters were measured by filtering each mixture with a 0.45 μm filter (ANPEL, Shanghai, China). NH4+-N was measured by the colorimetric method with Nessler’s reagent at 420 nm. NO3−-N was determined by the phenoldisulfonic acid method. NO2−-N was analyzed by the colorimetric method with N-(1-naphthalene)-diaminoethane. TN and DTN were estimated using the alkaline potassium persulfate digestion-ultraviolet spectrophotometric method (DRB200, Hach, Loveland, CO, USA). COD was determined using the dichromate method. Methods for chemical quantification were referred to standard methods [31] if not otherwise mentioned.
2.4.3. C/N Ratio
A series of C/N ratios (1–20) based on DM1 were investigated to illustrate their effects on W30′s NO3−-N removal capacity under oxic condition. The required C/N ratios were achieved by changing the content of sodium citrate in the system. The initial NO3−-N concentration was set as 50 mg/L. The initial pH was 7.0. The initial inoculation volume was 1% (v/v). The cell culture was incubated at 30 °C for 24 h with shaking (150 rpm). Samples were taken and tested at 0 h and 24 h.
2.4.4. Carbon Sources
Five carbon sources including sucrose, sodium citrate, glucose, sodium acetate, and sodium bicarbonate based on DM1 were investigated to illustrate the effects of different carbon sources on W30′s NO3−-N removal capacity under oxic condition. The initial NO3−-N concentration was set as 50 mg/L. The C/N ratio was adjusted to 8. The initial pH was 7.0. The initial inoculation volume was 1% (v/v). The cell culture was incubated at 30 °C for 24 h with shaking (150 rpm). Samples were taken and tested at 0 h and 24 h.
2.4.5. Dissolved Oxygen
Six levels of dissolved oxygen (1.8, 3.1, 4.2, 5.9, 6.1, and 6.2 mg/L) based on DM1 were investigated to illustrate their effects on W30′s NO3−-N removal capacity under different DO concentrations. The different levels of dissolved oxygen in the media were achieved using various rotation speeds (from 30 rpm to 180 rpm) of a thermostat vibrating incubator (MQL-61R, Shanghai Minquan Instrument Co., Ltd., Shanghai, China) [11,32]. The initial NO3−-N concentration was set as 50 mg/L. The C/N ratio was adjusted to 8. The initial pH was 7.0. The initial inoculation volume was 1% (v/v). The cell culture was incubated at 30 °C for 24 h. Samples were taken and tested at 0 h and 24 h.
2.4.6. Initial pH
A series of initial pH values (3–12) based on DM1 were investigated to illustrate their effects on W30′s NO3−-N removal capacity under oxic condition. The required pH values were achieved by adjusting the buffering ratio of 0.5 mol/L NaOH and 0.5 mol/L HCl in the system. The initial NO3−-N concentration was set as 50 mg/L. The C/N ratio was adjusted to 8. The initial inoculation volume was 1% (v/v). The cell culture was incubated at 30 °C for 24 h with shaking (150 rpm). Samples were taken and tested at 0 h and 24 h.
2.4.7. Temperature
A series of temperatures (15–45 °C) based on DM1 were investigated to illustrate their effects on W30′s NO3−-N removal capacity under oxic condition. The temperature range (15–45 °C) with a 5 °C ascending was achieved by setting the incubation temperature of the thermostat vibrating incubator (MQL-61R, Shanghai Minquan Instrument Co., Ltd., Shanghai, China). The initial NO3−-N concentration was set as 50 mg/L. The C/N ratio was adjusted to 8. The initial pH was 7.0. The initial inoculation volume was 1% (v/v). The cell culture was incubated for 24 h with shaking (150 rpm). Samples were taken and tested at 0 h and 24 h.
2.4.8. Inoculation Volume
A series of inoculation volumes based on DM1 were investigated to illustrate their effects on W30′s NO3−-N removal capacity under oxic conditions. The different inoculation volumes were achieved by adjusting the amount of the cell culture amendment. The series of inoculation volume was set as 0.1%, 0.2%, 0.5%, 1.0%, and 1.5% (v/v). The initial NO3−-N concentration was set as 50 mg/L. The C/N ratio was adjusted to 8. The initial pH was 7.0. The cell culture was incubated at 30 °C for 24 h with shaking (150 rpm). Samples were taken and tested at 0 h and 24 h.
2.5. Membrane Bioreactor (MBR) Effluent Assay
An assembled MBR with a biological treatment unit and a membrane unit for separation (pore size: 10 nm) was applied for the effluent assay. The aim of this study was to evaluate the feasibility of using isolate W30 for removing high N content wastewater after MBR treatment when the primary N removal efficiency was low. The influent was domestic wastewater. The daily wastewater treatment capacity of this MBR was about 1.62 m3. Continuous aeration (two hours) was followed by an hour of rest. The NO3−-N concentration in the effluent of the MBR was 58.65 ± 1.34 mg/L. Four treatments were conducted with the effluent to explore W30′s N removal capacity under oxic condition (6.1 mg/L DO) including (i) blank control (no W30 and carbon amendment); (ii) 1% (v/v) W30 suspension inoculation; (iii) 1% (v/v) W30 suspension inoculation and sodium citrate amendment, up to 400 mg/L carbon in the system; (iv) 1% (v/v) W30 suspension inoculation and sodium citrate amendment, up to 800 mg/L carbon in the system. Each treatment had three replicates. The incubations were carried out at 30 °C for 24 h with shaking (150 rpm). Samples were taken and tested at 0 h and 24 h.
