In Situ Rumen Degradation Characteristics and Bacterial Colonization of Corn Silages Differing in Ferulic and p-Coumaric Acid Contents
Abstract
1. Introduction
2. Materials and Methods
2.1. Preparation of Corn Silage
2.2. Sampling and Analytical Methods
2.3. Determination of Extracted Phenolic Acids
2.4. In Situ Rumen Incubation
2.5. Bacterial Community Analysis
2.5.1. Collection of Solid Attached Microbiota and DNA Extraction
2.5.2. PCR Amplification, Illumina Sequencing of 16S rRNA Gene, and Sequence Analysis
2.6. Calculations and Statistical Analysis
3. Results
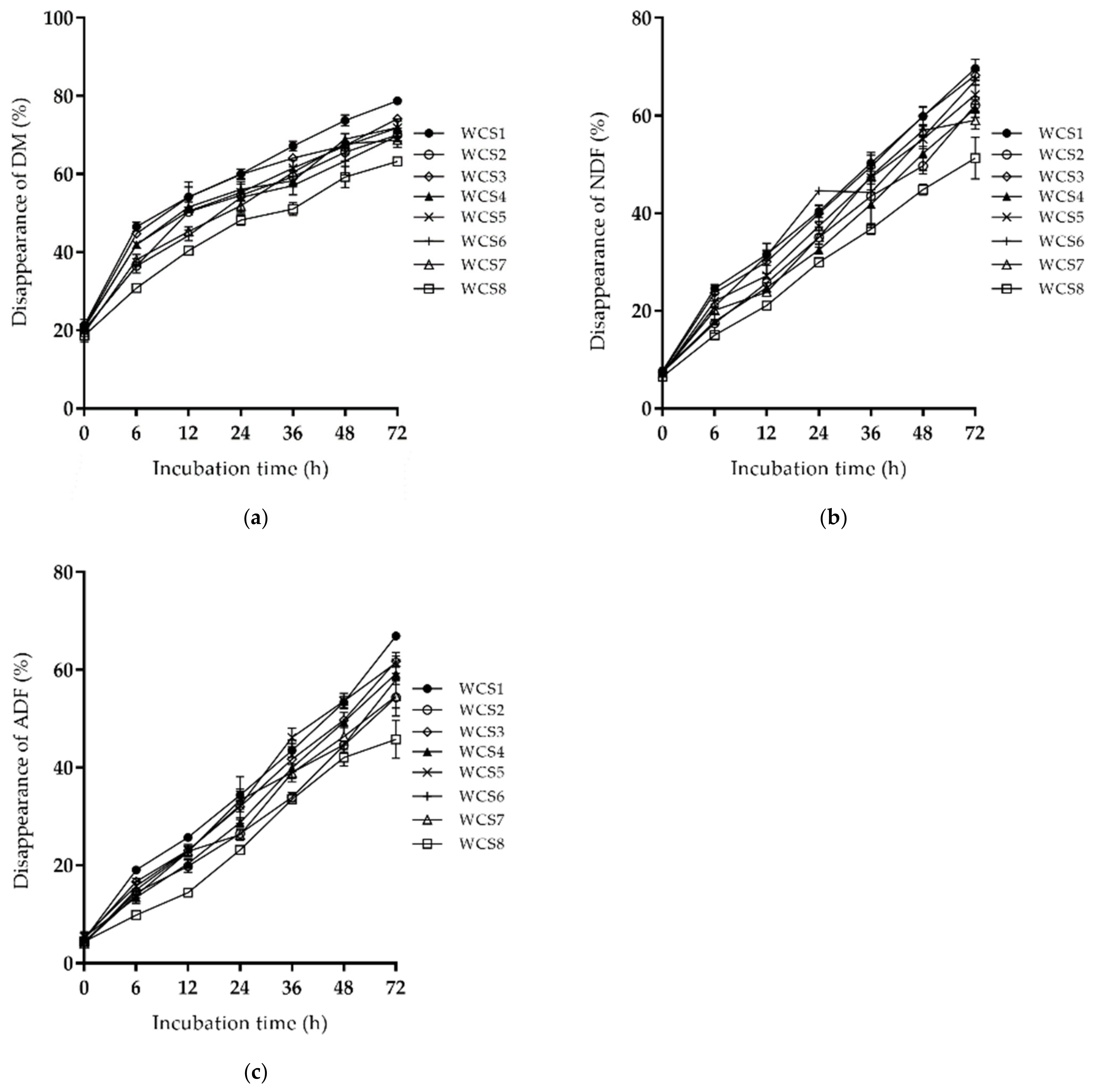
3.1. Chemical Composition and Rumen Degradation of Corn Silages
3.2. Relationships of Cell Wall Composition with Rumen Degradation
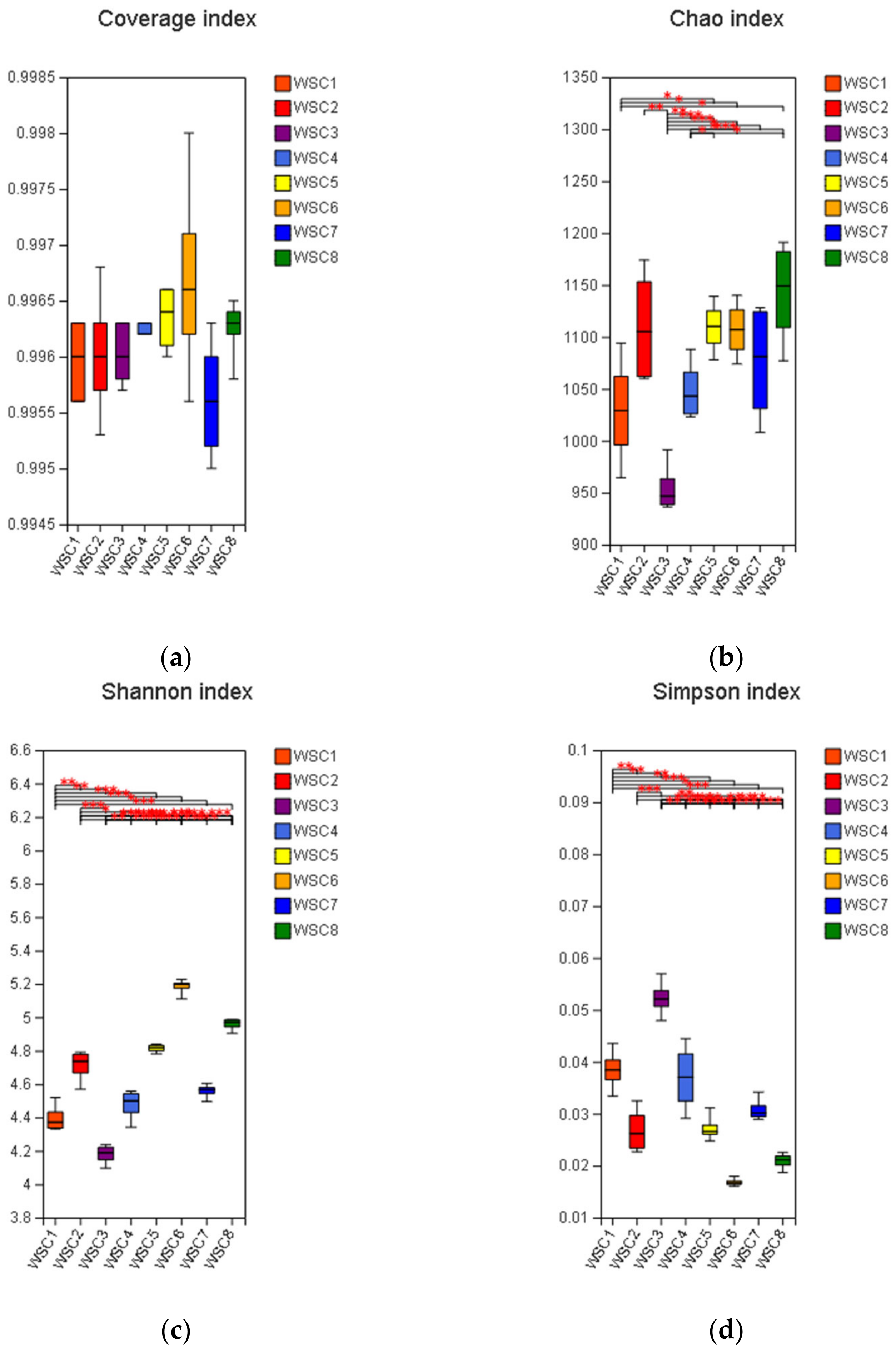
3.3. 16S rRNA Gene Sequencing
3.4. Diversity of the Bacterial Microbiota Attached to Different Corn Silages after 24 h of Rumen Fermentation
3.5. Community Composition of Corn Silage-Attached Microbes
3.6. The Correlationships between Attached Microbiota and Corn Silage Cell Wall Contents
4. Discussion
4.1. Chemical Composition of Corn Silages
4.2. The Relationships between the Contents of Phenolic Acids and Digestibility
4.3. In Situ Ruminal Release of Phenolic Acids
4.4. Rumen Microbial Colonization of Corn Silages with Different Phenolic Acid Contents
4.5. Relationships between Plant Cell Wall Contents and Forage-Attached Microbial Communities
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Item 1 | WCS1 | WCS2 | WCS3 | WCS4 | WCS5 | WCS6 | WCS7 | WCS8 | SEM |
|---|---|---|---|---|---|---|---|---|---|
| DM | |||||||||
| a | 24.66 a | 21.17 c | 22.24 bc | 23.35 ab | 22.81 abc | 21.70 bc | 20.87 c | 18.53 d | 0.539 |
| b | 50.88 a | 46.31 b | 47.16 b | 46.40 b | 45.97 cb | 47.24 b | 45.94 bc | 44.00 c | 0.598 |
| c(h−1) | 0.065 bc | 0.065 cb | 0.088 a | 0.080 ab | 0.065 bc | 0.057 cd | 0.047 d | 0.044 d | 0.0049 |
| a + b | 61.27 a | 54.64 d | 59.37 b | 56.02 c | 56.28 c | 5.35 d | 53.95 d | 47.92 e | 0.423 |
| ED | 75.55 a | 67.49 bc | 69.64 b | 68.38 b | 68.79 b | 68.94 b | 69.80 b | 63.79 c | 1.209 |
| NDF | |||||||||
| a | 11.08 a | 8.24 cde | 10.21 ab | 9.10 bcd | 9.56 bc | 9.91 ab | 7.95 ed | 7.65 e | 0.393 |
| b | 68.85 a | 66.87 ab | 69.07 a | 68.128 ab | 65.04 abc | 63.22 bc | 61.42 c | 61.34 c | 1.473 |
| c(h−1) | 0.025 | 0.021 | 0.025 | 0.02 | 0.025 | 0.026 | 0.027 | 0.019 | 0.002 |
| a + b | 45.50 a | 39.32 e | 44.69 ab | 39.11 e | 41.88 cd | 43.15 bc | 40.01 de | 33.26 f | 0.6274 |
| ED | 45.50 a | 39.32 e | 44.69 ab | 39.11 e | 41.88 cd | 43.15 bc | 40.01 de | 33.26 f | 0.627 |
| ADF | |||||||||
| a | 7.22 a | 6.19 b | 6.88 ab | 6.21 b | 5.25 c | 5.48 c | 6.55 ab | 3.53 d | 0.212 |
| b | 85.11 a | 69.53 bc | 87.05 a | 74.66 b | 68.82 bc | 62.15 c | 67.18 bc | 65.37 c | 2.047 |
