A Practical Bioinformatics Workflow for Routine Analysis of Bacterial WGS Data
Abstract
1. Introduction
2. Materials and Methods
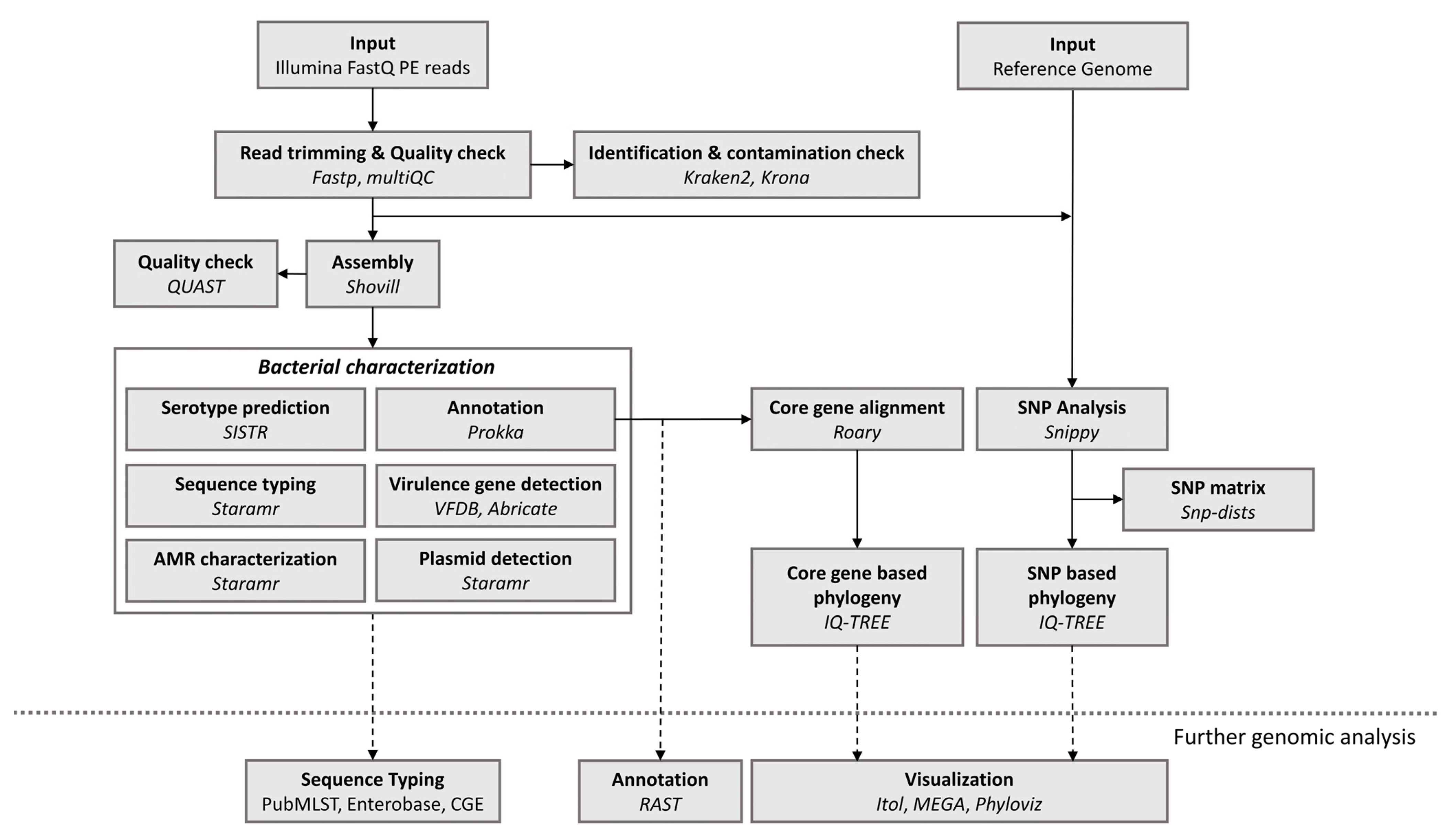
2.1. Bioinformatics Workflow
2.1.1. Data Processing and Quality Control
2.1.2. Strain Genotyping, AMR and Plasmid Replicon Detection
2.1.3. Virulence-Associated Gene Detection
2.1.4. Genome Annotation
2.1.5. Phylogenetic Analysis
2.1.6. Implementation and Availability
2.2. Validation of the Workflow
2.2.1. Validation Dataset
2.2.2. Validation Strategy
2.3. Further Genomic Analysis
3. Results
3.1. Validation of the Workflow
3.2. Data processing and Quality Control
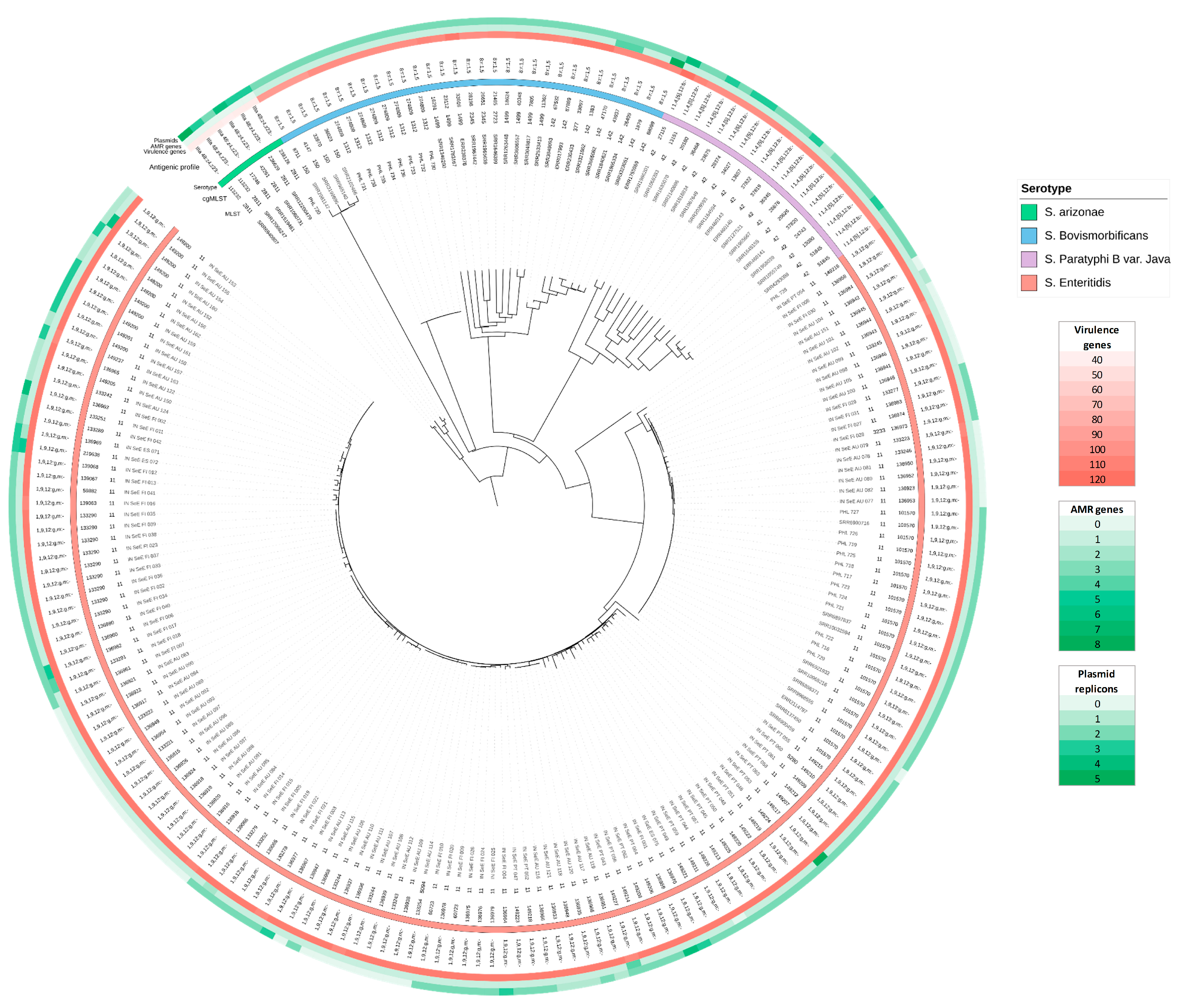
3.3. Strain Characterisation
3.4. Phylogenetic Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bogaerts, B.; Nouws, S.; Verhaegen, B.; Denayer, S.; van Braekel, J.; Winand, R.; Fu, Q.; Crombé, F.; Piérard, D.; Marchal, K.; et al. Validation Strategy of a Bioinformatics Whole Genome Sequencing Workflow for Shiga Toxin-Producing Escherichia coli Using a Reference Collection Extensively Characterized with Conventional Methods. Microb. Genom. 2021, 7, mgen000531. [Google Scholar] [CrossRef] [PubMed]
- Ronholm, J.; Nasheri, N.; Petronella, N.; Pagotto, F. Navigating Microbiological Food Safety in the Era of Whole-Genome Sequencing. Clin. Microbiol. Rev. 2016, 29, 837–857. [Google Scholar] [CrossRef] [PubMed]
- Allard, M.W.; Bell, R.; Ferreira, C.M.; Gonzalez-Escalona, N.; Hoffmann, M.; Muruvanda, T.; Ottesen, A.; Ramachandran, P.; Reed, E.; Sharma, S.; et al. Genomics of Foodborne Pathogens for Microbial Food Safety. Curr. Opin. Biotechnol. 2018, 49, 224–229. [Google Scholar] [CrossRef] [PubMed]
