Unravelling Formaldehyde Metabolism in Bacteria: Road towards Synthetic Methylotrophy
Abstract
1. Introduction
2. Understanding and Modification of Formaldehyde Dissimilation Pathways for the Optimization of Synthetic Methylotrophy
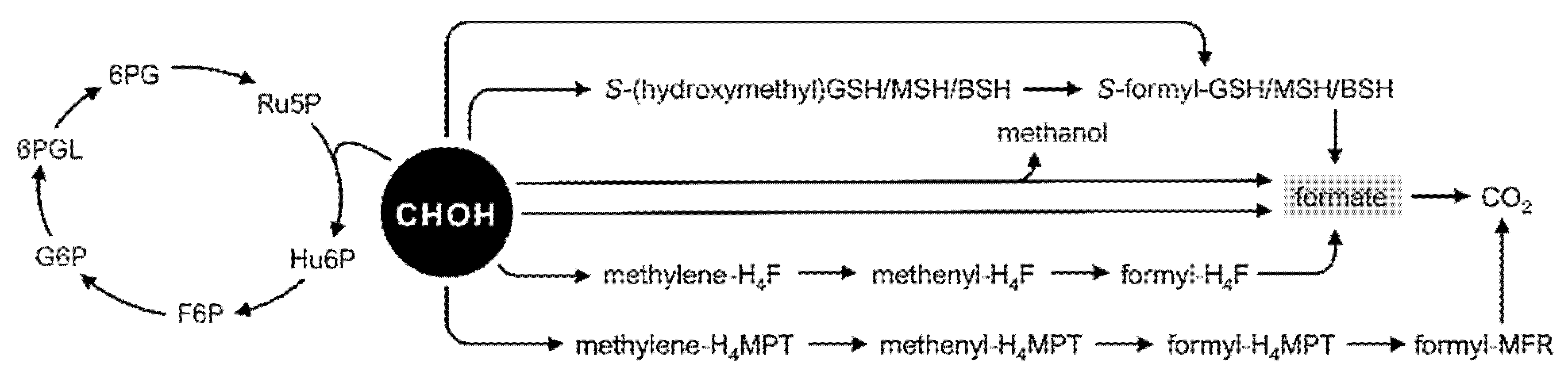
2.1. Formaldehyde Dissimilatory Pathways in Native Methylotrophs
2.2. Modification of Formaldehyde Dissimilation Pathways in Nonmethylotrophic Bacteria Is a Prerequisite for Synthetic Methylotrophy
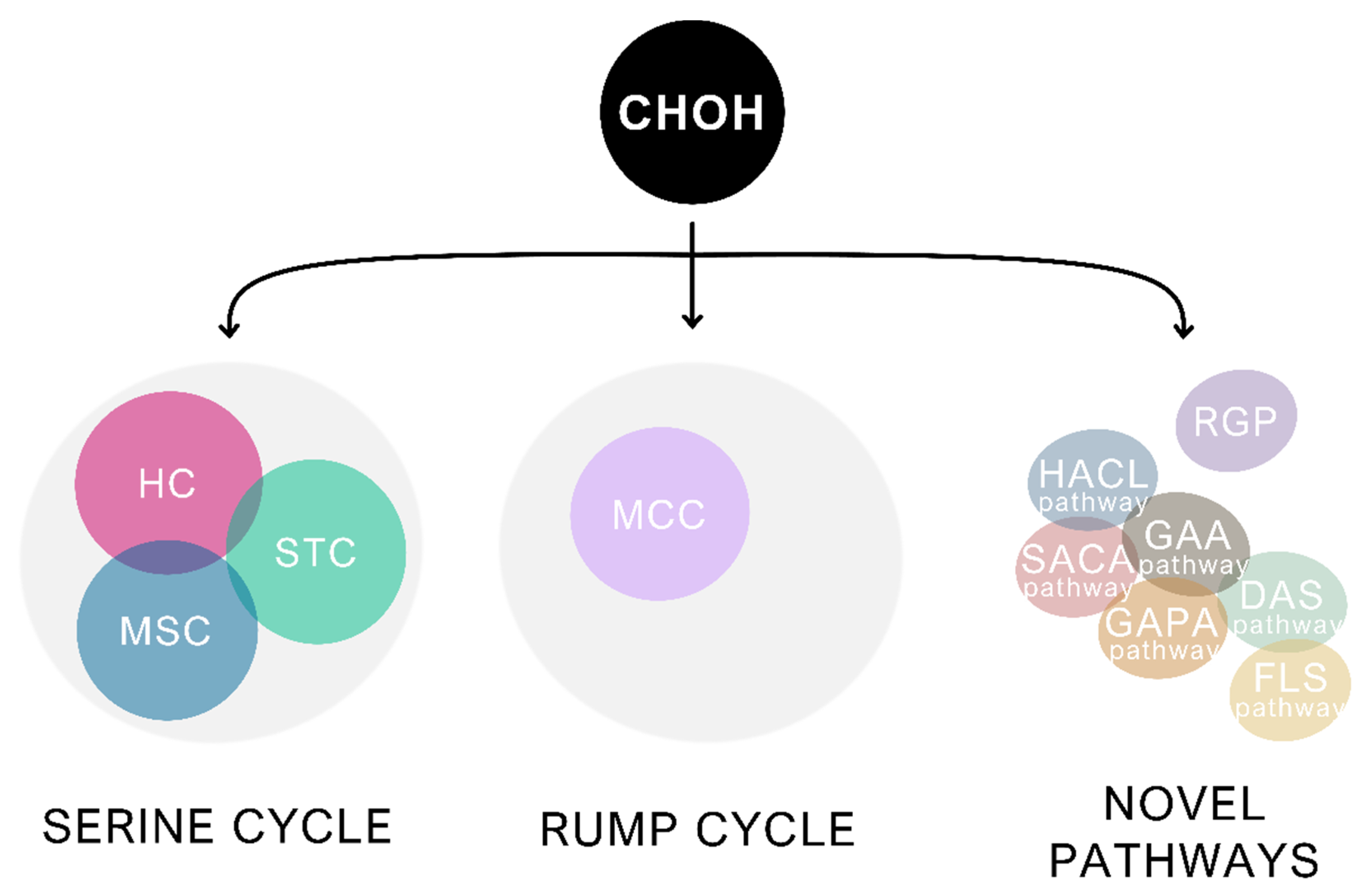
3. Formaldehyde Assimilation in Methylotrophic Bacteria Is an Inspiration for the Creation of Synthetic Methylotrophs
3.1. The RuMP Cycle and Its Adaptation to Synthetic Methylotrophy
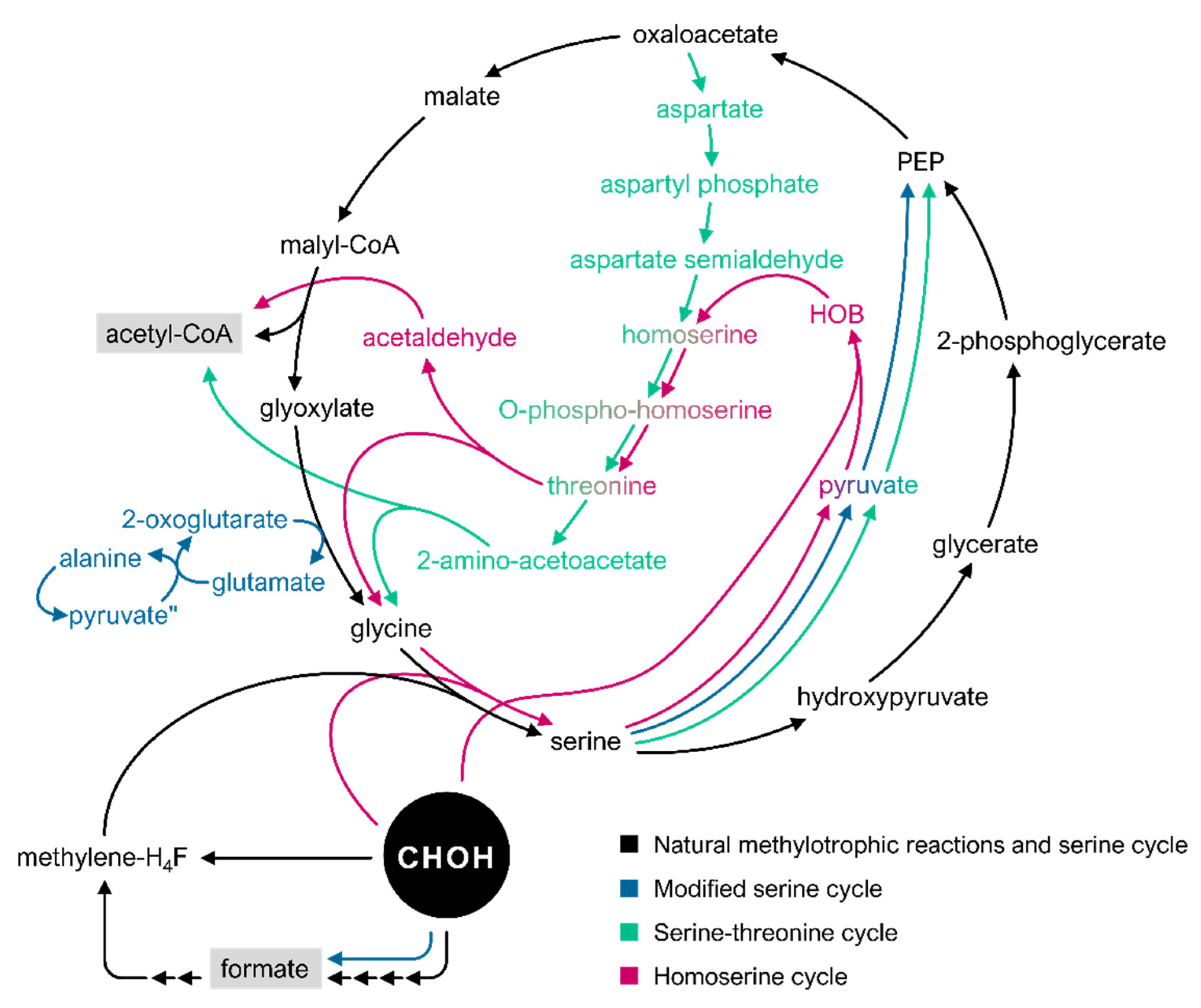
3.2. The Serine Cycle and Its Derivatives
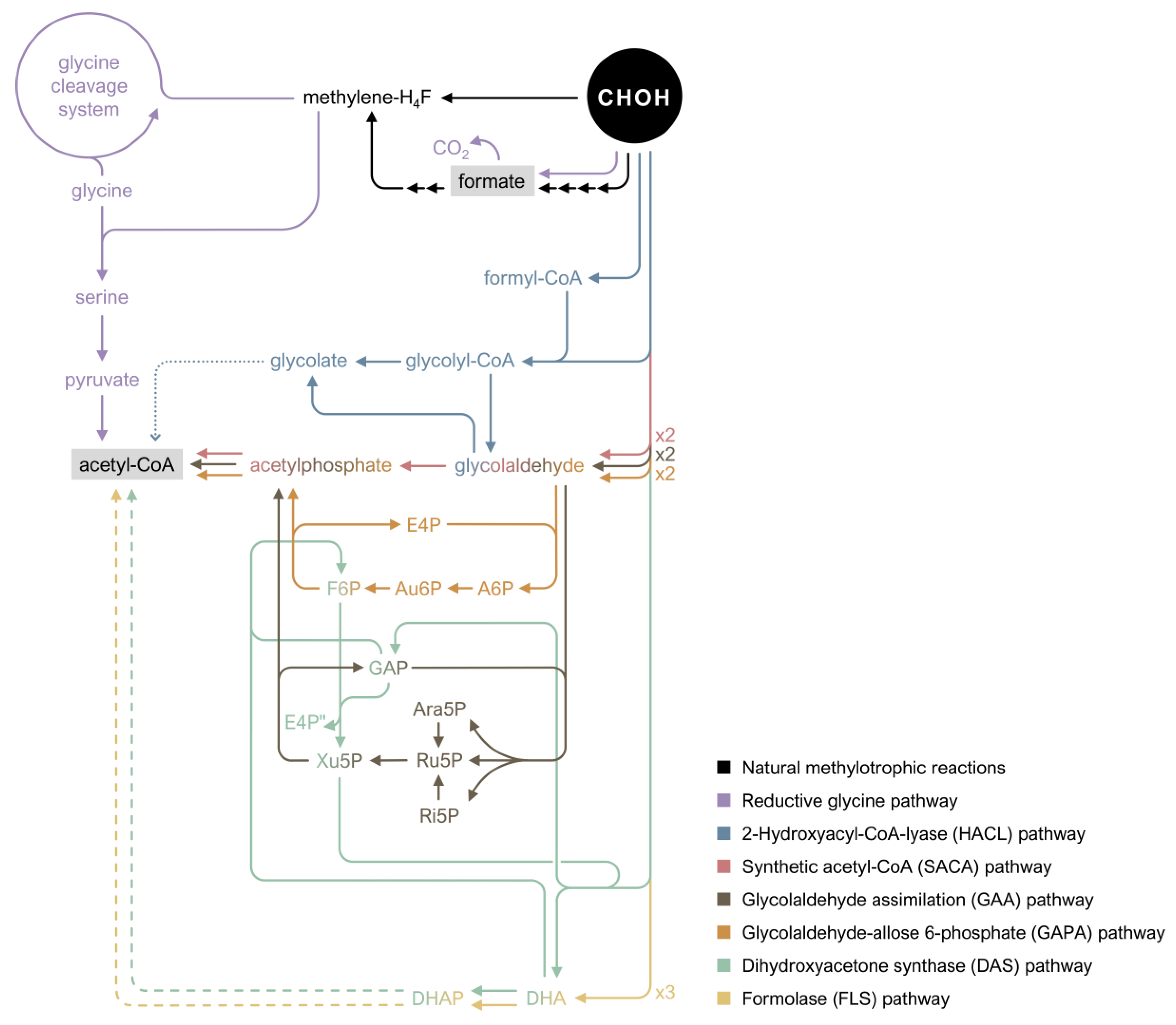
3.3. Novel Pathways for Assimilation of Formaldehyde
4. The Understanding of Formaldehyde Metabolism Regulation as a Support to Push Synthetic Methylotrophy
5. Concluding Remarks and Future Perspectives
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| Metabolites | |
| 6PG | 6-phosphogluconate |
| 6PGL | 6-phospho-glucono-1,5-lactone |
| A6P | 2R,3R-stereo allose 6-phosphate |
| Ara5P | arabinose 5-phosphate |
| Au6P | d-allulose 6-phosphate |
| BSH | bacillithiol |
| CHOH | formaldehyde |
| DHA | dihydroxyacetone |
| DHAP | dihydroxyacetone phosphate |
| E4P | erythrose 4-phosphate |
| F6P | fructose 6-phosphate |
| FBP | fructose 1,6-bisphosphate |
| G6P | glucose 6-phosphate |
| GAP | glyceraldehyde 3-phosphate |
| GAPA | glycolaldehyde-allose 6-phosphate |
| GSH | glutathione |
| H4F | tetrahydrofolate |
| H4MPT | tetrahydromethanopterin |
| HOB | 4-hydroxy-2-oxobutanoate |
| Hu6P | hexulose 6-phosphate |
| Ln | lanthanide |
| MSH | mycothiol |
| PEP | phosphoenolpyruvate |
| PQQ | pyrroloquinoline quinone |
| Ri5P | ribose 5-phosphate |
| RuBP | ribulose bisphosphate |
| Ru5P | ribulose 5-phosphate |
| S7P | sedoheptulose 7-phosphate |
| SBP | sedoheptulose 1,7-bisphosphate |
| TCA | tricarboxylic acid |
| Xu5P | xylulose 5-phosphate |
| XuMP | xylulose monophosphate |
| Proteins | |
| Agt | alanine-glyoxylate transaminase |
| Ald | acetaldehyde dehydrogenase |
| AlsE | d-allulose-6-phosphate 3-epimerase |
| AspC | aspartate aminotransferase |
| CyaA | adenylate cyclase |
| DAS | dihydroxyacetone synthase |
| DL-Faldh | dye-linked dehydrogenase |
| EfgA | enhanced formaldehyde growth protein A |
| Fae | formaldehyde-activating enzyme |
| Faldh | NAD(P)+-dependent formaldehyde dehydrogenase |
| FBPa | 1,6-bisphosphate aldolase |
| Fch | methenyl-H4F cyclohydrolase |
| Fdh | formate dehydrogenase |
| Fgh | S-formyl-GSH hydrolase |
| FlhR | response regulator FlhR |
| FlhS | signal regulator FlhS |
| FLS | formolase |
| Fsa | F6P aldolase |
| Ftl | formate-tetrahydrofolate ligase |
| Gals | glycolaldehyde synthase |
| GD-Faldh | NAD-GSH-dependent formaldehyde dehydrogenase |
| Gfa | glutathione-dependent formaldehyde-activating enzyme |
| HACL | 2-hydroxyacyl-CoA-lyase |
| Hal | HOB aldolase |
| Hat | HOB aminotransferase |
| HOB | 4-hydroxy-2-oxobutanoate |
| Hpr | hydroxypyruvate reductase |
| Hps | 3-hexulose-6-phosphate synthase |
| Hsk | homoserine kinase |
| Icd | isocitrate dehydrogenase |
| Kbl | 2-amino-3-ketobutyrate CoA ligase |
| KdsD | Ara5P isomerase |
| LtaE | threonine aldolase |
| Madh | methylamine dehydrogenase |
| Maldh | malate dehydrogenase |
| Mcl | malyl-CoA lyase |
| Mdh | methanol dehydrogenase |
| MdtA | methylene-H4F dehydrogenase |
| Mtk | malate thiokinase |
| Phi | 6-phospho-3-hexuloisomerase |
| Pkt | phosphoketolase |
| Pps | phosphoenolpyruvate synthetase |
| Pta | acetyltransferase |
| RhmA | 2-keto-3-deoxy-L-rhamnonate aldolase |
| Rpe | Ru5P epimerase |
| Rpi | Ri5P isomerase |
| RpiB | allose 6-phosphate isomerase/ribose 5-phosphate isomerase B |
| Sal | serine aldolase |
| SASP | acid-soluble spore proteins |
| SBPase | sedoheptulose-1,7-bisphosphatase |
| Sdh | serine dehydratase |
| Shmt | hydroxymethyltransferase |
| Ta | transaldolase |
| Tdh | threonine dehydrogenase |
| Tkt | transketolase |
| Ts | threonine synthase |
| Pathways | |
| GAA pathway | glycolaldehyde assimilation pathway |
| HC | homoserine cycle |
| MCC | methanol condensation cycle |
| MSC | modified serine cycle |
| NOG | nonoxidative glycolysis |
| PPP | pentose phosphate pathway |
| RGP | reductive glycine pathway |
| RuMP cycle | ribulose monophosphate cycle |
| SACA pathway | synthetic acetyl-CoA pathway |
| STC | Serine–threonine cycle |
| TCA cycle | tricarboxylic acid cycle |
| Other | |
| ALE | adaptive laboratory evolution |
| EMRA | ensemble modelling for robustness analysis |
| FBA | flux balance analysis |
References
- Yurimoto, H.; Kato, N.; Sakai, Y. Assimilation, Dissimilation, and Detoxification of Formaldehyde, a Central Metabolic Intermediate of Methylotrophic Metabolism. Chem. Rec. 2005, 5, 367–375. [Google Scholar] [CrossRef]
- Lowe, D.C.; Schmidt, U. Formaldehyde (HCHO) Measurements in the Nonurban Atmosphere. J. Geophys. Res. 1983, 88, 10844–10858. [Google Scholar] [CrossRef]
- Luecken, D.J.; Hutzell, W.T.; Strum, M.L.; Pouliot, G.A. Regional Sources of Atmospheric Formaldehyde and Acetaldehyde, and Implications for Atmospheric Modeling. Atmos. Environ. 2012, 47, 477–490. [Google Scholar] [CrossRef]
- Hun, D.E.; Corsi, R.L.; Morandi, M.T.; Siegel, J.A. Formaldehyde in Residences: Long-Term Indoor Concentrations and Influencing Factors. Indoor Air 2010, 20, 196–203. [Google Scholar] [CrossRef]
- Gandhi, N.N.; Barrett-Wilt, G.; Steele, J.L.; Rankin, S.A. Lactobacillus casei Expressing Methylglyoxal Synthase Causes Browning and Heterocyclic Amine Formation in Parmesan Cheese Extract. J. Dairy Sci. 2019, 102, 100–112. [Google Scholar] [CrossRef]
- Kamps, J.J.A.G.; Hopkinson, R.J.; Schofield, C.J.; Claridge, T.D.W. How Formaldehyde Reacts with Amino Acids. Commun. Chem. 2019, 2, 1–14. [Google Scholar] [CrossRef]
- Auerbach, C.; Moutschen-Dahmen, M.; Moutschen, J. Genetic and Cytogenetical Effects of Formaldehyde and Related Compounds. Mutat. Res. 1977, 39, 317–361. [Google Scholar] [CrossRef]
- Becerra, M.C.; Páez, P.L.; Laróvere, L.E.; Albesa, I. Lipids and DNA Oxidation in Staphylococcus aureus as a Consequence of Oxidative Stress Generated by Ciprofloxacin. Mol. Cell. Biochem. 2006, 285, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Imlay, J.A.; Chin, S.M.; Linn, S. Toxic DNA Damage by Hydrogen Peroxide through the Fenton Reaction in Vivo and in Vitro. Science. 1988, 240, 640–642. [Google Scholar] [CrossRef] [PubMed]
- Sobles, F.H. Organic Peroxides and Mutagenic Effects in Drosophila: Mutagenicity of Dihydroxydimethyl Peroxide and the Mutagenic Effects of Formaldehyde. Nature 1956, 177, 979–980. [Google Scholar] [CrossRef]
- Song, Z.B.; Xiao, S.Q.; You, L.; Wang, S.S.; Tan, H.; Li, K.Z.; Chen, L.M. C1 Metabolism and the Calvin Cycle Function Simultaneously and Independently during HCHO Metabolism and Detoxification in Arabidopsis thaliana Treated with HCHO Solutions. Plant, Cell Environ. 2013, 36, 1490–1506. [Google Scholar] [CrossRef]
- Kelly, D.P.; Dewar, M.K.; Johns, R.B.; Wei-Let, S.; Yates, J.F. Cross-Linking of Amino Acids by Formaldehyde. Preparation and 13C NMR Spectra of Model Compounds. Adv. Exp. Med. Biol. 1977, 86A, 641–647. [Google Scholar]