| c(h−1) | 0.016 de | 0.016 cd | 0.013 e | 0.017 cd | 0.023 b | 0.030 a | 0.019 c | 0.016 cd | 0.0008 |
| a + b | 93.03 a | 75.32 bc | 95.60 a | 80.49 b | 71.82 cd | 63.56 e | 73.15 cd | 68.90 de | 1.467 |
| ED | 40.91 a | 33.18 e | 38.31 bc | 36.15 cd | 39.02 ab | 36.29 cd | 34.16 de | 28.66 f | 0.749 |

| Item 1 | WCS1 | WCS2 | WCS3 | WCS4 | WCS5 | WCS6 | WCS7 | WCS8 | SEM |
|---|---|---|---|---|---|---|---|---|---|
| FAest | |||||||||
| a | 22.15 | 17.46 | 18.59 | 21.2 | 20.23 | 18.04 | 18.17 | 18.12 | 1.242 |
| b | 59.79 a | 47.20 cd | 47.04 cd | 54.02 b | 46.55 cd | 51.099 bc | 42.55 d | 37.52 e | 1.226 |
| c(h-1) | 0.063 ab | 0.064 ab | 0.075 a | 0.053 ab | 0.061 ab | 0.062 ab | 0.039 b | 0.041 b | 0.0078 |
| a + b | 83.06 a | 64.50 ed | 70.90 bc | 69.74 bc | 66.79 cd | 72.86 b | 61.10 e | 61.63 e | 1.596 |
| ED | 66.92 a | 52.06 c | 56.97 b | 55.13 b | 52.97 c | 55.22 b | 45.90 d | 44.25 d | 0.673 |
| pCAest | |||||||||
| a | 13.18 ab | 10.85 d | 13.98 a | 12.76 ab | 12.47 abc | 11.75 bc | 11.99 bc | 11.18 cd | 0.435 |
| b | 75.50 a | 58.91 bc | 66.36 ab | 52.88 c | 58.37 bc | 67.40 ab | 54.12 c | 48.78 c | 3.305 |
| c(h-1) | 0.009 | 0.012 | 0.011 | 0.011 | 0.0124 | 0.01 | 0.012 | 0.011 | 0.0013 |
| a + b | 88.67 a | 69.95 c | 80.33 ab | 65.82 c | 71.03 bc | 78.97 ab | 65.95 c | 60.05 c | 3.33 |
| ED | 33.19 a | 30.05 b | 33.94 a | 29.71 b | 32.76 a | 32.33 a | 29.37 b | 25.83 c | 0.695 |
References
- Bai, C.; Wang, C.; Sun, L.; Xu, H.; Jiang, Y.; Na, N.; Yin, G.; Liu, S.; Xue, Y. Dynamics of Bacterial and Fungal Communities and Metabolites During Aerobic Exposure in Whole-Plant Corn Silages with Two Different Moisture Levels. Front. Microbiol. 2021, 12, 663895. [Google Scholar] [CrossRef] [PubMed]
- Badhan, A.; Jin, L.; Wang, Y.; Han, S.; Kowalczys, K.; Brown, D.; Ayala, C.; Latoszek-Green, M.; Miki, B.; Tsang, A.; et al. Expression of a fungal ferulic acid esterase in alfalfa modifies cell wall digestibility. Biotechnol. Biofuels 2014, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, R.D.; Rancour, D.M.; Marita, J.M. Grass Cell Walls: A Story of Cross-Linking. Front. Plant Sci. 2017, 7, 2056. [Google Scholar] [CrossRef]
- Casler, M.D.; Jung, H.-J.G. Selection and evaluation of smooth bromegrass clones with divergent lignin or etherified ferulic acid concentration. Crop Sci. 1999, 39, 1866–1873. [Google Scholar] [CrossRef]
- Jung, H.o.G.; Vogel, K.P. Lignification of switchgrass (Panicum virgatum) and big bluestem (Andropogon gerardii) plant parts during maturation and its effect on fibre degradability. J. Sci. Food Agric. 1992, 59, 169–176. [Google Scholar] [CrossRef]
- Grabber, J.H.; Mertens, D.R.; Kim, H.; Funk, C.; Lu, F.; Ralph, J. Cell wall fermentation kinetics are impacted more by lignin content and ferulate cross-linking than by lignin composition. J. Sci. Food Agric. 2009, 89, 122–129. [Google Scholar] [CrossRef]