- Gerner-Smidt, P.; Besser, J.; Concepción-Acevedo, J.; Folster, J.P.; Huffman, J.; Joseph, L.A.; Kucerova, Z.; Nichols, M.C.; Schwensohn, C.A.; Tolar, B. Whole Genome Sequencing: Bridging One-Health Surveillance of Foodborne Diseases. Front. Public Health 2019, 7, 172. [Google Scholar] [CrossRef] [PubMed]
- Ashton, P.M.; Nair, S.; Peters, T.M.; Bale, J.A.; Powell, D.G.; Painset, A.; Tewolde, R.; Schaefer, U.; Jenkins, C.; Dallman, T.J.; et al. Identification of Salmonella for Public Health Surveillance Using Whole Genome Sequencing. PeerJ 2016, 4, e1752. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, B.; Winand, R.; Fu, Q.; van Braekel, J.; Ceyssens, P.J.; Mattheus, W.; Bertrand, S.; de Keersmaecker, S.C.J.; Roosens, N.H.C.; Vanneste, K. Validation of a Bioinformatics Workflow for Routine Analysis of Whole-Genome Sequencing Data and Related Challenges for Pathogen Typing in a European National Reference Center: Neisseria meningitidis as a Proof-of-Concept. Front. Microbiol. 2019, 10, 362. [Google Scholar] [CrossRef]
- Hendriksen, R.S.; Pedersen, S.K.; Leekitcharoenphon, P.; Malorny, B.; Borowiak, M.; Battisti, A.; Franco, A.; Alba, P.; Carfora, V.; Ricci, A.; et al. Final Report of ENGAGE—Establishing Next Generation Sequencing Ability for Genomic Analysis in Europe. EFSA Support. Publ. 2018, 15, 1431E. [Google Scholar] [CrossRef]
- Llarena, A.; Ribeiro-Gonçalves, B.F.; Nuno Silva, D.; Halkilahti, J.; Machado, M.P.; da Silva, M.S.; Jaakkonen, A.; Isidro, J.; Hämäläinen, C.; Joenperä, J.; et al. INNUENDO: A Cross-sectoral Platform for the Integration of Genomics in the Surveillance of Food-borne Pathogens. EFSA Support. Publ. 2018, 15, 1498E. [Google Scholar] [CrossRef]