- Metz, B.; Kersten, G.F.A.; Baart, G.J.E.; De Jong, A.; Meiring, H.; Ten Hove, J.; Van Steenbergen, M.J.; Hennink, W.E.; Crommelin, D.J.A.; Jiskoot, W. Identification of Formaldehyde-Induced Modifications in Proteins: Reactions with Insulin. Bioconjug. Chem. 2006, 17, 815–822. [Google Scholar] [CrossRef]
- Kawanishi, M.; Matsuda, T.; Yagi, T. Genotoxicity of Formaldehyde: Molecular Basis of DNA Damage and Mutation. Front. Environ. Sci. 2014, 2, 36. [Google Scholar] [CrossRef]
- Metz, B.; Kersten, G.F.A.; Hoogerhout, P.; Brugghe, H.F.; Timmermans, H.A.M.; De Jong, A.; Meiring, H.; Hove, J.; Hennink, W.E.; Crommelin, D.J.A.; et al. Identification of Formaldehyde-Induced Modifications in Proteins: Reactions with Model Peptides. J. Biol. Chem. 2004, 279, 6235–6243. [Google Scholar] [CrossRef] [PubMed]
- Wilson, S.M.; Gleisten, M.P.; Donohue, T.J. Identification of Proteins Involved in Formaldehyde Metabolism by Rhodobacter sphaeroides. Microbiology 2008, 154, 296–305. [Google Scholar] [CrossRef]
- Summers, R.M.; Louie, T.M.; Yu, C.L.; Gakhar, L.; Louie, K.C.; Subramanian, M. Novel, Highly Specific N-Demethylases Enable Bacteria to Live on Caffeine and Related Purine Alkaloids. J. Bacteriol. 2012, 194, 2041–2049. [Google Scholar] [CrossRef] [PubMed]
- Venkatesagowda, B.; Dekker, R.F.H. Microbial Demethylation of Lignin: Evidence of Enzymes Participating in the Removal of Methyl/Methoxyl Groups. Enzyme Microb. Technol. 2021, 147, 1–26. [Google Scholar] [CrossRef]
- Chen, N.H.; Djoko, K.Y.; Veyrier, F.J.; McEwan, A.G. Formaldehyde Stress Responses in Bacterial Pathogens. Front. Microbiol. 2016, 7, 1–17. [Google Scholar] [CrossRef]
- Anthony, C. The Biochemistry of Methylotrophs; Academic Press: London, UK; New York, NY, USA, 1982. [Google Scholar]
- Heux, S.; Brautaset, T.; Vorholt, J.A.; Wendisch, V.F.; Portais, J.C. Synthetic Methylotrophy: Past, Present, and Future. In Methane Biocatalysis: Paving the Way to Sustainability; Springer International Publishing: Cham, Switzerland, 2018; pp. 133–151. [Google Scholar]
- Arfman, N.; Watling, E.M.; Clement, W.; Van Oosterwijk, R.J.; de Vries, G.E.; Harder, W.; Attwood, M.M.; Dijkhuizen, L. Methanol Metabolism in Thermotolerant Methylotrophic Bacillus Strains Involving a Novel Catabolic NAD-Dependent Methanol Dehydrogenase as a Key Enzyme. Arch Microbiol 1989, 152, 280–288. [Google Scholar] [CrossRef]
- Kremp, F.; Volker, M. Methanol and Methyl Group Conversion in Acetogenic Bacteria: Biochemistry, Physiology and Application. FEMS Microbiol. Rev. 2020, 45, 1–22. [Google Scholar] [CrossRef]
- Lee, J.A.; Riazi, S.; Nemati, S.; Bazurto, J.V.; Vasdekis, A.E.; Ridenhour, B.J.; Remien, C.H.; Marx, C.J. Microbial Phenotypic Heterogeneity in Response to a Metabolic Toxin: Continuous, Dynamically Shifting Distribution of Formaldehyde Tolerance in Methylobacterium extorquens Populations. PLoS Genet. 2019, 15, e1008458. [Google Scholar] [CrossRef] [PubMed]
- Gao, B.; Zhao, N.; Deng, J.; Gu, Y.; Jia, S.; Hou, Y.; Lv, X.; Liu, L. Constructing a Methanol-Dependent Bacillus subtilis by Engineering the Methanol Metabolism. J. Biotechnol. 2022, 343, 128–137. [Google Scholar] [CrossRef]
- Koopman, F.W.; De Winde, J.H.; Ruijssenaars, H.J. C1 Compounds as Auxiliary Substrate for Engineered Pseudomonas putida S12. Appl. Microbiol. Biotechnol. 2009, 83, 705–713. [Google Scholar] [CrossRef]
- Chen, F.Y.H.; Jung, H.W.; Tsuei, C.Y.; Liao, J.C. Converting Escherichia coli to a Synthetic Methylotroph Growing Solely on Methanol. Cell 2020, 182, 933–946.e14. [Google Scholar] [CrossRef] [PubMed]
- Kalyuzhnaya, M.G.; Xing, X.-H. Methane Biocatalysis: Paving the Way to Sustainability; Kalyuzhnaya, M.G., Xing, X.-H., Eds.; Springer International Publishing AG part of Springer Nature: Cham, Switzerland, 2018. [Google Scholar]
- Vorholt, J.A. Cofactor-Dependent Pathways of Formaldehyde Oxidation in Methylotrophic Bacteria. Arch. Microbiol. 2002, 178, 239–249. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, T.; Wu, S.; Wu, M.; Xin, F.; Dong, W.; Ma, J.; Zhang, M.; Jiang, M. Guidance for Engineering of Synthetic Methylotrophy Based on Methanol Metabolism in Methylotrophy. RSC Adv. 2017, 7, 4083–4091. [Google Scholar] [CrossRef]
- Zhu, T.; Zhao, T.; Bankefa, O.E.; Li, Y. Engineering Unnatural Methylotrophic Cell Factories for Methanol-Based Biomanufacturing: Challenges and Opportunities. Biotechnol. Adv. 2020, 39, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Pfeifenschneider, J.; Brautaset, T.; Wendisch, V.F. Methanol as Carbon Substrate in the Bio-Economy: Metabolic Engineering of Aerobic Methylotrophic Bacteria for Production of Value-Added Chemicals. Biofuels Bioprod. Biorefining 2017, 11, 719–731. [Google Scholar] [CrossRef]
- Tuyishime, P.; Sinumvayo, J.P. Novel Outlook in Engineering Synthetic Methylotrophs and Formatotrophs: A Course for Advancing C1-Based Chemicals Production. World J. Microbiol. Biotechnol. 2020, 36, 1–16. [Google Scholar] [CrossRef]
- Guo, F.; Zhang, S.; Jiang, Y.; Xu, H.; Xin, F.; Zhang, W.; Jiang, M. Bioconversion of Methanol by Synthetic Methylotrophy. In Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar] [CrossRef]
- Jiang, W.; Villamor, D.H.; Peng, H.; Chen, J.; Liu, L.; Haritos, V.; Ledesma-amaro, R. Metabolic Engineering Strategies to Enable Microbial Utilization of C1 Feedstocks. Nat. Chem. Biol. 2021, 17, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Gregory, G.J.; Bennett, R.K.; Papoutsakis, E.T. Recent Advances toward the Bioconversion of Methane and Methanol in Synthetic Methylotrophs. Metab. Eng. 2021. ePublished ahead of print. [Google Scholar] [CrossRef]
- Yang, D.; Park, S.Y.; Park, Y.S.; Eun, H.; Lee, S.Y. Metabolic Engineering of Escherichia coli for Natural Product Biosynthesis. Trends Biotechnol. 2020, 38, 745–765. [Google Scholar] [CrossRef] [PubMed]
- Wolf, S.; Becker, J.; Tsuge, Y.; Kawaguchi, H.; Kondo, A.; Marienhagen, J.; Bott, M.; Wendisch, V.F.; Wittmann, C. Advances in Metabolic Engineering of Corynebacterium glutamicum to Produce High-Value Active Ingredients for Food, Feed, Human Health, and Well-Being. Essays Biochem. 2021, 65, 197–212. [Google Scholar]
- Vorholt, J.A.; Chistoserdova, L.; Stolyar, S.M.; Thauer, R.K.; Lidstrom, M.E. Distribution of Tetrahydromethanopterin-Dependent Enzymes in Methylotrophic Bacteria and Phylogeny of Methenyl Tetrahydromethanopterin Cyclohydrolases. J. Bacteriol. 1999, 181, 5750–5757. [Google Scholar] [CrossRef] [PubMed]
- Ras, J.; Van Ophem, P.W.; Reijnders, W.N.M.; Van Spanning, R.J.M.; Duine, J.A.; Stouthamer, A.H.; Harms, N. Isolation, Sequencing, and Mutagenesis of the Gene Encoding NAD- and Glutathione-Dependent Formaldehyde Dehydrogenase (GD-FALDH) from Paracoccus denitrificans, in Which GD-FALDH Is Essential for Methylotrophic Growth. J. Bacteriol. 1995, 177, 247–251. [Google Scholar] [CrossRef]
- Vorholt, J.A.; Chistoserdova, L.; Lidstrom, M.E.; Thauer, R.K. The NADP-Dependent Methylene Tetrahydromethanopterin Dehydrogenase in Methylobacterium extorquens AM1. J. Bacteriol. 1998, 180, 5351–5356. [Google Scholar] [CrossRef] [PubMed]
- Pomper, B.K.; Vorholt, J.A.; Chistoserdova, L.; Lidstrom, M.E.; Thauer, R.K. A Methenyl Tetrahydromethanopterin Cyclohydrolase and a Methenyl Tetrahydrofolate Cyclohydrolase in Methylobacterium extorquens AM1. Eur. J. Biochem. 1999, 261, 475–480. [Google Scholar] [CrossRef] [PubMed]