- Jung, H.J.G. Forage Lignins and Their Effects on Fiber Digestibility. Agron. J. 1989, 81, 33–38. [Google Scholar] [CrossRef]
- Cao, B.-B.; Wang, R.; Bo, Y.-K.; Bai, S.; Yang, H.-J. In situ rumen digestibility of ester-linked ferulic and p-coumaric acids in crop stover or straws in comparison with alfalfa and Chinese wild ryegrass hays. Anim. Feed Sci. Technol. 2016, 212, 27–34. [Google Scholar] [CrossRef]
- Wong, D.W.S.; Chan, V.J.; Liao, H.; Zidwick, M.J. Cloning of a novel feruloyl esterase gene from rumen microbial metagenome and enzyme characterization in synergism with endoxylanases. J. Ind. Microbiol. Biotechnol. 2013, 40, 287–295. [Google Scholar] [CrossRef]
- Ralph, J.; Quideau, S.; Grabber, J.H.; Hatfield, R.D. Identification and synthesis of new ferulic acid dehydrodimers present in grass cell walls. J. Chem. Soc. Perkin Trans. 1994, 1, 3485–3498. [Google Scholar] [CrossRef]
- Lu, F.; Ralph, J. Detection and Determination of p -Coumaroylated Units in Lignins. J. Agric. Food Chem. 1999, 47, 1988–1992. [Google Scholar] [CrossRef]
- Grabber, J.H.; Hatfield, R.D.; Ralph, J.; Zon, J.; Amrhein, N. Ferulate Cross-Linking in Cell-Walls Isolated from Maize Cell-Suspensions. Phytochemistry 1995, 40, 1077–1082. [Google Scholar] [CrossRef]
- Besle, J.M.; Cornu, A.s.; Jouany, J.I. Roles of structural phenylpropanoids in forage cell wall digestion. J. Sci. Food Agric. 1994, 64, 171–190. [Google Scholar] [CrossRef]
- Cao, B.B.; Wang, R.; Yang, H.J.; Jiang, L.S. In situ ruminal degradation of phenolic acid, cellulose and hemicellulose in crop brans and husks differing in ferulic and p-coumaric acid patterns. J. Agric. Sci. 2015, 153, 1312–1320. [Google Scholar] [CrossRef]
- Bergman, E.N. Energy contributions of volatile fatty acids from the gastrointestinal tract in various species. Physiol. Rev. 1990, 70, 567–590. [Google Scholar] [CrossRef]
- Wang, M.; Wang, H.; Zheng, H.; Dewhurst, R.; Roehe, R. A knowledge-driven network-based analytical framework for the identification of rumen metabolites. IEEE Trans. NanoBiosci. 2020, 19, 518–526. [Google Scholar] [CrossRef]
- Wang, W.K.; Wang, Y.L.; Li, W.J.; Wu, Q.C.; Yang, K.L.; Li, S.L.; Yang, H.J. In situ rumen degradation characteristics and bacterial colonization of whole cottonseed, cottonseed hull and cottonseed meal with different gossypol content. AMB Express 2021, 11, 91. [Google Scholar] [CrossRef]
- Craig, W.M.; Broderick, G.A.; Ricker, D.B. Quantitation of microorganisms associated with the particulate phase of ruminal ingesta. J. Nutr. 1987, 117, 56–62. [Google Scholar] [CrossRef]
- Gharechahi, J.; Vahidi, M.F.; Ding, X.-Z.; Han, J.-L.; Salekdeh, G.H. Temporal changes in microbial communities attached to forages with different lignocellulosic compositions in cattle rumen. FEMS Microbiol. Ecol. 2020, 96, fiaa069. [Google Scholar] [CrossRef]