- Joensen, K.G.; Tetzschner, A.M.M.; Iguchi, A.; Aarestrup, F.M.; Scheutz, F. Rapid and Easy in Silico Serotyping of Escherichia coli Isolates by Use of Whole-Genome Sequencing Data. J. Clin. Microbiol. 2015, 53, 2410–2426. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Carattoli, A.; Zankari, E.; Garciá-Fernández, A.; Larsen, M.V.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. In Silico Detection and Typing of Plasmids Using Plasmidfinder and Plasmid Multilocus Sequence Typing. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [PubMed]
- Deng, X.; den Bakker, H.C.; Hendriksen, R.S. Genomic Epidemiology: Whole-Genome-Sequencing-Powered Surveillance and Outbreak Investigation of Foodborne Bacterial Pathogens. Annu. Rev. Food Sci. Technol. 2016, 7, 353–374. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Mohamed, K.; Fan, Y.; Achtman, M. The EnteroBase User’s Guide, with Case Studies on Salmonella Transmissions, Yersinia pestis Phylogeny, and Escherichia Core Genomic Diversity. Genome Res. 2020, 30, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Jalili, V.; Afgan, E.; Gu, Q.; Clements, D.; Blankenberg, D.; Goecks, J.; Taylor, J.; Nekrutenko, A. The Galaxy Platform for Accessible, Reproducible and Collaborative Biomedical Analyses: 2020 Update. Nucleic Acids Res. 2021, 48, W395–W402. [Google Scholar] [CrossRef]
- Gangiredla, J.; Rand, H.; Benisatto, D.; Payne, J.; Strittmatter, C.; Sanders, J.; Wolfgang, W.J.; Libuit, K.; Herrick, J.B.; Prarat, M.; et al. GalaxyTrakr: A Distributed Analysis Tool for Public Health Whole Genome Sequence Data Accessible to Non-Bioinformaticians. BMC Genom. 2021, 22, 114. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An Ultra-Fast All-in-One FASTQ Preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved Metagenomic Analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Ondov, B.D.; Bergman, N.H.; Phillippy, A.M. Interactive Metagenomic Visualization in a Web Browser.Pdf. BMC Bioinformatics 2011, 12, 385. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality Assessment Tool for Genome Assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Bharat, A.; Petkau, A.; Avery, B.P.; Chen, J.; Folster, J.; Carson, C.A.; Kearney, A.; Nadon, C.; Mabon, P.; Thiessen, J.; et al. Correlation between Phenotypic and In Silico Detection of Antimicrobial Resistance in Salmonella enterica in Canada Using Staramr. Microorganisms 2022, 10, 292. [Google Scholar] [CrossRef] [PubMed]
- Bortolaia, V.; Kaas, R.S.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.F.; et al. ResFinder 4.0 for Predictions of Phenotypes from Genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, C.E.; Kruczkiewicz, P.; Laing, C.R.; Lingohr, E.J.; Gannon, V.P.J.; Nash, J.H.E.; Taboada, E.N. The Salmonella In Silico Typing Resource (SISTR): An Open Web-Accessible Tool for Rapidly Typing and Subtyping Draft Salmonella Genome Assemblies. PLoS ONE 2016, 11, e0147101. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and Refined Dataset for Big Data Analysis—10 Years On. Nucleic Acids Res. 2016, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid Large-Scale Prokaryote Pan Genome Analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Pightling, A.W.; Pettengill, J.B.; Luo, Y.; Baugher, J.D.; Rand, H.; Strain, E. Interpreting Whole-Genome Sequence Analyses of Foodborne Bacteria for Regulatory Applications and Outbreak Investigations. Front. Microbiol. 2018, 9, 1482. [Google Scholar] [CrossRef]
- ISO 6579-1:2017; Microbiology of the Food chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp. International Organization for Standardization: Geneva, Switzerland, 2017.