- Goenrich, M.; Bursy, J.; Hübner, E.; Linder, D.; Schwartz, A.C.; Vorholt, J.A. Purification and Characterization of the Methylene Tetrahydromethanopterin Dehydrogenase MtdB and the Methylene Tetrahydrofolate Dehydrogenase FolD from Hyphomicrobium zavarzinii ZV580. Arch. Microbiol. 2002, 177, 299–303. [Google Scholar] [CrossRef]
- Chistoserdova, L.; Gomelsky, L.; Vorholt, J.A.; Gomelsky, M.; Tsygankov, Y.D.; Lidstrom, M.E. Analysis of Two Formaldehyde Oxidation Pathways in Methylobacillus flagellatus KT, a Ribulose Monophosphate Cycle Methylotroph. Microbiology 2000, 146, 233–238. [Google Scholar] [CrossRef]
- Müller, J.E.N.; Meyer, F.; Litsanov, B.; Kiefer, P.; Vorholt, J.A. Core Pathways Operating during Methylotrophy of Bacillus methanolicus MGA3 and Induction of a Bacillithiol-Dependent Detoxification Pathway upon Formaldehyde Stress. Mol. Microbiol. 2015, 98, 1089–1100. [Google Scholar] [CrossRef]
- Heggeset, T.M.B.; Krog, A.; Balzer, S.; Wentzel, A.; Ellingsen, T.E.; Brautaseta, T. Genome Sequence of Thermotolerant Bacillus methanolicus: Features and Regulation Related to Methylotrophy and Production of L-Lysine and L-Glutamate from Methanol. Appl. Environ. Microbiol. 2012, 78, 5170–5181. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.E.N.; Litsanov, B.; Bortfeld-Miller, M.; Trachsel, C.; Grossmann, J.; Brautaset, T.; Vorholt, J.A. Proteomic Analysis of the Thermophilic Methylotroph Bacillus methanolicus MGA3. Proteomics 2014, 14, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Marx, C.J.; Lidstrom, M.E. Development of an Insertional Expression Vector System for Methylobacterium extorquens AM1 and Generation of Null Mutants Lacking MtdA and/or Fch. Microbiology 2004, 150, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Crowther, G.J.; Kosály, G.; Lidstrom, M.E. Formate as the Main Branch Point for Methylotrophic Metabolism in Methylobacterium extorquens AM1. J. Bacteriol. 2008, 190, 5057–5062. [Google Scholar] [CrossRef] [PubMed]
- Vorholt, J.A.; Marx, C.J.; Lidstrom, M.E.; Thauer, R.K. Novel Formaldehyde-Activating Enzyme in Methylobacterium extorquens AM1 Required for Growth on Methanol. J. Bacteriol. 2000, 182, 6645–6650. [Google Scholar] [CrossRef]
- Yanpirat, P.; Nakatsuji, Y.; Hiraga, S.; Fujitani, Y.; Izumi, T.; Masuda, S.; Mitsui, R.; Nakagawa, T.; Tani, A. Lanthanide-Dependent Methanol and Formaldehyde Oxidation in Methylobacterium aquaticum Strain 22A. Microorganisms 2020, 8, 822. [Google Scholar] [CrossRef]
- Ten, L.N.; Li, W.; Salah, N.; Myung, E.; Kim, K.; Yeol, S.; Alejandro, L. Methylobacterium segetis Sp. Nov., a Novel Member of the Family Methylobacteriaceae Isolated from Soil on Jeju Island. Arch. Microbiol. 2020, 202, 747–754. [Google Scholar] [CrossRef]
- Chou, H.-H.; Chiu, H.-C.; Delaney, N.F.; Segrè, D.; Marx, C.J. Diminishing Returns Epistasis among Beneficial Mutations Decelerates Adaptation. Science. 2011, 332, 1190–1192. [Google Scholar] [CrossRef]
- Harms, N.; Ras, J.; Koning, S.; Reijnders, W.N.M.; Stouthamer, A.H.; Van Spanning, R.J.M. Genetics of C1 Metabolism Regulation in Paracoccus Denitrificans. In Microbial Growth on C1 Compounds; Springer: Dordrecht, The Netherlands, 1996; pp. 126–132. [Google Scholar]
- Goenrich, M.; Bartoschek, S.; Hagemeier, C.H.; Griesinger, C.; Vorholt, J.A. A Glutathione-Dependent Formaldehyde-Activating Enzyme (Gfa) from Paracoccus denitrificans Detected and Purified via Two-Dimensional Proton Exchange NMR. J. Biol. Chem. 2002, 277, 3069–3072. [Google Scholar] [CrossRef]
- Marx, C.J.; Chistoserdova, L.; Lidstrom, M.E. Formaldehyde-Detoxifying Role of the Tetrahydromethanopterin-Linked Pathway in Methylobacterium extorquens AM1. J. Bacteriol. 2003, 185, 7160–7168. [Google Scholar] [CrossRef] [PubMed]
- Barber, R.D.; Rott, M.A.; Donohue, T.J. Characterization of a Glutathione-Dependent Formaldehyde Dehydrogenase from Rhodobacter sphaeroides. J. Bacteriol. 1996, 178, 1386–1393. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barber, R.D.; Donohue, T.J. Function of a Glutathione-Dependent Formaldehyde Dehydrogenase in Rhodobacter sphaeroides Formaldehyde Oxidation and Assimilation. Biochemistry 1998, 37, 530–537. [Google Scholar] [CrossRef] [PubMed]
- Harms, N.; Ras, J.; Reijnders, W.N.M.; Van Spanning, R.J.M.; Stouthamer, A.H. S-Formylglutathione Hydrolase of Paracoccus denitrificans Is Homologous to Human Esterase D: A Universal Pathway for Formaldehyde Detoxification? J. Bacteriol. 1996, 178, 6296–6299. [Google Scholar] [CrossRef]
- Sahm, H.; Cox, R.B.; Quayle, J.R. Metabolism of Methanol by Rhodopseudomonas acidophila. J. Gen. Microbiol. 1976, 94, 313–322. [Google Scholar] [CrossRef] [PubMed]
- Misset-Smits, M.; Van Ophem, P.W.; Sakuda, S.; Duine, J.A. Mycothiol, 1-O-(2′-[N-Acetyl-L-Cysteinyl]Amido-2′-Deoxy-α-D-Glucopyranosyl)-D-Myo -Inositol, Is the Factor of NAD/Factor-Dependent Formaldehyde Dehydrogenase. FEBS Lett. 1997, 409, 221–222. [Google Scholar] [CrossRef]
- Eggeling, L.; Sahm, H. A Trimeric Enzyme Requiring a Cofactor and Active with Alcohols. Eur. J. Biochem. 1985, 150, 129–134. [Google Scholar] [CrossRef]
- van Ophem, P.W.; van Beeumen, J.; Duine, J.A. NAD-linked, Factor-dependent Formaldehyde Dehydrogenase or Trimeric, Zinc-containing, Long-chain Alcohol Dehydrogenase from Amycolatopsis methanolica. Eur. J. Biochem. 1992, 206, 511–518. [Google Scholar] [CrossRef]
- Klein, C.R.; Kesseler, F.P.; Perrei, C.; Frank, J.; Duine, J.A.; Schwartz, A.C. A Novel Dye-Linked Formaldehyde Dehydrogenase with Some Properties Indicating the Presence of a Protein-Bound Redox-Active Quinone Cofactor. Biochem. J. 1994, 301, 289–295. [Google Scholar] [CrossRef]
- Zahn, J.A.; Bergmann, D.J.; Boyd, J.M.; Kunz, R.C.; DiSpirito, A.A. Membrane-Associated Quinoprotein Formaldehyde Dehydrogenase from Methylococcus capsulatus Bath. J. Bacteriol. 2001, 183, 6832–6840. [Google Scholar] [CrossRef]
- Schmidt, S.; Christen, P.; Kiefer, P.; Vorholt, J.A. Functional Investigation of Methanol Dehydrogenase-like Protein XoxF in Methylobacterium extorquens AM1. Microbiology 2010, 156, 2575–2586. [Google Scholar] [CrossRef] [PubMed]
- Pol, A.; Barends, T.R.M.; Dietl, A.; Khadem, A.F.; Eygensteyn, J.; Jetten, M.S.M.; Op den Camp, H.J.M. Rare Earth Metals Are Essential for Methanotrophic Life in Volcanic Mudpots. Environ. Microbiol. 2014, 16, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Good, N.M.; Moore, R.S.; Suriano, C.J.; Martinez-Gomez, N.C. Contrasting in Vitro and in Vivo Methanol Oxidation Activities of Lanthanide-Dependent Alcohol Dehydrogenases XoxF1 and ExaF from Methylobacterium extorquens AM1. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef]
- Skovran, E.; Palmer, A.D.; Rountree, A.M.; Good, N.M.; Lidstrom, M.E. XoxF Is Required for Expression of Methanol Dehydrogenase in Methylobacterium extorquens AM1. J. Bacteriol. 2011, 193, 6032–6038. [Google Scholar] [CrossRef]
- De Zwart, J.M.M.; Nelisse, P.N.; Kuenen, J.G. Isolation and Characterization of Methylophaga sulfidovorans Sp. Nov.: An Obligately Methylotrophic, Aerobic, Dimethylsulfide Oxidizing Bacterium from a Microbial Mat. FEMS Microbiol. Ecol. 1996, 20, 261–270. [Google Scholar] [CrossRef]