- Gharechahi, J.; Vahidi, M.; Bahram, M.; Han, J.; Ding, X.; Salekdeh, G. Metagenomic analysis reveals a dynamic microbiome with diversified adaptive functions to utilize high lignocellulosic forages in the cattle rumen. ISME J. 2021, 15, 1108–1120. [Google Scholar] [CrossRef]
- Várnai, A.; Costa, T.H.; Faulds, C.B.; Milagres, A.M.; Siika-Aho, M.; Ferraz, A. Effects of enzymatic removal of plant cell wall acylation (acetylation, p-coumaroylation, and feruloylation) on accessibility of cellulose and xylan in natural (non-pretreated) sugar cane fractions. Biotechnol. Biofuels 2014, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Association of Official Agricultural Chemists. Official Methods of Analysis; Association of Official Agricultural Chemists: Washington, DC, USA, 1990. [Google Scholar]
- Van Soest, P.V.; Robertson, J.B.; Lewis, B.A. Methods for Dietary Fiber, Neutral Detergent Fiber, and Nonstarch Polysaccharides in Relation to Animal Nutrition. J. Dairy Sci. 1991, 74, 3583–3597. [Google Scholar] [CrossRef]
- Cao, B.B.; Jin, X.; Yang, H.J.; Li, S.L.; Jiang, L.S. Microbial release of ferulic and p-coumaric acids from forages and their digestibility in lactating cows fed total mixed rations with different forage combinations. J. Sci. Food Agric. 2016, 96, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Larue, R.; Yu, Z.; Parisi, V.; Egan, A.; Morrison, M. Novel microbial diversity adherent to plant biomass in the herbivore gastrointestinal tract, as revealed by ribosomal intergenic spacer analysis and rrs gene sequencing. Environ. Microbiol. 2005, 7, 530–543. [Google Scholar] [CrossRef] [PubMed]
- Whitehouse, N.L.; Olson, V.M.; Schwab, C.G.; Chesbro, W.R.; Cunningham, K.D.; Lykos, T. Improved techniques for dissociating particle-associated mixed ruminal microorganisms from ruminal digesta solids. J. Anim. Sci. 1994, 72, 1335–1343. [Google Scholar] [CrossRef][Green Version]
- Evans, C.C.; Lepard, K.J.; Kwak, J.W.; Stancukas, M.C.; Laskowski, S.; Dougherty, J.; Moulton, L.; Glawe, A.; Wang, Y.; Leone, V. Exercise Prevents Weight Gain and Alters the Gut Microbiota in a Mouse Model of High Fat Diet-Induced Obesity. PLoS ONE 2014, 9, e92193. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Ørskov, E.-R.; McDonald, I. The estimation of protein degradability in the rumen from incubation measurements weighted according to rate of passage. J. Agric. Sci. 1979, 92, 499–503. [Google Scholar] [CrossRef]
- Negi, S. Theoretical estimation of rumen outflow rate from time-course rumen fermentation observations. J. Appl. Anim. Res. 1995, 7, 35–47. [Google Scholar] [CrossRef]
- Wang, Y.; Shahidi, F.; Ho, C.T. Metabolism of dietary phenolic acids. Spec. Publ. R. Soc. Chem. 2013, 178–204. [Google Scholar] [CrossRef]