2.6. Data Analysis and Statistic
The removal efficiency (RE) and the removal rate (RR) of N compounds under different conditions were calculated according to the following formula [11]:
In which, C0 h and Ct h were the initial and final concentrations of each N compound such as NH4+-N, NO3−-N, and NO2−-N at 0 h and t h, respectively.
The cellular N content (biomass produced) was calculated by subtracting cellular-N at 0 h from cellular-N at 24 h [11,33]:
Gaseous N content (N loss) was calculated by subtracting TN at 24 h from TN at 0 h as the following formula, or, the produced Gas-N can be also calculated from the parameter DTN:
The denitrification and assimilation efficiency of W30 on N transformation was calculated by the following formula:
TN removal efficiency was estimated by summing denitrification and assimilation efficiency as the following formula:
One-way analysis of variance (ANOVA) with Tukey method was used to evaluate the statistical significance of environmental constraints on W30′s N removal capacity under oxic condition. Data are shown as mean ± standard deviation.
2.7. Data Availability
The sequenced partial 16S rRNA gene of Pannonibacter sp. isolate W30 was deposited in GenBank under accession number KT380575.1. The sequenced functional gene series nosZ, nirK, norB, narZ, and narH were deposited in GenBank under accession numbers from OK431598 to OK431602.
3. Results
3.1. Isolation and Identification of Aerobic Heterotrophic Nitrogen Removal Bacterial Isolate W30
A total of 13 potential HN-AD bacteria were isolated from the 10−1 dilution of the cultured Liangshui River sample. One of the aerobic denitrifiers, isolate W30, could form a yellowish, circular, and convex colony with a smooth surface and entire edge on a LB plate. The colony was opaque and sticky (Figure 1A). The cell of W30 showed an average length of 1.24 ± 0.37 μm. Cells were non-spore-forming rods (Figure 1B,C).
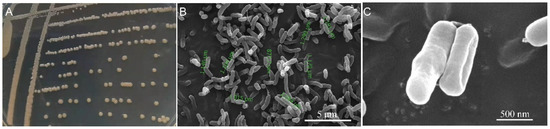
Figure 1.
Morphology of isolate W30: (A) colonies on LB solid medium; (B) scanning electron micrograph of W30 cells. Green marks showed the measured cell lengths; (C) detailed morphology of individual W30 cells.
The sequenced partial 16S rRNA gene of W30 (1353 bp) was nearly identical (99.78%) to that of the type strain Pannonibacter phragmitetus C6-19 (NR_028009.1) (Figure 2), which belongs to the class Alphaproteobacteria. The phylogenetic tree based on evolutionary distance (neighbor-joining) showed that the HN-AD microorganisms were quite diverse, distributed among at least 13 different families. The genus Pannonibacter comprised several species such as P. phragmitetus, P. indicus, and P. carbonis. Although isolate W30 was grouped within Pannonibacter and close to Pannonibacter phragmitetus, it detached slightly from the internal clusters of the genus. As observed in the phylogenetic tree (Figure 2), outside the genus Pannonibacter, the closest species to W30 was Polymorphum gilvum, which is a potential HN-AD bacterium belonging to the Alphaproteobacteria class that is able to reduce nitrite to N2 based on the related functional genes in the KEGG database. Therefore, the isolate W30 was primitively designated as Pannonibacter sp. W30. Furthermore, the genetic relationship between W30 and other reported typical HN-AD bacteria were also exhibited, such as Agrobacterium tumefaciens LAD9 [34], Alcaligenes faecalis NR [35], Klebsiella pneumoniae CF-S9 [36], Acinetobacter junii strain YB [25,37], Pseudomonas stutzeri YZN-001 [20], and Bacillus methylotrophicus strain L7 [38]. Various denitrification-related genes were detected in the genomes of known HN-AD microbes in the phylogenetic tree including narG(Z)HI, napAB, nirK, nirS, norBC, and nosZ. An entire series of reduction genes targeting substances from nitrate to N2O were detected in Stappiaceae (https://www.genome.jp/pathway/pphr00910/, accessed on 1 November 2021), Burkholderiaceae (https://www.genome.jp/pathway/reh00910/, accessed on 1 November 2021), and Pseudomonadaceae (https://www.genome.jp/pathway/psz00910/, accessed on 1 November 2021), which belong to Proteobacteria only. Other HN-AD microbes showed no full capacity to reduce all the intermediates in denitrification.
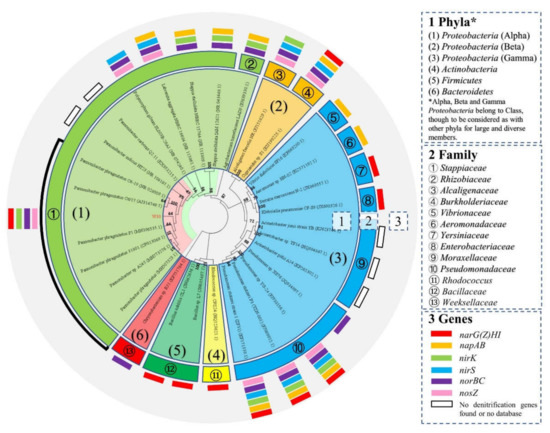
Figure 2.