- Delépine, B.; López, M.G.; Carnicer, M.; Vicente, C.M.; Wendisch, V.F.; Heux, S. Charting the Metabolic Landscape of the Facultative Methylotroph Bacillus methanolicus. mSystems 2020, 5, e00745-20. [Google Scholar] [CrossRef] [PubMed]
- Chistoserdova, L.; Kalyuzhnaya, M.G.; Lidstrom, M.E. The Expanding World of Methylotrophic Metabolism. Annu. Rev. Microbiol. 2009, 63, 477–499. [Google Scholar] [CrossRef] [PubMed]
- Vargas, D.; Hageman, S.; Gulati, M.; Nobile, C.J.; Rawat, M. S-Nitrosomycothiol Reductase and Mycothiol Are Required for Survival under Aldehyde Stress and Biofilm Formation in Mycobacterium smegmatis. IUBMB Life 2016, 68, 621–628. [Google Scholar] [CrossRef]
- Mitsui, R.; Kusano, Y.; Yurimoto, H.; Sakai, Y.; Kato, N.; Tanaka, M. Formaldehyde Fixation Contributes to Detoxification for Growth of a Nonmethylotroph, Burkholderia cepacia TM1, on Vanillic Acid. Appl. Environ. Microbiol. 2003, 69, 6128–6132. [Google Scholar] [CrossRef]
- Tanaka, N.; Kusakabe, Y.; Ito, K.; Yoshimoto, T.; Nakamura, K.T. Crystal Structure of Glutathione-Independent Formaldehyde Dehydrogenase. Chem. Biol. Interact. 2003, 143–144, 211–218. [Google Scholar] [CrossRef]
- Marx, C.J.; Miller, J.A.; Chistoserdova, L.; Lidstrom, M.E. Multiple Formaldehyde Oxidation/Detoxification Pathways in Burkholderia Fungorum LB400. J. Bacteriol. 2004, 186, 2173–2178. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Chen, S.; Wang, D.; Zhang, W.; Wang, S.; Ding, J.; Wang, Y.; Cai, L.; Ran, X.; Wang, X.; et al. Structure of Formaldehyde Dehydrogenase from Pseudomonas aeruginosa: The Binary Complex with the Cofactor NAD +. Struct. Biol. Cryst. Commun. 2013, 69, 967–972. [Google Scholar] [CrossRef]
- Ando, M.; Yoshtmoto, T.; Ogushl, S.; Rikitake, K.; Shibata, S.; Tsuru, D. Formaldehyde Dehydrogenase from Pseudomonas putida: Purification and Some Properties. J. Biochem. 1979, 85, 1165–1172. [Google Scholar]
- Kato, N.; Shirakawa, K.; Kobayashi, H.; Sakazawa, C. The Dismutation of Aldehydes by a Bacterial Enzyme. Agric. Biol. Chem. 1983, 47, 39–46. [Google Scholar]
- Bystrykh, L.V.; Govorukhina, N.I.; Van Ophem, P.W.; Hektor, H.J.; Dijkhuizen, L.; Duine, J.A. Formaldehyde Dismutase Activities in Gram-Positive Bacteria Oxidizing Methanol. J. Gen. Microbiol. 1993, 139, 1979–1985. [Google Scholar] [CrossRef]
- Nguyen, T.T.H.; Eiamphungporn, W.; Mäder, U.; Liebeke, M.; Lalk, M.; Hecker, M.; Helmann, J.D.; Antelmann, H. Genome-Wide Responses to Carbonyl Electrophiles in Bacillus subtilis: Control of the Thiol-Dependent Formaldehyde Dehydrogenase AdhA and Cysteine Proteinase YraA by the MerR-Family Regulator YraB (AdhR). Mol. Microbiol. 2009, 71, 876–894. [Google Scholar]
- Loshon, C.A.; Genest, P.C.; Setlow, B.; Setlow, P. Formaldehyde Kills Spores of Bacillus subtilis by DNA Damage and Small, Acid-Soluble Spore Proteins of the α/β-Type Protect Spores against This DNA Damage. J. Appl. Microbiol. 1999, 87, 8–14. [Google Scholar] [CrossRef]
- Moeller, R.; Vlašić, I.; Reitz, G.; Nicholson, W.L. Role of Altered RpoB Alleles in Bacillus subtilis Sporulation and Spore Resistance to Heat, Hydrogen Peroxide, Formaldehyde, and Glutaraldehyde. Arch. Microbiol. 2012, 194, 759–767. [Google Scholar] [CrossRef]
- Newton, G.L.; Rawat, M.; La Clair, J.J.; Jothivasan, V.K.; Budiarto, T.; Hamilton, C.J.; Claiborne, A.; Helmann, J.D.; Fahey, R.C. Bacillithiol Is an Antioxidant Thiol Produced in Bacilli. Nat. Chem. Biol. 2009, 5, 625–627. [Google Scholar] [CrossRef]
- Yasueda, H.; Kawahara, Y.; Sugimoto, S.I. Bacillus subtilis yckG and yckF Encode Two Key Enzymes of the Ribulose Monophosphate Pathway Used by Methylotrophs, and yckH Is Required for Their Expression. J. Bacteriol. 1999, 181, 7154–7160. [Google Scholar] [CrossRef] [PubMed]
- Yurimoto, H.; Hirai, R.; Matsuno, N.; Yasueda, H.; Kato, N.; Sakai, Y. HxlR, a Member of the DUF24 Protein Family, Is a DNA-Binding Protein That Acts as a Positive Regulator of the Formaldehyde-Inducible hxlAB Operon in Bacillus subtilis. Mol. Microbiol. 2005, 57, 511–519. [Google Scholar] [CrossRef]
- Gaballa, A.; Antelmann, H.; Hamilton, C.J.; Helmann, J.D. Regulation of Bacillus subtilis Bacillithiol Biosynthesis Operons by Spx. Microbiol. 2013, 159, 2025–2035. [Google Scholar] [CrossRef]
- Witthoff, S.; Schmitz, K.; Niedenführ, S.; Nöh, K.; Noack, S.; Bott, M.; Marienhagen, J. Metabolic Engineering of Corynebacterium glutamicum for Methanol Metabolism. Appl. Environ. Microbiol. 2015, 81, 2215–2225. [Google Scholar] [CrossRef]
- Hennig, G.; Haupka, C.; Brito, L.F.; Rückert, C.; Cahoreau, E.; Heux, S.; Wendisch, V.F. Methanol-Essential Growth of Corynebacterium glutamicum: Adaptive Laboratory Evolution Overcomes Limitation Due to Methanethiol Assimilation Pathway. Int. J. Mol. Sci. 2020, 21, 3617. [Google Scholar] [CrossRef] [PubMed]
- Witthoff, S.; Mühlroth, A.; Marienhagen, J.; Bott, M. C1 Metabolism in Corynebacterium glutamicum: An Endogenous Pathway for Oxidation of Methanol to Carbon Dioxide. Appl. Environ. Microbiol. 2013, 79, 6974–6983. [Google Scholar] [CrossRef] [PubMed]
- Lessmeier, L.; Hoefener, M.; Wendisch, V.F. Formaldehyde Degradation in Corynebacterium glutamicum Involves Acetaldehyde Dehydrogenase and Mycothiol-Dependent Formaldehyde Dehydrogenase. Microbiology 2013, 159, 2651–2662. [Google Scholar] [CrossRef]
- Witthoff, S.; Eggeling, L.; Bott, M.; Polen, T. Corynebacterium glutamicum Harbours a Molybdenum Cofactor-Dependent Formate Dehydrogenase Which Alleviates Growth Inhibition in the Presence of Formate. Microbiology 2012, 158, 2428–2439. [Google Scholar] [CrossRef]
- Leßmeier, L.; Pfeifenschneider, J.; Carnicer, M.; Heux, S.; Portais, J.C.; Wendisch, V.F. Production of Carbon-13-Labeled Cadaverine by Engineered Corynebacterium glutamicum Using Carbon-13-Labeled Methanol as Co-Substrate. Appl. Microbiol. Biotechnol. 2015, 99, 10163–10176. [Google Scholar] [CrossRef]
- Tuyishime, P.; Wang, Y.; Fan, L.; Zhang, Q.; Li, Q.; Zheng, P.; Sun, J.; Ma, Y. Engineering Corynebacterium glutamicum for Methanol-Dependent Growth and Glutamate Production. Metab. Eng. 2018, 49, 220–231. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Liu, Y.; Liu, J.; Guo, Y.; Fan, L.; Ni, X.; Zheng, X.; Wang, M.; Zheng, P.; Sun, J.; et al. MACBETH: Multiplex Automated Corynebacterium glutamicum Base Editing Method. Metab. Eng. 2018, 47, 200–210. [Google Scholar] [CrossRef]
- Gutheil, W.G.; Holmquist, B.; Vallee, B.L. Purification, Characterization, and Partial Sequence of the Glutathione-Dependent Formaldehyde Dehydrogenase from Escherichia coli: A Class III Alcohol Dehydrogenase. Biochemistry 1992, 31, 475–481. [Google Scholar] [CrossRef]
- Gutheil, W.G.; Kasimoglu, E.; Nicholson, P.C. Induction of Glutathione-Dependent Formaldehyde Dehydrogenase Activity in Escherichia coli and Hemophilus influenza. Biochem. Biophys. Res. Commun. 1997, 238, 693–696. [Google Scholar] [CrossRef]
- Herring, C.D.; Blattner, F.R. Global Transcriptional Effects of a Suppressor TRNA and the Inactivation of the Regulator FrmR. J. Bacteriol. 2004, 186, 6714–6720. [Google Scholar] [CrossRef]
- Gonzalez, C.F.; Proudfoot, M.; Brown, G.; Korniyenko, Y.; Mori, H.; Savchenko, A.V.; Yakunin, A.F. Molecular Basis of Formaldehyde Detoxification: Characterization of Two S-Formylglutathione Hydrolases from Escherichia coli, FrmB and YeiG. J. Biol. Chem. 2006, 281, 14514–14522. [Google Scholar] [CrossRef]