- Raffrenato, E.; Fievisohn, R.; Cotanch, K.W.; Grant, R.J.; Chase, L.E.; Van Amburgh, M.E. Effect of lignin linkages with other plant cell wall components on in vitro and in vivo neutral detergent fiber digestibility and rate of digestion of grass forages. J. Dairy Sci. 2017, 100, 8119–8131. [Google Scholar] [CrossRef]
- Hartley, R.D. p-Coumaric and ferulic acid components of cell walls of ryegrass and their relationships with lignin and digestibility. J. Sci. Food Agric. 2010, 23, 1347–1354. [Google Scholar] [CrossRef]
- Casler, M. Breeding forage crops for increased nutritional value. Adv. Agron. 2001, 7, 51–107. [Google Scholar] [CrossRef]
- Rodrigues, M.A.M.; Guedes, C.M.; Cone, J.W.; Van Gelder, A.H.; Ferreira, L.M.M.; Sequeira, C.A. Effects of phenolic acid structures on meadow hay digestibility. Anim. Feed Sci. Technol. 2007, 136, 297–311. [Google Scholar] [CrossRef]
- Mandebvu, P.; West, J.W.; Hill, G.M.; Gates, R.N.; Hatfield, R.D.; Mullinix, B.G.; Parks, A.H.; Caudle, A.B. Comparison of Tifton 85 and Coastal bermudagrasses for yield, nutrient traits, intake, and digestion by growing beef steers. J. Anim. Sci. 1999, 77, 1572. [Google Scholar] [CrossRef]
- Jung, H.; Shalita-Jones, S.C. Variation in the extractability of esterified p-coumaric and ferulic acids from forage cell walls. J. Agric. Food Chem. 1990, 38, 397–402. [Google Scholar] [CrossRef]
- Giada, M. Food phenolic compounds: Main classes, sources and their antioxidant power. Oxid. Stress Chronic Degener. Dis.: Role Antioxid. 2013, 2013, 87–112. [Google Scholar] [CrossRef]
- Argillier, O.; Barrière, Y.; Lila, M.; Jeanneteau, F.; Gélinet, K.; Menanteau, V. Genotypic variation in phenolic components of cell-walls in relation to the digestibility of maize stalks. Agronomie 1996, 16, 123–130. [Google Scholar] [CrossRef]
- Lam, T.; Iiyama, K.; Stone, B.A. Cinnamic acid bridges between cell wall polymers in wheat and Phalaris internodes. Phytochemistry 1992, 31, 1179–1183. [Google Scholar] [CrossRef]
- Hu, Q.P.; Xu, J.G. Profiles of carotenoids, anthocyanins, phenolics, and antioxidant activity of selected color waxy corn grains during maturation. J. Agric. Food Chem. 2011, 59, 2026–2033. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.G.; Casler, M.D. Maize Stem Tissues: Impact of Development on Cell Wall Degradability. Crop Sci. 2006, 46, 1801–1809. [Google Scholar] [CrossRef]
- Iiyama, K.; Lam, T.; Stone, B.A. Covalent Cross-Links in the Cell Wall. Plant Physiol. 1994, 104, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.G.; Mertens, D.R.; Phillips, R.L. Effect of reduced ferulate-mediated lignin/arabinoxylan cross-linking in corn silage on feed intake, digestibility, and milk production. J. Dairy Sci. 2011, 94, 5124–5137. [Google Scholar] [CrossRef] [PubMed]