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- World Health Organization. Estimating the Burden of Foodborne Diseases: A Practical Handbook for Countries: A Guide for Planning, Implementing and Reporting Country-Level Burden of Foodborne Disease; World Health Organization: Geneva, Switzerland, 2021. [Google Scholar]
- Rantsiou, K.; Kathariou, S.; Winkler, A.; Skandamis, P.; Saint-Cyr, M.J.; Rouzeau-Szynalski, K.; Amézquita, A. Next Generation Microbiological Risk Assessment: Opportunities of Whole Genome Sequencing (WGS) for Foodborne Pathogen Surveillance, Source Tracking and Risk Assessment. Int. J. Food Microbiol. 2018, 287, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Pearce, M.E.; Alikhan, N.F.; Dallman, T.J.; Zhou, Z.; Grant, K.; Maiden, M.C.J. Comparative Analysis of Core Genome MLST and SNP Typing within a European Salmonella Serovar Enteritidis Outbreak. Int. J. Food Microbiol. 2018, 274, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Bae, D.; Cheng, C.M.; Khan, A.A. Characterization of Extended-Spectrum β-Lactamase (ESBL) Producing Non-Typhoidal Salmonella (NTS) from Imported Food Products. Int. J. Food Microbiol. 2015, 214, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Rossen, J.W.A.; Friedrich, A.W.; Moran-Gilad, J. Practical Issues in Implementing Whole-Genome-Sequencing in Routine Diagnostic Microbiology. Clin. Microbiol. Infect. 2018, 24, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Villa, L.; García-Fernández, A.; Fortini, D.; Carattoli, A. Replicon Sequence Typing of IncF Plasmids Carrying Virulence and Resistance Determinants. J. Antimicrob. Chemother. 2010, 65, 2518–2529. [Google Scholar] [CrossRef]
- Lyu, N.; Feng, Y.; Pan, Y.; Huang, H.; Liu, Y.; Xue, C.; Zhu, B.; Hu, Y. Genomic Characterization of Salmonella enterica Isolates From Retail Meat in Beijing, China. Front. Microbiol. 2021, 12, 636332. [Google Scholar] [CrossRef]
- Egorova, A.; Mikhaylova, Y.; Saenko, S.; Tyumentseva, M.; Tyumentsev, A.; Karbyshev, K.; Chernyshkov, A.; Manzeniuk, I.; Akimkin, V.; Shelenkov, A. Comparative Whole-Genome Analysis of Russian Foodborne Multidrug-Resistant Salmonella infantis Isolates. Microorganisms 2022, 10, 89. [Google Scholar] [CrossRef]
- Carriço, J.A.; Rossi, M.; Moran-Gilad, J.; van Domselaar, G.; Ramirez, M. A Primer on Microbial Bioinformatics for Nonbioinformaticians. Clin. Microbiol. Infect. 2018, 24, 342–349. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Ribeiro-Gonçalves, B.; Francisco, A.P.; Vaz, C.; Ramirez, M.; Carriço, J.A. PHYLOViZ Online: Web-Based Tool for Visualization, Phylogenetic Inference, Analysis and Sharing of Minimum Spanning Trees. Nucleic Acids Res. 2016, 44, W246–W251. [Google Scholar] [CrossRef]
- Petit, R.A.; Read, T.D. Bactopia: A Flexible Pipeline for Complete Analysis of Bacterial Genomes. mSystems 2020, 5, e00190-20. [Google Scholar] [CrossRef]
- Deneke, C.; Brendebach, H.; Uelze, L.; Borowiak, M.; Malorny, B.; Tausch, S.H. Species-Specific Quality Control, Assembly and Contamination Detection in Microbial Isolate Sequences with Aquamis. Genes 2021, 12, 644. [Google Scholar] [CrossRef]
- Schwengers, O.; Hoek, A.; Fritzenwanker, M.; Falgenhauer, L.; Hain, T.; Chakraborty, T.; Goesmann, A. ASA3P: An Automatic and Scalable Pipeline for the Assembly, Annotation and Higher-Level Analysis of Closely Related Bacterial Isolates. PLoS Comput. Biol. 2020, 16, e1007134. [Google Scholar] [CrossRef]