Phylogenetic relationship of isolate W30 within the Bacterial domain based on comparison of partial 16S ribosomal RNA gene sequences. W30′s sequence was aligned to representative sequences from the GenBank databases. Phylogenetic analysis was performed with the MEGA software. The phylogenetic tree was constructed based on 16S ribosomal RNA genes in MEGA by evolutionary distance (neighbor-joining). The information on known functional genes associated with the representative sequences were added in the tree. Accession numbers shown for the comparison sequences were obtained from GenBank. Bootstrap value was 1000 replications. The red rectangle denotes the alpha subunit for nitrate reductase I (narG) and nitrate reductase II (narZ) for Gram-positive and -negative bacteria with other subunits.
3.2. Isolate W30′s Capacity on N Removal and Constraints under Oxic Condition
3.2.1. The Aerobic Denitrification Potential of Isolate W30
The entire series of denitrification genes were successfully amplified from W30′s genome (Figure 3A). However, the amplification results of the periplasmic nitrate reductases gene napA and nitrite reductase nirS were negatively supported by further sequencing. The results were in line with the N metabolism pathway of Pannonibacter phragmitetus 31801 (GenBank: CP013068) shown in KEGG (Figure 3B). Therefore, a denitrification pathway (NO3−-N → NO2−-N → NO → N2O → N2) of Pannonibacter sp. W30 under oxic conditions was proposed (Figure 3C).
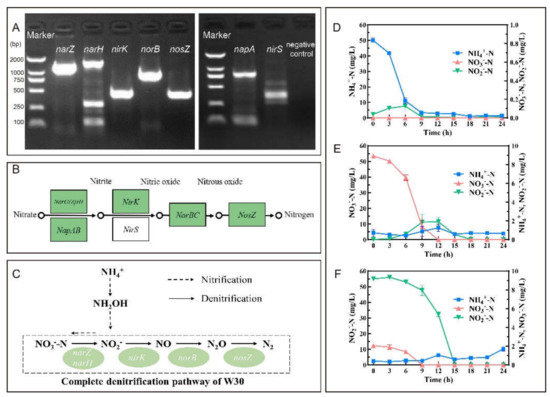
Figure 3.
Isolate W30′s potential and capacity on N removal. (A): The amplification of denitrification genes of isolate W30. (B): Reference denitrification pathway of Pannonibacter phragmitetus 31801 (GenBank: CP013068) from KEGG database (www.genome.jp/pathway/pphr00910/, accessed on 1 November 2021). (C): Proposed denitrification pathway of isolate W30. Nitrogen removal characteristic of isolate W30 in media of (D) HNM, (E) DM1, and (F) DM2. Symbols: ■, NH4+-N; ▲, NO3−-N; ▼, NO2−-N. Values represent the mean ± SD (standard deviation) of three replicates.
3.2.2. HN-AD Performance of Isolate W30
Isolate W30 could heterotrophically remove NH4+-N in HNM with an average rate of 5.18 mg/L/h in 9 h (Figure 3D). The removal rate was 1.34-fold higher than that of Ochrobactrum anthropic LJ81 (3.85 mg/L/h) [32] and 2.26-fold higher than that of Vibrio diabolicus SF16 (2.29 mg/L/h) [15]. The removal efficiency of NH4+-N was 97.11% with a linear average rate of 10.21 mg/L/h during 3-6 h. At the same time, a NO2−-N anomaly around 0.105 ± 0.005 mg/L (3 h) and 0.127 ± 0.058 mg/L (6 h) appeared but rapidly decreased therefrom. In addition, no obvious amount of NO3−-N appeared during isolate W30-initiated heterotrophic nitrification.
Isolate W30′s ability of aerobic denitrification was evaluated using DM1 and DM2 with KNO3 and NaNO2 as the sole N source, respectively. The concentration of NO3−-N was drastically depleted under the oxic condition from 53.43 ± 0.73 mg/L to a concentration below the detection limit in 12 h (Figure 3E). The linear average removal rate was 10.46 mg/L/h during 6–9 h which was much higher than that of Klebsiella pneumonia CF-S9 (8.64 mg/L/h) [30]. Moreover, the removal efficiency could reach 100%. NO2−-N appeared at 6 h and reached 1.913 ± 0.448 mg/L at 12 h, which was in parallel to the linear depletion of NO3−-N. Aerobic utilization of NO2−-N by isolate W30 was also observed (Figure 3F). The linear average depletion rate was 10.77 mg/L/h during 9-12 h and only a trace amount of NO2−-N remained in the system at 24 h (≤0.016 mg/L). The removal efficiency of NO2−-N by isolate W30 in 24 h was 99.98%.
Through HN-AD, isolate W30 achieved high TN removal efficiency with various N sources (Table 1). When (NH4)2SO4 was the sole N source in HNM, the highest TN removal efficiency could reach 91.34%. Relative to NO3−-N (8.74%) and NO2−-N (10.31%), more gaseous N was prone to occur when NH4+-N (26.64%) was the N source. Assimilation was the dominant driver for N depletion in all the systems. There is no marked difference in the denitrification of KNO3 and NaNO2 in DM1 and DM2.