- Casler, M.D.; Jung, H.-J.G. Relationships of fibre, lignin, and phenolics to in vitro fibre digestibility in three perennial grasses. Anim. Feed Sci. Technol. 2006, 125, 151–161. [Google Scholar] [CrossRef]
- Wang, Y.L.; Wang, W.K.; Wu, Q.C.; Yang, H.J. The release and catabolism of ferulic acid in plant cell wall by rumen microbes: A review. Anim. Nutr. 2022, 9, 335–344. [Google Scholar] [CrossRef]
- Jung, H.-J.G.; Fahey, G.C.; Merchen, N.R. Effects of ruminant digestion and metabolism on phenolic monomers of forages. Br. J. Nutr. 1983, 50, 637–651. [Google Scholar] [CrossRef]
- O’Neill, F.; Christov, L.; Botes, P.; Prior, B. Rapid and simple assay for feruloyl and p-coumaroyl esterases. World J. Microbiol. Biotechnol. 1996, 12, 239–242. [Google Scholar] [CrossRef]
- Vahidi, M.F.; Gharechahi, J.; Behmanesh, M.; Ding, X.Z.; Salekdeh, G.H. Diversity of microbes colonizing forages of varying lignocellulose properties in the sheep rumen. PeerJ 2021, 8, e10463. [Google Scholar] [CrossRef]
- Akin, D.E.; Borneman, W.S.; Rigsby, L.L.; Martin, S.A. p-Coumaroyl and feruloyl arabinoxylans from plant cell walls as substrates for ruminal bacteria. Appl. Environ. Microbiol. 1993, 59, 644–647. [Google Scholar] [CrossRef]
- Kim, B.; Shin, J.; Guevarra, R.; Lee, J.; Kim, D.; Seol, K.; Lee, J.; Kim, H.; Isaacson, R. Deciphering Diversity Indices for a Better Understanding of Microbial Communities. J. Microbiol. Biotechnol. 2017, 27, 2089–2093. [Google Scholar] [CrossRef]
- Liu, J.; Zhang, M.; Xue, C.; Zhu, W.; Mao, S. Characterization and comparison of the temporal dynamics of ruminal bacterial microbiota colonizing rice straw and alfalfa hay within ruminants. J Dairy Sci 2016, 99, 9668–9681. [Google Scholar] [CrossRef]
- Thoetkiattikul, H.; Mhuantong, W.; Laothanachareon, T.; Tangphatsornruang, S.; Pattarajinda, V.; Eurwilaichitr, L.; Champreda, V. Comparative analysis of microbial profiles in cow rumen fed with different dietary fiber by tagged 16S rRNA gene pyrosequencing. Curr. Microbiol. 2013, 67, 130–137. [Google Scholar] [CrossRef]
- Kim, M.; Morrison, M.; Yu, Z. Status of the phylogenetic diversity census of ruminal microbiomes. FEMS Microbiol. Ecol. 2011, 76, 49–63. [Google Scholar] [CrossRef]
- Huo, W.; Zhu, W.; Mao, S. Impact of subacute ruminal acidosis on the diversity of liquid and solid-associated bacteria in the rumen of goats. World J. Microbiol. Biotechnol. 2014, 30, 669–680. [Google Scholar] [CrossRef]
- Van Gylswyk, N. Succiniclasticum ruminis gen. nov., sp. nov., a ruminal bacterium converting succinate to propionate as the sole energy-yielding mechanism. Int. J. Syst. Bacteriol. 1995, 45, 297–300. [Google Scholar] [CrossRef]
- Kim, H.; Park, T.; Kwon, I.; Seo, J. Specific inhibition of Streptococcus bovis by endolysin LyJH307 supplementation shifts the rumen microbiota and metabolic pathways related to carbohydrate metabolism. J. Anim. Sci. Biotechnol. 2021, 12, 93. [Google Scholar] [CrossRef]