- Mason, R.P.; Sanders, J.K.M.; Crawford, A.; Hunter, B.K. Formaldehyde Metabolism by Escherichia coli. Detection by in Vivo 13C NMR Sectroscopy of S-(Hydroxymethyl)Glutathione as a Transient Intracellular Intermediate. Biochemistry 1986, 25, 4504–4507. [Google Scholar] [CrossRef] [PubMed]
- De Simone, A.; Vicente, C.M.; Peiro, C.; Gales, L.; Bellvert, F.; Enjalbert, B.; Heux, S. Mixing and Matching Methylotrophic Enzymes to Design a Novel Methanol Utilization Pathway in E. coli. Metab. Eng. 2020, 61, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Zhang, T.; Song, M.; Dai, Z.; Zhang, S.; Xin, F.; Dong, W.; Ma, J.; Jiang, M. Metabolic Engineering of Escherichia coli for High Yield Production of Succinic Acid Driven by Methanol. ACS Synth. Biol. 2018, 7, 2803–2811. [Google Scholar] [CrossRef]
- Price, J.V.; Chen, L.; Whitaker, W.B.; Papoutsakis, E.; Chen, W. Scaffoldless Engineered Enzyme Assembly for Enhanced Methanol Utilization. Proc. Natl. Acad. Sci. USA 2016, 113, 12691–12696. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.K.; Agee, A.; Har, J.R.G.; von Hagel, B.; Antoniewicz, M.R.; Papoutsakis, E.T. Regulatory Interventions Improve the Biosynthesis of Limiting Amino Acids from Methanol Carbon to Improve Synthetic Methylotrophy in Escherichia coli. Biotechnol. Bioeng. 2021, 118, 43–57. [Google Scholar] [CrossRef]
- Bennett, R.K.; Agee, A.; Ren, J.; Har, G.; Von Hagel, B.; Siu, K.; Antoniewicz, M.R.; Papoutsakis, E.T. Triggering the Stringent Response Enhances Synthetic Methanol Utilization in Escherichia Coli. Metab. Eng. 2020, 61, 1–10. [Google Scholar] [CrossRef]
- Bennett, R.K.; Dillon, M.; Ren, J.; Har, G.; Agee, A.; Von Hagel, B.; Rohlhill, J.; Antoniewicz, M.R.; Papoutsakis, E.T. Engineering Escherichia coli for Methanol-Dependent Growth on Glucose for Metabolite Production. Metab. Eng. 2020, 60, 45–55. [Google Scholar] [CrossRef]
- Wang, J.; Jian, X.; Xing, X.; Zhang, C.; Fei, Q. Empowering a Methanol-Dependent Escherichia coli via Adaptive Evolution Using a High-Throughput Microbial Microdroplet Culture System. Front. Bioeng. Biotechnol. 2020, 8, 570. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.K.; Gregory, G.J.; Gonzalez, J.E.; Ren, J.; Har, G.; Antoniewicz, M.R.; Papoutsakis, E.T. Improving the Methanol Tolerance of an Escherichia coli Methylotroph via Adaptive Laboratory Evolution Enhances Synthetic Methanol Utilization. Front. Microbiol. 2021, 12, 638426. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.K.; Dzvova, N.; Dillon, M.; Jones, S.; Hestmark, K.; Zhu, B.; Helman, N.; Greenfield, D.; Clarke, E.; Papoutsakis, E.T. Expression of Soluble Methane Monooxygenase in Escherichia coli Enables Methane Conversion. bioRxiv 2021. [Google Scholar]
- He, H.; Höper, R.; Dodenhöft, M.; Marlière, P.; Bar-Even, A. An Optimized Methanol Assimilation Pathway Relying on Promiscuous Formaldehyde-Condensing Aldolases in E. coli. Metab. Eng. 2020, 60, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Ren, J.; Zhou, L.; Li, Z.; Chen, L.; Zeng, A. An Aldolase-Catalyzed New Metabolic Pathway for the Assimilation of Formaldehyde and Methanol to Synthesize 2—Keto-4- Hydroxybutyrate and 1,3-Propanediol in Escherichia coli. ACS Synth. Biol. 2019, 8, 2483–2493. [Google Scholar] [CrossRef]
- Gonzalez, J.E.; Bennett, R.K.; Papoutsakis, E.T.; Antoniewicz, M.R. Methanol Assimilation in Escherichia coli Is Improved by Co-Utilization of Threonine and Deletion of Leucine-Responsive Regulatory Protein. Metab. Eng. 2018, 45, 67–74. [Google Scholar] [CrossRef]
- Wang, X.; Wang, Y.; Liu, J.; Li, Q.; Zhang, Z.; Zheng, P.; Lu, F. Biological Conversion of Methanol by Evolved Escherichia coli Carrying a Linear Methanol Assimilation Pathway. Bioresour. Bioprocess 2017, 4, 41. [Google Scholar] [CrossRef]
- Chou, A.; Clomburg, J.M.; Qian, S.; Gonzalez, R. 2-Hydroxyacyl-CoA Lyase Catalyzes Acyloin Condensation for One-Carbon Bioconversion. Nat. Chem. Biol. 2019, 15, 900–906. [Google Scholar] [CrossRef] [PubMed]
- Woolston, B.M.; King, J.R.; Reiter, M.; Van Hove, B.; Stephanopoulos, G. Improving Formaldehyde Consumption Drives Methanol Assimilation in Engineered E. coli. Nat. Commun. 2018, 9, 1–12. [Google Scholar] [CrossRef]
- Chen, C.T.; Chen, F.Y.H.; Bogorad, I.W.; Wu, T.Y.; Zhang, R.; Lee, A.S.; Liao, J.C. Synthetic Methanol Auxotrophy of Escherichia coli for Methanol-Dependent Growth and Production. Metab. Eng. 2018, 49, 257–266. [Google Scholar] [CrossRef] [PubMed]
- Meyer, F.; Keller, P.; Hartl, J.; Gröninger, O.G.; Kiefer, P.; Vorholt, J.A. Methanol-Essential Growth of Escherichia coli. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Rohlhill, J.; Gerald Har, J.R.; Antoniewicz, M.R.; Papoutsakis, E.T. Improving Synthetic Methylotrophy via Dynamic Formaldehyde Regulation of Pentose Phosphate Pathway Genes and Redox Perturbation. Metab. Eng. 2020, 57, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Bennett, R.K.; Gonzalez, J.E.; Whitaker, W.B.; Antoniewicz, M.R.; Papoutsakis, E.T. Expression of Heterologous Non-Oxidative Pentose Phosphate Pathway from Bacillus methanolicus and Phosphoglucose Isomerase Deletion Improves Methanol Assimilation and Metabolite Production by a Synthetic Escherichia coli Methylotroph. Metab. Eng. 2018, 45, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Whitaker, W.B.; Jones, J.A.; Bennett, R.K.; Gonzalez, J.E.; Vernacchio, V.R.; Collins, S.M.; Palmer, M.A.; Schmidt, S.; Antoniewicz, M.R.; Koffas, M.A.; et al. Engineering the Biological Conversion of Methanol to Specialty Chemicals in Escherichia coli. Metab. Eng. 2017, 39, 49–59. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.E.N.; Meyer, F.; Litsanov, B.; Kiefer, P.; Potthoff, E.; Heux, S.; Quax, W.J.; Wendisch, V.F.; Brautaset, T.; Portais, J.C.; et al. Engineering Escherichia coli for Methanol Conversion. Metab. Eng. 2015, 28, 190–201. [Google Scholar] [CrossRef] [PubMed]
- Mitsui, R.; Sakai, Y.; Yasueda, H.; Kato, N. A Novel Operon Encoding Formaldehyde Fixation: The Ribulose Monophosphate Pathway in the Gram-Positive Facultative Methylotrophic Bacterium Mycobacterium gastri MB19. J. Bacteriol. 2000, 182, 944–948. [Google Scholar] [CrossRef] [PubMed]
- Alves, A.M.C.R.; Euverink, G.J.W.; Hektor, H.J.; Hessels, G.I.; Van der Vlag, J.; Vrijbloed, J.W.; Hondmann, D.; Visser, J.; Dijkhuizen, L. Enzymes of Glucose and Methanol Metabolism in the Actinomycete Amycolatopsis methanolica. J. Bacteriol. 1994, 176, 6827–6835. [Google Scholar] [CrossRef] [PubMed]
- Ferenci, T.; Strom, T.; Quayle, J.R. Purification and Properties of 3 Hexulose Phosphate Synthase and Phospho 3 Hexuloisomerase from Methylococcus capsulatus. Biochem. J. 1974, 144, 477–486. [Google Scholar] [CrossRef] [PubMed]
- Hazeu, W.; de Bruyn, J.C.; van Dijken, J.P. Nocardia Sp. 239, a Facultative Methanol Utilizer with the Ribulose Monophosphate Pathway of Formaldehyde Fixation. Arch. Microbiol. 1983, 135, 205–210. [Google Scholar]
- Sakai, Y.; Mitsui, R.; Katayama, Y.; Yanase, H.; Kato, N. Organization of the Genes Involved in the Ribulose Monophosphate Pathway in an Obligate Methylotrophic Bacterium, Methylomonas aminofaciens 77a. FEMS Microbiol. Lett. 1999, 176, 125–130. [Google Scholar] [CrossRef]