Table 1.
W30′s N removal capacity under oxic condition.
3.2.3. NO3−-N Removal Constraints of Isolate W30
The investigation on the effect of C/N ratio showed that the higher the ratio, the greater the N removal efficiency, until the ratio reached 14 (Figure 4A). The isolate W30 was able to function normally with C/N ratios ranging from 3 to 20. With a C/N of 3, isolate W30 was able to remove 16.96 ± 2.52% NO3−-N. When the ratio was 1, little N (2.36 ± 0.77%) was removed. As the C/N ratio increased to 14, W30 achieved the greatest NO3−-N removal (98.62 ± 1.40%). Increasing the C/N ratio from 14 did not enhance NO3−-N removal efficiency while cell density continually increased.
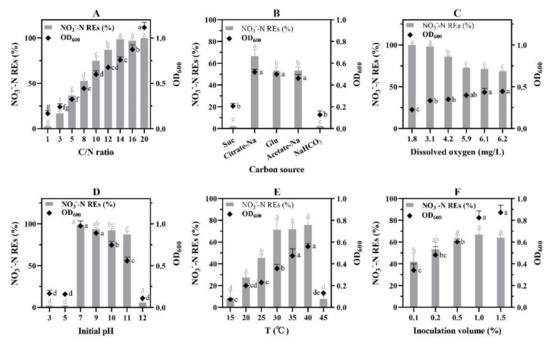
Figure 4.
Species-specific constraints on NO3−-N removal mediated by W30. (A) C/N ratio; (B) carbon sources, Suc and Glu denoted sucrose and glucose, respectively; (C) dissolved oxygen; (D) initial pH; (E) temperature; (F) inoculation volume. Values with different letters indicate being significantly different at p < 0.05.
Different carbon sources resulted in different NO3−-N removal efficiencies for isolate W30 (Figure 4B). The highest removal efficiency was achieved by using sodium citrate, which was 66.57 ± 6.53%. Glucose and sodium acetate showed no statistical significance on NO3−-N removal, which were 52.20 ± 4.0% and 53.27 ± 3.19%, respectively. Sucrose and sodium bicarbonate were not the preferential carbon sources for W30 to reduce NO3−-N in 24 h. The NO3−-N removal efficiencies were only 1.80 ± 0.46% and 2.10 ± 0.46%, respectively.
Six dissolved oxygen concentrations were achieved by adjusting the rotation speed from 30 to 180 rpm representing 1.8, 3.1, 4.2, 5.9, 6.1, and 6.2 mg/L DO (Figure 4C). The highest NO3−-N removal efficiency was achieved at anoxic/hypoxic conditions (1.8 and 3.1 mg/L DO) with no statistical significance. Following that, the NO3−-N removal efficiency decreased as dissolved oxygen concentrations increased from hypoxic condition to oxic condition (5.9 mg/L DO) significantly. Finally, a medium NO3−-N removal efficiency occurred at oxic conditions (DO ≥ 5.9 mg/L).
Isolate W30 could not reduce NO3−-N at low pH (Figure 4D). The NO3−-N removal efficiencies were only 2.21 ± 0.87% and 1.67 ± 0.46% at pH 3 and 5, respectively. The optimal pH range allowing isolate W30 to proliferate was from 7 to 11 when the maximum NO3−-N removal efficiency reached 98.35 ± 1.03% at neutral pH.
A linear relationship between temperatures and NO3−-N removal efficiencies was observed from 15 °C to 30 °C (Figure 4E). The highest NO3−-N removal efficiency was achieved at 40 °C. However, the nitrate removal ability decreased markedly when W30 was cultured at 45 °C (7.80 ± 2.90%) and 15 °C (8.45 ± 6.31%).
NO3−-N removal efficiencies were proportional to the initial inoculation volumes from 0.1% to 0.5%, whereas no statistical significance was observed from 0.5% to 1.5% (Figure 4F).
3.3. Pilot MBR Effluent Test
The major N species in the effluent of the pilot MBR was NO3−-N (58.65 ± 1.34 mg/L) with a trace amount of NH4+-N and NO2−-N. Isolate W30 showed a high capacity for NO3−-N removal in the effluent under oxic condition if a sufficient carbon source was supplied (Table 2). A total of 99.3% NO3−-N could be reduced in 24 h with 800 mg/L carbon from sodium citrate.

Table 2.
Batch treatment results of MBR effluent by isolate W30.