- Liu, J.; Bian, G.; Zhu, W.; Mao, S. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front. Microbiol. 2015, 6, 167. [Google Scholar] [CrossRef]
- Xiong, Y.; Guo, C.; Wang, L.; Chen, F.; Dong, X.; Li, X.; Ni, K.; Yang, F. Effects of paper mulberry silage on the growth performance, rumen microbiota and muscle fatty acid composition in hu lambs. Fermentation 2021, 7, 286. [Google Scholar] [CrossRef]
- Goodrich, J.K.; Waters, J.L.; Poole, A.C.; Sutter, J.L.; Koren, O.; Blekhman, R.; Beaumont, M.; Treuren, W.V.; Knight, R.; Bell, J.T.; et al. Human genetics shape the gut microbiome. Cell 2014, 159. [Google Scholar] [CrossRef]
- Kabel, M.A.; Yeoman, C.J.; Han, Y.; Dodd, D.; Abbas, C.A.; De Bont, J.A.; Mackie, R.I. Biochemical characterization and relative expression levels of multiple carbohydrate esterases of the xylanolytic rumen bacterium Prevotella ruminicola 23 grown on an ester-enriched substrate. Appl. Environ. Microbiol. 2011, 77, 5678–5681. [Google Scholar] [CrossRef] [PubMed]
- Jami, E.; Mizrahi, I. Composition and Similarity of Bovine Rumen Microbiota across Individual Animals. PLoS ONE 2012, 7, e33306. [Google Scholar] [CrossRef] [PubMed]
- Petri, R.M.; Schwaiger, T.; Penner, G.B.; Beauchemin, K.A.; Forster, R.J.; McKinnon, J.J.; McAllister, T.A. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl. Environ. Microbiol. 2013, 79, 3744–3755. [Google Scholar] [CrossRef] [PubMed]
- Firrman, J.; Liu, L.; Zhang, L.; Arango Argoty, G.; Wang, M.; Tomasula, P.; Kobori, M.; Pontious, S.; Xiao, W. he effect of quercetin on genetic expression of the commensal gut microbes Bifidobacterium catenulatum, Enterococcus caccae and Ruminococcus gauvreauii. Anaerobe 2016. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; He, Y.; Li, H.; Wu, F.; Qiu, Q.; Niu, W.; Gao, Z.; Su, H.; Cao, B. Rumen fermentation, intramuscular fat fatty acid profiles and related rumen bacterial populations of Holstein bulls fed diets with different energy levels. Appl. Microbiol. Biotechnol. 2019, 103, 4931–4942. [Google Scholar] [CrossRef]
- Li, X.; Jensen, R.; Højberg, O.; Canibe, N.; Jensen, B. Olsenella scatoligenes sp. nov., a 3-methylindole- (skatole) and 4-methylphenol- (p-cresol) producing bacterium isolated from pig faeces. Int. J. Syst. Evol. Microbiol. 2015, 65, 1227–1233. [Google Scholar] [CrossRef]
- Ddc, T.; Göker, M.; Held, B.; Lucas, S.; Nolan, M.; Yasawong, M.; Rio, G.D.; Tice, H.; Cheng, J.; Bruce, D. Complete genome sequence of Olsenella uli type strain (VPI D76D-27CT). Stand. Genom. Sci. 2010, 3, 76–84. [Google Scholar] [CrossRef]
- Huws, S.A.; Edwards, J.E.; Creevey, C.J.; Rees Stevens, P.; Lin, W.; Girdwood, S.E.; Kingston-Smith, A.H. Temporal dynamics of the metabolically active rumen 1 bacteria colonising fresh perennial ryegrass. FEMS Microbiol. Ecol. 2016, 92, fiv137. [Google Scholar] [CrossRef]