- Yanase, H.; Ikeyama, K.; Mitsui, R.; Ra, S.; Kita, K.; Sakai, Y.; Kato, N. Cloning and Sequence Analysis of the Gene Encoding 3-Hexulose-6-Phosphate Synthase from the Methylotrophic Bacterium, Methylomonas aminofaciens 77a, and Its Expression in Escherichia coli. FEMS Microbiol. Lett. 1996, 135, 201–205. [Google Scholar] [CrossRef]
- Chistoserdova, L.; Lapidus, A.; Han, C.; Goodwin, L.; Saunders, L.; Brettin, T.; Tapia, R.; Gilna, P.; Lucas, S.; Richardson, P.M.; et al. Genome of Methylobacillus flagellatus, Molecular Basis for Obligate Methylotrophy, and Polyphyletic Origin of Methylotrophy. J. Bacteriol. 2007, 189, 4020–4027. [Google Scholar] [CrossRef]
- Hendrickson, E.L.; Beck, D.A.C.; Wang, T.; Lidstrom, M.E.; Hackett, M.; Chistoserdova, L. Expressed Genome of Methylobacillus flagellatus as Defined through Comprehensive Proteomics and New Insights into Methylotrophy. J. Bacteriol. 2010, 192, 4859–4867. [Google Scholar] [CrossRef]
- Whitaker, W.B.; Sandoval, N.R.; Bennett, R.K.; Fast, A.G.; Papoutsakis, E.T. Synthetic Methylotrophy: Engineering the Production of Biofuels and Chemicals Based on the Biology of Aerobic Methanol Utilization. Curr. Opin. Biotechnol. 2015, 33, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Dijkhuizen, L.; Levering, P.R.; Vries, G.E. Methane and Methanol Utilizers: The Physiology and Biochemistry of Aerobic Methanol-Utilizing Gram-Negative Bacteria. In Biotechnology Handbooks; Colin Murrell, J., Dalton, H., Eds.; Springer: Boston, MA, USA, 1992; pp. 149–181. [Google Scholar]
- Stolzenberger, J.; Lindner, S.N.; Persicke, M.; Brautaset, T.; Wendisch, V.F. Characterization of Fructose 1,6-Bisphosphatase and Sedoheptulose 1,7-Bisphosphatase from the Facultative Ribulose Monophosphate Cycle Methylotroph Bacillus methanolicus. J. Bacteriol. 2013, 195, 5112–5122. [Google Scholar] [CrossRef]
- Jakobsen, Ø.M.; Benichou, A.; Flickinger, M.C.; Valla, S.; Ellingsen, T.E.; Brautaset, T. Upregulated Transcription of Plasmid and Chromosomal Ribulose Monophosphate Pathway Genes Is Critical for Methanol Assimilation Rate and Methanol Tolerance in the Methylotrophic Bacterium Bacillus methanolicus. J. Bacteriol. 2006, 188, 3063–3072. [Google Scholar] [CrossRef] [PubMed]
- Pfeifenschneider, J.; Markert, B.; Stolzenberger, J.; Brautaset, T.; Wendisch, V.F. Transaldolase in Bacillus methanolicus: Biochemical Characterization and Biological Role in Ribulose Monophosphate Cycle. BMC Microbiol. 2020, 1–13. [Google Scholar] [CrossRef]
- Krog, A.; Heggeset, T.M.B.; Müller, J.E.N.; Kupper, C.E.; Schneider, O.; Vorholt, J.A.; Ellingsen, T.E.; Brautaset, T. Methylotrophic Bacillus methanolicus Encodes Two Chromosomal and One Plasmid Born NAD+ Dependent Methanol Dehydrogenase Paralogs with Different Catalytic and Biochemical Properties. PLoS ONE 2013, 8, e59188. [Google Scholar] [CrossRef]
- Fan, L.; Wang, Y.; Tuyishime, P.; Gao, N.; Li, Q.; Zheng, P.; Sun, J.; Ma, Y. Engineering Artificial Fusion Proteins for Enhanced Methanol Bioconversion. ChemBioChem 2018, 19, 2465–2471. [Google Scholar] [CrossRef] [PubMed]
- Orita, I.; Sakamoto, N.; Kato, N. Bifunctional Enzyme Fusion of 3-Hexulose-6-Phosphate Synthase and 6-Phospho-3-Hexuloisomerase. Appl. Microbiol. Biotechnol. 2007, 76, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fan, L.; Tuyishime, P.; Liu, J.; Zhang, K.; Gao, N.; Zhang, Z.; Ni, X.; Feng, J.; Yuan, Q.; et al. Adaptive Laboratory Evolution Enhances Methanol Tolerance and Conversion in Engineered Corynebacterium glutamicum. Commun. Biol. 2020, 3, 1–15. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Edlich-Muth, C.; Lindner, S.N.; Bar-Even, A. Ribulose Monophosphate Shunt Provides Nearly All Biomass and Energy Required for Growth of E. coli. ACS Synth. Biol. 2018, 7, 1601–1611. [Google Scholar] [CrossRef]
- Lee, Y.; Lafontaine, J.G.; Liao, J.C. Ensemble Modeling for Robustness Analysis in Engineering Non-Native Metabolic Pathways. Metab. Eng. 2014, 25, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Keller, P.; Noor, E.; Meyer, F.; Reiter, M.A.; Anastassov, S.; Kiefer, P.; Vorholt, J.A. Methanol-Dependent Escherichia coli Strains with a Complete Ribulose Monophosphate Cycle. Nat. Commun. 2020, 11, 5430. [Google Scholar] [CrossRef] [PubMed]
- Leßmeier, L.; Wendisch, V.F. Identification of Two Mutations Increasing the Methanol Tolerance of Corynebacterium glutamicum. BMC Microbiol. 2015, 15, 1–11. [Google Scholar] [CrossRef]
- Wang, X.; Wang, X.; Lu, X.; Ma, C.; Chen, K.; Ouyang, P. Methanol Fermentation Increases the Production of NAD(P)H-Dependent Chemicals in Synthetic Methylotrophic Escherichia coli. Biotechnol. Biofuels 2019, 12, 17. [Google Scholar] [CrossRef]
- Bogorad, I.W.; Chen, C.T.; Theisen, M.K.; Wu, T.Y.; Schlenz, A.R.; Lam, A.T.; Liao, J.C. Building Carbon-Carbon Bonds Using a Biocatalytic Methanol Condensation Cycle. Proc. Natl. Acad. Sci. USA 2014, 111, 15928–15933. [Google Scholar] [CrossRef]
- Bogorad, I.W.; Lin, T.S.; Liao, J.C. Synthetic Non-Oxidative Glycolysis Enables Complete Carbon Conservation. Nature 2013, 502, 693–697. [Google Scholar] [CrossRef]
- Henard, C.A.; Freed, E.F.; Guarnieri, M.T. Phosphoketolase Pathway Engineering for Carbon-Efficient Biocatalysis. Curr. Opin. Biotechnol. 2015, 36, 183–188. [Google Scholar] [CrossRef]
- Henard, C.A.; Smith, H.K.; Guarnieri, M.T. Phosphoketolase Overexpression Increases Biomass and Lipid Yield from Methane in an Obligate Methanotrophic Biocatalyst. Metab. Eng. 2017, 41, 152–158. [Google Scholar] [CrossRef]
- He, H.; Noor, E.; Ramos-Parra, P.A.; García-valencia, L.E.; Patterson, J.A.; de la Garza, R.I.D.; Hanson, A.D.; Bar-Even, A. In Vivo Rate of Formaldehyde Condensation with Tetrahydrofolate. Metabolites 2020, 10, 65. [Google Scholar] [CrossRef] [PubMed]
- Harder, W.; Quayle, J.R. Aspects of Glycine and Serine Biosynthesis during Growth of Pseudomonas AM1 on C Compounds. Biochem. J. 1971, 121, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Vuilleumier, S.; Chistoserdova, L.; Lee, M.-C.; Bringel, F.; Lajus, A.; Zhou, Y.; Gourion, B.; Barbe, V.; Chang, J.; Cruveiller, S.; et al. Methylobacterium Genome Sequences: A Reference Blueprint to Investigate Microbial Metabolism of C1 Compounds from Natural and Industrial Sources. PLoS ONE 2009, 4, e5584. [Google Scholar] [CrossRef]
- Yu, H.; Liao, J.C. A Modified Serine Cycle in Escherichia coli Coverts Methanol and CO2 to Two-Carbon Compounds. Nat. Commun. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Yishai, O.; Goldbach, L.; Tenenboim, H.; Lindner, S.N.; Bar-Even, A. Engineered Assimilation of Exogenous and Endogenous Formate in Escherichia coli. ACS Synth. Biol. 2017, 6, 1722–1731. [Google Scholar] [CrossRef]
- Vieira, G.; Carnicer, M.; Portais, J. FindPath: A Matlab Solution for in Silico Design of Synthetic Metabolic Pathways. Bioinformatics 2014, 30, 2986–2988. [Google Scholar] [CrossRef][Green Version]
- Cotton, C.A.R.; Claassens, N.J.; Benito-Vaquerizo, S.; Bar-Even, A. Renewable Methanol and Formate as Microbial Feedstocks. Curr. Opin. Biotechnol. 2020, 62, 168–180. [Google Scholar] [CrossRef]
- Bar-Even, A. Formate Assimilation: The Metabolic Architecture of Natural and Synthetic Pathways. Biochemistry 2016, 55, 3851–3863. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lindner, S.N.; Aslan, S.; Yishai, O.; Wenk, S.; Schann, K.; Bar-Even, A. Growth of E. coli on Formate and Methanol via the Reductive Glycine Pathway. Nat. Chem. Biol. 2020, 16, 538–545. [Google Scholar] [CrossRef]