4. Discussion
The development of new wastewater treatment technologies has been influenced by the discovery that HN-AD microorganisms can simultaneously nitrify NH4+-N and denitrify NO3−-N. A number of HN-AD mechanisms and pathways have been identified, including denitrification mechanisms such as nitrate–nitrite reduction or hydroxylamine reduction, and others as incomplete N removal and organic N removal [7]. There is an increasing recognition that the HN-AD process is species-specific. The characteristics and constraints of diverse HN-AD microorganisms play a pivotal role in N removal efficiency. Among all the known HN-AD microbes, several potential Alphaproteobacteria HN-AD microbes have been identified, such as Polymorphum gilvum SL003B-26A1, Labrenzia aggregata NBRC 16684, Stappia stellulata NBRC15764, Stappia stellulata LAM 12621, and Agrobacterium tumefaciens LAD9. With the exception of norBC, which was found in most Alphaproteobacteria HN-AD strains, all other reductase genes involved in denitrification differed among Alphaproteobacteria species. In our study, the negative amplification of napA of the periplasmic nitrate reductases, a specific enzyme in aerobic denitrification, indirectly verified that membrane-bound nitrate reductases might prevail in Pannonibacter. The membrane-bound nitrate reductase gene narG was amplified from both W30 and Pannonibacter phragmitetus B1 [39]. Since O2 inhibits the activity of membrane-bound nitrate reductases [40], W30′s ability of NO3−-N removal under oxic conditions may involve additional mechanisms. The nitrite reductase gene nirK (467 bp) and the nitrous oxide reductase gene nosZ (447 bp) were successfully amplified, indicating that W30 could use nitrite as an intermediate electron acceptor for aerobic denitrification [39] and nitrous oxide could be reduced to N2 during the process. It is noteworthy that among a few known HN-AD microbes belonging to Alphaproteobacteria including Paracoccus, Pannonibacter, and Agrobacterium, Pannonibacter shows an advantage to develop a felicitous technology for the treatment of N-laden wastewater at the full-scale level. According to the N metabolism pathway in KEGG, the type strain Agrobacterium sp. RAC06 has no nosZ in its genome, which may lead to the accumulation of nitrous oxide, a major scavenger of stratospheric ozone. In addition, nirS rather than nirK is in the genome of Paracoccus denitrificans [8]. nirS encodes a homodimer cytochrome cd1-containing nitrite reductase while nirK encodes a copper-containing nitrite reductase [41,42]. The iron-based nitrite reductase nirS may be sensitive to environmental pH, which limits its application in wastewater treatments. Indeed, it was reported that a greater loss in nirS abundance occurred when the environmental pH was below 4.7 [43].
Isolate W30 could remove ammonia with a high rate and efficiency (97.11%) by two processes, assimilation into cellular nitrogen and aerobic denitrification (AD). Furthermore, nitrite was found to be a nitrification intermediate during this latter process as it was produced in trace amounts at the initial 6 h and then quickly consumed. This indicated that isolate W30 could transform ammonia to nitrite by heterotrophic nitrification (HN) and then use it for aerobic denitrification (AD). In parallel, no obvious amount of nitrate was detected during isolate W30-mediated HN. However, it was reported that a known HN-AD bacterium Acinetobacter sp. ND7 produced 4.7 mg/L of nitrate from ammonia at the initial 8 h of heterotrophic nitrification, but it delayed till 24 h to consume it completely [11]. Therefore, it seems that W30 can rapidly transform ammonia under oxic conditions by HN-AD with the advantage of non-accumulate nitrate in the environment. Isolate W30′s ability of aerobic denitrification (AD) was also evaluated and it was a half to that reported for other HNAD microorganisms when ammonia was the initial N source [11]. However, removal efficiency of nitrite and nitrate by isolate W30 in 24 h (99.98% and 100%, respectively) was mainly obtained through assimilation. Thus, we have found that when KNO3 and NaNO2 were the initial N sources, isolate W30 was apt to store them as cellular nitrogen.
Despite most Alphaproteobacteria being oligotrophs, isolate W30 seemed to benefit from high C/N ratios during aerobic NO3−-N removal. This result is in line with that of other aerobic denitrifiers such as Citrobacter diversus [44] and Cupriavidus sp. S1 [30]. An earlier study found that oligotrophic and eutrophic bacteria were interchangeable, depending on both the specific nutrients available and their concentrations [45]. In this context, isolate W30, an Alphaproteobacteria, could adapt to a broad and flexible range of C/N ratios. Moreover, W30 was verified a strict heterotroph through the NaHCO3 test. Meanwhile, sucrose was verified not a favorite carbon source for W30. W30′s aerobic N removal efficiency decreased as dissolved oxygen concentration increased. Similarly, an aerobic denitrification bacterium Citrobacter diversus showed the highest denitrification rate when dissolved oxygen was low [44]. For isolate W30, however, a significant NO3−-N removal efficiency could still be achieved even at oxic conditions, indicating W30 is an aerobic bacterium that could tolerate high O2 concentrations. In addition, W30 exhibited a series of high NO3−-N removal efficiencies from neutral to alkaline pHs. The result is consistent with that of known HN-AD microbes such as Pannonibacter phragmitetus F1 [46] and a strain of Pannonibacter isolated from a Hungarian soda lake [47]. It is believed that acidic pH may affect free ammonia and ammonia monooxygenase in the environment, although explicit mechanisms need to be further explored [38]. Moreover, high NO3−-N removal efficiencies up to 75.73 ± 6.23% were achieved by W30 at temperatures from 30 °C to 40 °C. As a comparison, the NO3−-N removal efficiency of Acinetobacter sp. ND7 reached only 6.69 ± 0.66% at 40 °C [11]. Our result also verified that 0.5% inoculation volume (initial OD600 = 0.305) was sufficient for initiating the NO3−-N removal process. Furthermore, Pannonibacter sp. W30 achieved nearly full NO3−-N removal in effluent from the pilot MBR. The C/N ratio, however, seemed critical for isolate W30 to tackle high N content wastewater at the full-scale level.