- Sserwadda, I.; Mboowa, G. Rmap: The Rapid Microbial Analysis Pipeline for Eskape Bacterial Group Whole-Genome Sequence Data. Microb. Genom. 2021, 7, 000583. [Google Scholar] [CrossRef]
- Quijada, N.M.; Rodríguez-Lázaro, D.; Eiros, J.M.; Hernández, M. TORMES: An Automated Pipeline for Whole Bacterial Genome Analysis. Bioinformatics 2019, 35, 4207–4212. [Google Scholar] [CrossRef]
- Jagtap, P.D.; Johnson, J.E.; Onsongo, G.; Sadler, F.W.; Murray, K.; Wang, Y.; Shenykman, G.M.; Bandhakavi, S.; Smith, L.M.; Griffin, T.J. Flexible and Accessible Workflows for Improved Proteogenomic Analysis Using the Galaxy Framework. J. Proteome Res. 2014, 13, 5898–5908. [Google Scholar] [CrossRef]
- Cock, P.J.A.; Grüning, B.A.; Paszkiewicz, K.; Pritchard, L. Galaxy Tools and Workflows for Sequence Analysis with Applications in Molecular Plant Pathology. PeerJ 2013, 1, e167. [Google Scholar] [CrossRef]
- Thanki, A.S.; Soranzo, N.; Haerty, W.; Davey, R.P. GeneSeqToFamily: A Galaxy Workflow to Find Gene Families Based on the Ensembl Compara GeneTrees Pipeline. Gigascience 2018, 7, giy005. [Google Scholar] [CrossRef]
- Thang, M.W.C.; Chua, X.Y.; Price, G.; Gorse, D.; Field, M.A. Metadegalaxy: Galaxy Workflow for Differential Abundance Analysis of 16S Metagenomic Data [Version 1; Peer Review: 1 Approved, 1 Approved with Reservations]. F1000Res 2019, 8, 726. [Google Scholar] [CrossRef]
- Wee, S.K.; Yap, E.P.H. GALAXY Workflow for Bacterial Next-Generation Sequencing De Novo Assembly and Annotation. Curr. Protoc. 2021, 1, e242. [Google Scholar] [CrossRef]
- Bogaerts, B.; Delcourt, T.; Soetaert, K.; Boarbi, S.; Ceyssens, P.J.; Winand, R.; van Braekel, J.; de Keersmaecker, S.C.J.; Roosens, N.H.C.; Marchal, K.; et al. A Bioinformatics Whole-Genome Sequencing Workflow for Clinical Mycobacterium tuberculosis Complex Isolate Analysis, Validated Using a Reference Collection Extensively Characterized with Conventional Methods and in Silico Approaches. J. Clin. Microbiol. 2021, 59, e00202-21. [Google Scholar] [CrossRef]
- Brown, E.; Dessai, U.; Mcgarry, S.; Gerner-Smidt, P. Use of Whole-Genome Sequencing for Food Safety and Public Health in the United States. Foodborne Pathog. Dis. 2019, 16, 441–450. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atxaerandio-Landa, A.; Arrieta-Gisasola, A.; Laorden, L.; Bikandi, J.; Garaizar, J.; Martinez-Malaxetxebarria, I.; Martinez-Ballesteros, I. A Practical Bioinformatics Workflow for Routine Analysis of Bacterial WGS Data. Microorganisms 2022, 10, 2364. https://doi.org/10.3390/microorganisms10122364
Atxaerandio-Landa A, Arrieta-Gisasola A, Laorden L, Bikandi J, Garaizar J, Martinez-Malaxetxebarria I, Martinez-Ballesteros I. A Practical Bioinformatics Workflow for Routine Analysis of Bacterial WGS Data. Microorganisms. 2022; 10(12):2364. https://doi.org/10.3390/microorganisms10122364
Chicago/Turabian StyleAtxaerandio-Landa, Aitor, Ainhoa Arrieta-Gisasola, Lorena Laorden, Joseba Bikandi, Javier Garaizar, Irati Martinez-Malaxetxebarria, and Ilargi Martinez-Ballesteros. 2022. "A Practical Bioinformatics Workflow for Routine Analysis of Bacterial WGS Data" Microorganisms 10, no. 12: 2364. https://doi.org/10.3390/microorganisms10122364
APA StyleAtxaerandio-Landa, A., Arrieta-Gisasola, A., Laorden, L., Bikandi, J., Garaizar, J., Martinez-Malaxetxebarria, I., & Martinez-Ballesteros, I. (2022). A Practical Bioinformatics Workflow for Routine Analysis of Bacterial WGS Data. Microorganisms, 10(12), 2364. https://doi.org/10.3390/microorganisms10122364