- Siegel, J.B.; Smith, A.L.; Poust, S.; Wargacki, A.J.; Bar-Even, A.; Louw, C.; Shen, B.W.; Eiben, C.B.; Tran, H.M.; Noor, E.; et al. Computational Protein Design Enables a Novel One-Carbon Assimilation Pathway. Proc. Natl. Acad. Sci. USA 2015, 112, 3704–3709. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Yuan, Q.; Yang, X.; Liu, P.; Cheng, Y.; Luo, J.; Liu, H.; Yao, Y.; Sun, H.; Cai, T.; et al. Non-Natural Aldol Reactions Enable the Design and Construction of Novel One-Carbon Assimilation Pathways in Vitro. Front. Microbiol. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Lu, X.; Liu, Y.; Yang, Y.; Wang, S.; Wang, Q.; Wang, X.; Yan, Z.; Cheng, J.; Liu, C.; Yang, X.; et al. Constructing a Synthetic Pathway for Acetyl-Coenzyme A from One-Carbon through Enzyme Design. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]
- Yang, X.; Yuan, Q.; Luo, H.; Li, F.; Mao, Y.; Zhao, X. Systematic Design and in Vitro Validation of Novel One-Carbon Assimilation Pathways. Metab. Eng. 2019, 56, 142–153. [Google Scholar] [CrossRef]
- Jiang, H.; Lu, X.; Liu, Y.; Yang, Y.; Lu, L.; Yang, S.; Gou, J. Enzyme for Synthesizing Hydroxyl Acetaldehyde and/or 1, 3-Dihydroxyacetone by Catalyzing Formaldehyde and Applications Thereof. U.S. Patent Application 16/483,716; WO/2018/153306 A1, 2017. [Google Scholar]
- Schneider, S.; Gutiørrez, M.; Sandalova, T.; Schneider, G.; Clapøs, P.; Sprenger, G.A.; Samland, A.K. Redesigning the Active Site of Transaldolase TalB from Escherichia coli: New Variants with Improved Affinity towards Nonphosphorylated Substrates. Chem. Bio. Chem. 2010, 11, 681–690. [Google Scholar] [CrossRef]
- Harms, N.; Reijnders, W.N.M.; Koning, S.; Van Spanning, R.J.M. Two-Component System That Regulates Methanol and Formaldehyde Oxidation in Paracoccus denitrificans. J. Bacteriol. 2001, 183, 664–670. [Google Scholar] [CrossRef][Green Version]
- Van Spanning, R.J.M.; De Vries, S.; Harms, N. Coping with Formaldehyde during C1 Metabolism of Paracoccus denitrificans. J. Mol. Catal. -B Enzym. 2000, 8, 37–50. [Google Scholar] [CrossRef]
- Nokhal, T.-H.; Schlegel, H.G. Taxonomic Study of Paracoccus denitrificans. Int. J. Syst. Bacteriol. 1983, 33, 26–37. [Google Scholar] [CrossRef]
- Ganesh, I.; Vidhya, S.; Eom, G.T.; Hong, S.H. Construction of Methanol-Sensing Escherichia coli by the Introduction of a Paracoccus denitrificans MxaY-Based Chimeric Two-Component System. J. Microbiol. Biotechnol. 2017, 27, 1106–1111. [Google Scholar] [CrossRef]
- Selvamani, V.; Maruthamuthu, M.K.; Arulsamy, K.; Eom, G.T.; Hong, S.H. Construction of Methanol Sensing Escherichia coli by the Introduction of Novel Chimeric MxcQZ/OmpR Two-Component System from Methylobacterium organophilum XX. Korean J. Chem. Eng. 2017, 34, 1734–1739. [Google Scholar] [CrossRef]
- Selvamani, V.; Ganesh, I.; Chae, S.; Maruthamuthu, M.K.; Hong, S.H. Engineering of Recombinant Escherichia coli towards Methanol Sensing Using Methylobacterium extroquens Two-Component Systems. Microbiol. Biotechnol. Lett. 2020, 48, 24–31. [Google Scholar] [CrossRef]
- Bazurto, J.V.; Nayak, D.D.; Ticak, T.; Davlieva, M.; Lee, J.A.; Hellenbrand, C.N.; Lambert, L.B.; Benski, O.J.; Quates, C.J.; Johnson, J.L.; et al. EfgA Is a Conserved Formaldehyde Sensor That Leads to Bacterial Growth Arrest in Response to Elevated Formaldehyde. PLOS Biol. 2021, 19, 1–42. [Google Scholar] [CrossRef] [PubMed]
- Bazurto, J.V.; Riazi, S.; Alton, S.D.; Deatherage, D.E.; Bruger, E.L.; Barrick, J.E.; Marx, C.J. Global Transcriptional Response of Methylorubrum extorquens to Formaldehyde Stress Expands the Role of EfgA and Is Distinct from Antibiotic Translational Inhibition. Microorganisms 2021, 9, 347. [Google Scholar] [CrossRef] [PubMed]
- Bazurto, J.V.; Bruger, E.L.; Lee, J.A.; Lambert, L.B. Formaldehyde-Responsive Proteins TtmR and EfgA Reveal a Trade-off between Formaldehyde Resistance and Efficient Transition to Methylotrophy in Methylorubrum extorquens. J. Bacteriol. 2021, 203, e00589-20. [Google Scholar] [CrossRef]
- Bozdag, A.; Komives, C.; Flickinger, M.C. Growth of Bacillus methanolicus in 2 M Methanol at 50 °C: The Effect of High Methanol Concentration on Gene Regulation of Enzymes Involved in Formaldehyde Detoxification by the Ribulose Monophosphate Pathway. J. Ind. Microbiol. Biotechnol. 2015, 42, 1027–1038. [Google Scholar] [CrossRef]
- Zhu, R.; Zhang, G.; Jing, M.; Han, Y.; Li, J.; Zhao, J.; Li, Y.; Chen, P.R. Genetically Encoded Formaldehyde Sensors Inspired by a Protein Intra-Helical Crosslinking Reaction. Nat. Commun. 2021, 12, 1–13. [Google Scholar] [CrossRef]
- Brown, N.L.; Stoyanov, J.V.; Kidd, S.P.; Hobman, J.L. The MerR Family of Transcriptional Regulators. FEMS Microbiol. Rev. 2003, 27, 145–163. [Google Scholar] [CrossRef]
- Kidd, S.P.; Potter, A.J.; Apicella, M.A.; Jennings, M.P.; McEwan, A.G. NmIR of Neisseria gonorrhoeae: A Novel Redox Responsive Transcription Factor from the MerR Family. Mol. Microbiol. 2005, 57, 1676–1689. [Google Scholar] [CrossRef]
- Stroeher, U.H.; Kidd, S.P.; Stafford, S.L.; Jennings, M.P.; Paton, J.C.; McEwan, A.G. A Pneumococcal MerR-like Regulator and S-Nitrosoglutathione Reductase Are Required for Systemic Virulence. J. Infect. Dis. 2007, 196, 1820–1826. [Google Scholar] [CrossRef]
- Auchter, M.; Arndt, A.; Eikmanns, B.J. Dual Transcriptional Control of the Acetaldehyde Dehydrogenase Gene ald of Corynebacterium glutamicum by RamA and RamB. J. Biotechnol. 2009, 140, 84–91. [Google Scholar] [CrossRef]
- Giedroc, D.P.; Arunkumar, A.I. Metal Sensor Proteins: Nature’s Metalloregulated Allosteric Switches. Dalt. Trans. 2007, No. 29, 3107–3120. [Google Scholar] [CrossRef]
- Higgins, K.A.; Giedroc, D. Insights into Protein Allostery in the Csor/Rcnr Family of Transcriptional Repressors. Chem. Lett. 2014, 43, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Denby, K.J.; Iwig, J.; Bisson, C.; Westwood, J.; Rolfe, M.D.; Sedelnikova, S.E.; Higgins, K.; Maroney, M.J.; Baker, P.J.; Chivers, P.T.; et al. The Mechanism of a Formaldehyde-Sensing Transcriptional Regulator. Sci. Rep. 2016, 6, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Osman, D.; Piergentili, C.; Chen, J.; Sayer, L.N.; Usón, I.; Huggins, T.G.; Robinson, N.J.; Pohl, E. The Effectors and Sensory Sites of Formaldehyde-Responsive Regulator FrmR and Metal-Sensing Variant. J. Biol. Chem. 2016, 291, 19502–19516. [Google Scholar] [CrossRef] [PubMed]
- Rohlhill, J.; Sandoval, N.R.; Papoutsakis, E.T. Sort-Seq Approach to Rngineering a Formaldehyde-Inducible Promoter for Dynamically Regulated Escherichia coli Growth on Methanol. ACS Synth. Biol. 2017, 6, 1584–1595. [Google Scholar] [CrossRef]

| Pathway 1 | Characteristic | Example Organism | References |
|---|---|---|---|
| H4F-dependent pathway | Linear formaldehyde dissimilation pathway, requires pterin cofactor H4F | B. methanolicus MGA3 | [45,46,47] |
| H4MPT-dependent pathway | Linear formaldehyde dissimilation pathway, requires pterin cofactor H4MPT | M. extorquens AM1, Methylobacterium organophilum XX, M. aquaticum 22A, Methylobacterium segetis 17J42-1T, Hyphomicrobium methylovorum GM2, Hyphomicrobium zavarzinii ZV580, Methylosinus trichosporium OB3b, M. capsulatus Bath, Methylococcus thermophilus IIIp, Methylomicrobium album BG8, Methylomonas rubra 15sh, M. flagellatus KT, Methylophilus methylotrophus AS | [39,41,42,43,44,51] |