5. Conclusions
An aerobic heterotrophic nitrogen removal Alphaproteobacteria Pannonibacter sp. W30 was isolated, identified, and characterized in this study. A potential HN-AD pathway of W30 was proposed. The effect of environmental constraints on aerobic NO3−-N removal was investigated. Based on the results, Pannonibacter W30 was proved to be able to perform N removal from water environments with high inorganic N removal efficiency. The study verified that Pannonibacter sp. W30 contains nirK, norB, and nosZ genes which encode a nitrite reductase, a nitric oxide reductase, and a nitrous oxide reductase. These enzymes can transform NO2−-N to NO, N2O, and N2, avoiding NO2−-N accumulation in the process. Pannonibacter sp. W30 also has a copper-containing nitrite reductase nirK, a relatively robust enzyme under low pH. Our results suggest that although Pannonibacter sp. W30 could be able to conduct aerobic denitrification, dissolved oxygen is still a limiting factor affecting this process. As an aerobic heterotrophic nitrogen removal bacterium, Pannonibacter sp. W30 adapts to a broad range of C/N ratios in wastewaters, which may be feasible for developing novel HN-AD technologies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/microorganisms10020235/s1, Table S1: Primer pairs related to aerobic denitrification gene amplification.
Author Contributions
Conceptualization, Z.B.; methodology, Z.B. and N.Z.; software, N.Z.; validation, N.Z. and Y.Z.; formal analysis, Y.Z.; investigation, N.Z.; resources, Z.B.; data curation, Y.Z.; writing—original draft preparation, N.Z.; writing—review and editing, X.Z., T.B. and S.W.; visualization, Y.Z.; supervision, X.Z., T.B. and S.W.; project administration, Z.B.; funding acquisition, Z.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Major Science and Technology Program for Water Pollution Control and Treatment of China (No. 2018ZX07110), the S&T Program of Hebei (No. 19227313D), the S&T Program of Huairou (No. CXF2K2020-4), and the International Partnership Program of Chinese Academy of Sciences (No. 121311KYSB20200017).
Data Availability Statement
The data presented in this study are openly available in the NCBI. The accession number has been listed in the article.
Acknowledgments
The authors would like to thank the Key Laboratory of Environmental Biotechnology of Research Center for Eco-Environmental Sciences, Chinese Academy of Sciences for their technical support.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Khardenavis, A.A.; Kapley, A.; Purohit, H.J. Simultaneous nitrification and denitrification by diverse Diaphorobacter sp. Appl. Microbiol. Biotechnol. 2007, 77, 403–409. [Google Scholar] [CrossRef] [PubMed]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.-F.; An, J.; Fu, G.-H.; Yang, X.-L. Isolation and characterization of an aerobic denitrifying Bacillus sp. YX-6 from shrimp culture ponds. Aquaculture 2011, 319, 188–193. [Google Scholar] [CrossRef]
- Alonso, Á.; Camargo, J.A. Effects of pulse duration and post-exposure period on the nitrite toxicity to a freshwater amphipod. Ecotoxicol. Environ. Saf. 2009, 72, 2005–2008. [Google Scholar] [CrossRef]
- Jin, P.; Chen, Y.; Xu, T.; Cui, Z.; Zheng, Z. Efficient nitrogen removal by simultaneous heterotrophic nitrifying-aerobic denitrifying bacterium in a purification tank bioreactor amended with two-stage dissolved oxygen control. Bioresour. Technol. 2019, 281, 392–400. [Google Scholar] [CrossRef]
- Tilak, K.S.; Lakshmi, S.J.; Susan, T.A. The toxicity of ammonia, nitrite and nitrate to the fish, Catla catla (Hamilton). J. Environ. Biol. 2002, 23, 147–149. [Google Scholar]
- Song, T.; Zhang, X.; Li, J.; Wu, X.; Feng, H.; Dong, W. A review of research progress of heterotrophic nitrification and aerobic denitrification microorganisms (HNADMs). Sci. Total Environ. 2021, 801, 149319. [Google Scholar] [CrossRef]
- Robertson, L.A.; Van Niel, E.W.J.; Torremans, R.A.M.; Kuenen, J.G. Simultaneous Nitrification and Denitrification in Aerobic Chemostat Cultures of Thiosphaera pantotropha. Appl. Environ. Microbiol. 1988, 54, 2812–2818. [Google Scholar] [CrossRef] [Green Version]
- Gao, H.; Schreiber, F.; Collins, G.; Jensen, M.M.; Kostka, J.E.; Lavik, G.; de Beer, D.; Zhou, H.-Y.; Kuypers, M.M.M. Aerobic denitrification in permeable Wadden Sea sediments. ISME J. 2009, 4, 417–426. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Li, W.; Zhang, D.; Qin, W. Ammonium removal by a novel oligotrophic Acinetobacter sp. Y16 capable of heterotrophic nitrification–aerobic denitrification at low temperature. Bioresour. Technol. 2013, 146, 44–50. [Google Scholar] [CrossRef]
- Xia, L.; Li, X.; Fan, W.; Wang, J. Heterotrophic nitrification and aerobic denitrification by a novel Acinetobacter sp. ND7 isolated from municipal activated sludge. Bioresour. Technol. 2020, 301, 122749. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; He, S.; Wu, C.; Du, D. Characteristics of heterotrophic nitrification and aerobic denitrification bacterium Acinetobacter sp. T1 and its application for pig farm wastewater treatment. J. Biosci. Bioeng. 2019, 127, 201–205. [Google Scholar] [CrossRef] [PubMed]
- HungSoo, J.; Mitsuyo, H.; Makoto, S. Piggery wastewater treatment using Alcaligenes faecalis strain No. 4 with heterotrophic nitrification and aerobic denitrification. Water Res. 2006, 40, 3029–3036. [Google Scholar]
- Chen, P.; Li, J.; Li, Q.X.; Wang, Y.; Li, S.; Ren, T.; Wang, L. Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp. CPZ24. Bioresour. Technol. 2012, 116, 266–270. [Google Scholar] [CrossRef]
- Duan, J.; Fang, H.; Su, B.; Chen, J.; Lin, J. Characterization of a halophilic heterotrophic nitrification–aerobic denitrification bacterium and its application on treatment of saline wastewater. Bioresour. Technol. 2015, 179, 421–428. [Google Scholar] [CrossRef]
- Kokufuta, E.; Shimohashi, M.; Nakamura, I. Simultaneously occurring nitrification and denitrification under oxygen gradient by polyelectrolyte complex-coimmobilizedNitrosomonas europaea andParacoccus denitrificans cells. Biotechnol. Bioeng. 1988, 31, 382–384. [Google Scholar] [CrossRef]
- Li, W. Study on Characteristics in the Removal Process of Ammonia Nitrogen and Nitrate Nitrogen by an Isolated Heterotrophic Nitrification-Aerobic Denitrification Strain Rhodococcus Sp. J. Environ. Prot. 2013, 4, 74–79. [Google Scholar] [CrossRef]
- Sun, Y.; Li, A.; Zhang, X.; Ma, F. Regulation of dissolved oxygen from accumulated nitrite during the heterotrophic nitrification and aerobic denitrification of Pseudomonas stutzeri T13. Appl. Microbiol. Biotechnol. 2014, 99, 3243–3248. [Google Scholar] [CrossRef]
- Zhou, M.; Ye, H.; Zhao, X. Ammonium removal by a novel heterotrophic nitrifying and aerobic denitrifying bacterium Pseudomonas stutzeri KTB from wastewater. Water Qual. Res. J. 2015, 50, 219–227. [Google Scholar] [CrossRef]
- Zhang, J.; Wu, P.; Hao, B.; Yu, Z. Heterotrophic nitrification and aerobic denitrification by the bacterium Pseudomonas stutzeri YZN-001. Bioresour. Technol. 2011, 102, 9866–9869. [Google Scholar] [CrossRef]
- Zhao, B.; He, Y.L.; Zhang, X.F. Nitrogen removal capability through simultaneous heterotrophic nitrification and aerobic denitrification by Bacillus sp. LY. Environ. Technol. 2010, 31, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Shoda, M.; Ishikawa, Y. Heterotrophic nitrification and aerobic denitrification of high-strength ammonium in anaerobically digested sludge by Alcaligenes faecalis strain No. 4. J. Biosci. Bioeng. 2014, 117, 737–741. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Li, J.; Zhang, L.H.; Yu, Y.; Zhu, Y.M. Simultaneous heterotrophic nitrification and aerobic denitrification at high concentrations of NaCl and ammonia nitrogen by Halomonas bacteria. Water Sci. Technol. 2017, 76, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, P.; Yarza, P.; Rapp, J.Z.; Glöckner, F.O. Expanding the World of Marine Bacterial and Archaeal Clades. Front. Microbiol. 2016, 6, 1524. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Ren, Y.-X.; Liang, X.; Zhao, S.-Q.; Wang, J.-P.; Xia, Z.-H. Nitrogen removal characteristics of a heterotrophic nitrifier Acinetobacter junii YB and its potential application for the treatment of high-strength nitrogenous wastewater. Bioresour. Technol. 2015, 193, 227–233. [Google Scholar] [CrossRef]
- Lv, P.; Luo, J.; Zhuang, X.; Zhang, D.; Bai, Z. Diversity of culturable aerobic denitrifying bacteria in the sediment, water and biofilms in Liangshui River of Beijing, China. Sci. Rep. 2017, 7, 10032. [Google Scholar] [CrossRef] [Green Version]
- Stanley, J.; Baquar, N.; Burnens, A. Molecular subtyping scheme for Salmonella panama. J. Clin. Microbiol. 1995, 33, 1206–1211. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Shwu-Ling, P.; Nyuk-Min, C.; Chei-Hsiang, C. Potential applications of aerobic denitrifying bacteria as bioagents in wastewater treatment. Bioresour. Technol. 1999, 68, 179–185. [Google Scholar]