| GSH-dependent pathway | Linear formaldehyde dissimilation pathway, requires thiol cofactor GSH | M. aquaticum 22A, P. denitrificans, R. sphaeroides, R. acidophila | [40,51,54,57,58,59,60] |
| MSH-dependent pathway | Linear formaldehyde dissimilation pathway, requires thiol cofactor MSH | A. methanolica, R. erythropolis | [61,62,63] |
| BSH-dependent pathway | Linear formaldehyde dissimilation pathway, requires thiol cofactor BSH | B. methanolicus MGA3 | [45] |
| DL-Faldh-mediated formaldehyde dissimilation process | Formaldehyde dissimilation process, relies on activity of DL-Faldh; membrane-associated in M. capsulatus Bath | H. zavarzinii ZV580, M. capsulatus Bath | [64,65] |
| PQQ-Ln-dependent formaldehyde dissimilation process | Formaldehyde oxidation by a PQQ-Ln-dependent Mdh (XoxF1) | M. extorquens AM1, M. aquaticum 22A, M. fumariolicum SolV | [51,66,67,68] |
| Dissimilatory variant of RuMP cycle | Cyclic formaldehyde dissimilation pathway | B. methanolicus MGA3, M. flagellatus KT, M. sulfidovorans | [22,44,70] |
| Pathway 1 | Characteristic | Example Organism | Reference |
|---|---|---|---|
| H4MPT-dependent pathway | Linear formaldehyde dissimilation pathway, requires pterin cofactor H4MPT | B. fungorum LB400 | [76] |
| GSH-dependent pathway | Linear formaldehyde dissimilation pathway, requires thiol cofactor GSH | E. coli, B. fungorum LB400 | [76,96,97,98] |
| BSH-dependent pathway | Linear formaldehyde dissimilation pathway, requires thiol cofactor BSH | B. subtilis | [81,84] |
| MSH-dependent pathway | Linear formaldehyde dissimilation pathway, requires thiol cofactor MSH | C. glutamicum, M. smegmatis | [73,90,91] |
| Faldh dissimilation process | Zinc-dependent formaldehyde oxidation pathway, relies on activity of Faldh that utilizes NAD+ as an electron acceptor | P. putida, P. aeruginosa, B. fungorum LB400 | [75,76,77,78] |
| Formaldehyde dismutase-mediated dissimilation process | Formaldehyde dissimilation based on the activity of formaldehyde dismutase, leading to the formation of equimolar amounts of methanol and formate | P. putida | [79] |
| Ald-mediated dissimilation process | Formaldehyde dissimilation through direct oxidation to formate by Ald | C. glutamicum | [38,90,91] |
| Dissimilatory variant of RuMP cycle | Cyclic formaldehyde dissimilation pathway | B. subtilis, B. cepacia | [74,85,86] |
| Pathway 1 | Characteristic | Example Organism | Reference |
|---|---|---|---|
| Native pathways | |||
| RuMP cycle | Cyclic formaldehyde assimilation pathway; formaldehyde enters the RuMP cycle through condensation with Ru5P | B. methanolicus MGA3, M. gastri MB19, Nocardia sp. 239, A. methanolica, M. capsulatus, M. aminofaciens 77a, M. flagellatus KT | [122,123,124,125,126,127,128,129,132,133,134] |
| Serine cycle | Cyclic formaldehyde assimilation pathway; formaldehyde enters the pathway through methylene-H4F | M. extorquens AM1, M. organophilum XX, H. methylovorum GM2, M. trichosporium OB3b | [20,39] |
| Modified pathways | |||
| MCC | Modified RuMP cycle; synthetic biocatalytic MCC; no carbon loss | Has not been applied in vivo yet | [144] |
| Modified serine cycle | Simplified variant of the serine cycle which uses one step for the oxidation of formaldehyde instead of four in the native serine pathway; avoids the use of the Hpr route by glyoxylate transamination with alanine to form glycine | E.coli | [151] |
| Serine–threonine cycle | Synthetic variant of the serine cycle; aims to avoid interference with central metabolic fluxes; circumvents the formation of hydroxypyruvate as intermediate; further recycling of glycine via the threonine biosynthesis and cleavage system | E. coli | [152] |
| Homoserine cycle | Modified variant of the serine cycle; glycine is directly condensed with formaldehyde to generate serine; aims to avoid the competition of flux between the pathway reactions and those of the central metabolism; reduction of thermodynamic disadvantages of the natural serine cycle; CO2 fixation is avoided | E. coli | [110] |
| Pathway 1 | Characteristic | Host Organism | Reference |
|---|---|---|---|
| Reductive glycine pathway | Linear route that can be divided into four modules; small overlaps with the central metabolism minimizes requirements in regulatory optimization | E. coli | [156] |
| HACL pathway | Synthetic pathway based on the ligation of formaldehyde with formyl-CoA; whole-cell biocatalysis of glycolate | E. coli | [114] |
| SACA pathway | Synthetic linear pathway based on condensation of two formaldehyde molecules using designed Gals | E. coli | [159] |
| FLS pathway | Synthetic pathway in which the computationally designed enzyme FLS catalyzes the carboligation of three formaldehyde molecules | E. coli | [113,157] |
| GAA pathway | Synthetic pathway based on computationally-predicted ATP-independent and carbon-conserving reactions; starts with condensation of two formaldehyde molecules using Gals | Has not been applied in vivo yet | [160] |
| DAS pathway | Synthetic pathway based on bacterial Mdh and yeast DAS identified via in silico modelling | E. coli | [101] |
| GAPA pathway | Synthetic pathway based on the introduction of non-natural aldolase reactions; starts with condensation of two formaldehyde molecules using Gals | Has not been applied in vivo yet | [158] |
| Regulator 1 | Regulated Processes | Example Organism | Reference |
|---|---|---|---|
| FlhRS | Production of formaldehyde (Mdh and Madh) or its consumption (GD-Faldh, Fgh) | P. denitrificans | [40,163,164,165] |
| HxlR | Hps-Phi in RuMP cycle (assimilatory or dissimilatory variant) | B. methanolicus MGA3, B. subtilis | [133,173] |
| TtmR | EfgA-mediated formaldehyde stress response | M. extorquens AM1 | [171] |
| AdhR | BSH-dependent formaldehyde dissimilation pathway | B. subtilis | [19] |
| FrmR | GSH-dependent formaldehyde dissimilation pathway composed of GD-Faldh and Fgh | E. coli | [98,99,178,179,180] |
| RamAB, GlxR | Ald-mediated formaldehyde dissimilation process | C. glutamicum | [91,177] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klein, V.J.; Irla, M.; Gil López, M.; Brautaset, T.; Fernandes Brito, L. Unravelling Formaldehyde Metabolism in Bacteria: Road towards Synthetic Methylotrophy. Microorganisms 2022, 10, 220. https://doi.org/10.3390/microorganisms10020220
Klein VJ, Irla M, Gil López M, Brautaset T, Fernandes Brito L. Unravelling Formaldehyde Metabolism in Bacteria: Road towards Synthetic Methylotrophy. Microorganisms. 2022; 10(2):220. https://doi.org/10.3390/microorganisms10020220
Chicago/Turabian StyleKlein, Vivien Jessica, Marta Irla, Marina Gil López, Trygve Brautaset, and Luciana Fernandes Brito. 2022. "Unravelling Formaldehyde Metabolism in Bacteria: Road towards Synthetic Methylotrophy" Microorganisms 10, no. 2: 220. https://doi.org/10.3390/microorganisms10020220
APA StyleKlein, V. J., Irla, M., Gil López, M., Brautaset, T., & Fernandes Brito, L. (2022). Unravelling Formaldehyde Metabolism in Bacteria: Road towards Synthetic Methylotrophy. Microorganisms, 10(2), 220. https://doi.org/10.3390/microorganisms10020220