- Sun, Z.; Lv, Y.; Liu, Y.; Ren, R. Removal of nitrogen by heterotrophic nitrification-aerobic denitrification of a novel metal resistant bacterium Cupriavidus sp. S1. Bioresour. Technol. 2016, 220, 142–150. [Google Scholar] [CrossRef]
- APHA; AWWA; WPCF. Standard Methods for the Examination of Water and Wastewater, 20th ed.; American Public Health Association: Washington, DC, USA; American Water Work Association: Denver, CO, USA; Water Environment Federation: Alexandria, VA, USA, 1998. [Google Scholar]
- Lei, X.; Jia, Y.; Chen, Y.; Hu, Y. Simultaneous nitrification and denitrification without nitrite accumulation by a novel isolated Ochrobactrum anthropic LJ81. Bioresour. Technol. 2019, 272, 442–450. [Google Scholar] [CrossRef] [PubMed]
- Rout, P.R.; Bhunia, P.; Dash, R.R. Simultaneous removal of nitrogen and phosphorous from domestic wastewater using Bacillus cereus GS-5 strain exhibiting heterotrophic nitrification, aerobic denitrification and denitrifying phosphorous removal. Bioresour. Technol. 2017, 244, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Ni, J. Ammonium removal by Agrobacterium sp. LAD9 capable of heterotrophic nitrification–aerobic denitrification. J. Biosci. Bioeng. 2012, 113, 619–623. [Google Scholar] [CrossRef]
- Zhao, B.; An, Q.; He, Y.L.; Guo, J.S. N2O and N2 production during heterotrophic nitrification by Alcaligenes faecalis strain NR. Bioresour. Technol. 2012, 116, 379–385. [Google Scholar] [CrossRef] [PubMed]
- Padhi, S.K.; Tripathy, S.; Sen, R.; Mahapatra, A.S.; Mohanty, S.; Maiti, N.K. Characterisation of heterotrophic nitrifying and aerobic denitrifying Klebsiella pneumoniae CF-S9 strain for bioremediation of wastewater. Int. Biodeterior. Biodegrad. 2013, 78, 67–73. [Google Scholar] [CrossRef]
- Ren, Y.-X.; Yang, L.; Liang, X. The characteristics of a novel heterotrophic nitrifying and aerobic denitrifying bacterium, Acinetobacter junii YB. Bioresour. Technol. 2014, 171, 1–9. [Google Scholar] [CrossRef]
- Zhang, Q.-L.; Liu, Y.; Ai, G.-M.; Miao, L.-L.; Zheng, H.-Y.; Liu, Z.-P. The characteristics of a novel heterotrophic nitrification–aerobic denitrification bacterium, Bacillus methylotrophicus strain L7. Bioresour. Technol. 2012, 108, 35–44. [Google Scholar] [CrossRef]
- Bai, H.; Liao, S.; Wang, A.; Huang, J.; Shu, W.; Ye, J. High-efficiency inorganic nitrogen removal by newly isolated Pannonibacter phragmitetus B1. Bioresour. Technol. 2019, 271, 91–99. [Google Scholar] [CrossRef]
- Wang, X.; Wang, W.; Zhang, Y.; Sun, Z.; Zhang, J.; Chen, G.; Li, J. Simultaneous nitrification and denitrification by a novel isolated Pseudomonas sp. JQ-H3 using polycaprolactone as carbon source. Bioresour. Technol. 2019, 288, 121506. [Google Scholar] [CrossRef]
- Braker, G.; Fesefeldt, A.; Witzel, K.P. Development of PCR primer systems for amplification of nitrite reductase genes (nirK and nirS) to detect denitrifying bacteria in environmental samples. Appl. Environ. Microbiol. 1998, 64, 3769–3775. [Google Scholar] [CrossRef] [Green Version]
- Heylen, K.; Gevers, D.; Vanparys, B.; Wittebolle, L.; Geets, J.; Boon, N.; De Vos, P. The incidence of nirS and nirK and their genetic heterogeneity in cultivated denitrifiers. Environ. Microbiol. 2006, 8, 2012–2021. [Google Scholar] [CrossRef] [PubMed]
- Herold, M.B.; Giles, M.E.; Alexander, C.; Baggs, L.; Daniell, T.J. Variable response of nirK and nirS containing denitrifier communities to long-term pH manipulation and cultivation. FEMS Microbiol. Lett. 2018, 365, fny035. [Google Scholar] [CrossRef]
- Huang, H.K.; Tseng, S.K. Nitrate reduction by Citrobacter diversus under aerobic environment. Appl. Microbiol. Biotechnol. 2001, 55, 90–94. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; MacLeod, R.A. Observations on the distinction between oligotrophic and eutrophic marine bacteria. Appl. Environ. Microbiol. 1984, 47, 1017–1022. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zhu, H.; Shutes, B.; Fu, B.; Yan, B.; Yu, X.; Wen, H.; Chen, X. Identification and denitrification characteristics of a salt-tolerant denitrifying bacterium Pannonibacter phragmitetus F1. AMB Express 2019, 9, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Borsodi, A.K.; Micsinai, A.; Kovacs, G.M.; Toth, E.; Schumann, P.; Kovács, A.L.; Böddi, B.; Márialigeti, K. Pannonibacter phragmitetus gen. nov., sp. nov., a novel alkalitolerant bacterium isolated from decomposing reed rhizomes in a Hungarian soda lake. Int. J. Syst. Evol. Microbiol. 2003, 53, 555–561. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).