- Kraatz, M.; Wallace, R.J.; Svensson, L. Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profusa. Int. J. Syst. Evol. Microbiol. 2011, 61, 795–803. [Google Scholar] [CrossRef]
- Domingo, M.; Huletsky, A.; Boissinot, M.; Bernard, K.; Picard, F.; Bergeron, M. Ruminococcus gauvreauii sp. nov., a glycopeptide-resistant species isolated from a human faecal specimen. Int. J. Syst. Evol. Microbiol. 2008, 58, 1393–1397. [Google Scholar] [CrossRef]
- Hua, C.; Tian, J.; Tian, P.; Cong, R.; Luo, Y.; Geng, Y.; Tao, S.; Ni, Y.; Zhao, R. Feeding a High Concentration Diet Induces Unhealthy Alterations in the Composition and Metabolism of Ruminal Microbiota and Host Response in a Goat Model. Front. Microbiol. 2017, 8, 138. [Google Scholar] [CrossRef]
- Jung, D.; Seo, D.; Kim, G.; Nam, Y.; Song, E.; Yoon, S.; Park, C. The effect of resistant starch (RS) on the bovine rumen microflora and isolation of RS-degrading bacteria. Appl. Microbiol. Biotechnol. 2018, 102, 4927–4936. [Google Scholar] [CrossRef]
- Pérez-Burillo, S.; Rajakaruna, S.; Pastoriza, S.; Paliy, O.; Rufián-Henares, J.Á. Bioactivity of food melanoidins is mediated by gut microbiota. Food Chem. 2020, 316, 126309. [Google Scholar] [CrossRef]


| Item 1 | WCS1 | WCS2 | WCS3 | WCS4 | WCS5 | WCS6 | WCS7 | WCS8 |
|---|---|---|---|---|---|---|---|---|
| DM (g/kg FM) | 97.0 | 95.6 | 95.5 | 91.9 | 94.8 | 94.3 | 94.5 | 98.5 |
| NDF (g/kg DM) | 377.4 | 378.6 | 407.5 | 452.8 | 477.3 | 492.9 | 511.3 | 531.4 |
| ADF (g/kg DM) | 197.9 | 270.3 | 225.7 | 283.0 | 271.4 | 283.5 | 312.1 | 301.1 |
| ADL (g/kg DM) | 14.2 | 16.6 | 22.5 | 29.4 | 29.2 | 31.3 | 37.5 | 45.5 |
| Phenolic acids (g/kg DM) | ||||||||
| pCAest | 6.08 | 8.21 | 7.91 | 7.39 | 6.41 | 7.1 | 7.36 | 8.23 |
| pCAeth | 0.69 | 1.80 | 0.92 | 1.42 | 1.44 | 1.22 | 2.17 | 2.06 |
| FAest | 5.45 | 3.63 | 5.1 | 4.01 | 3.33 | 3.61 | 3.57 | 2.9 |
| FAeth | 0.08 | 1.47 | 0.51 | 1.17 | 1.47 | 1.94 | 1.29 | 2.16 |
| pCA/FA ratio | 1.22 | 1.96 | 1.57 | 1.70 | 1.64 | 1.50 | 1.83 | 2.03 |
| Constant 1 | Phenolic Acid and Lignin Concentrations 2 | |||||||
|---|---|---|---|---|---|---|---|---|
| pCAest | pCAeth | FAest | FAeth | pCA/FA | NDF | ADF | ADL | |
| DM | ||||||||
| a | −0.75 * | −0.78 * | 0.73 * | −0.75 * | −0.83 ** | −0.62 | −0.70 | −0.73 * |
| b | −0.66 | −0.86 ** | 0.85 ** | −0.79 * | −0.89 ** | −0.68 | −0.83 * | −0.77 * |
| c (h−1) | −0.02 | −0.46 | 0.62 | −0.55 | −0.38 | −0.64 | −0.58 | −060 |
| a + b | −0.74 * | −0.86 ** | 0.83 * | −0.80 * | −0.90 ** | −0.68 | −0.80 * | −0.78 * |
| ED | −0.57 | −0.86 ** | 0.89 ** | −0.91 ** | −0.78 * | −0.76 * | −0.84 * | −0.78 * |
| NDF | ||||||||
| a | −0.68 | −0.87 ** | 0.80 * | −0.66 | −0.96 ** | −0.54 | −0.84 ** | −0.64 |
| b | −0.20 | −0.55 | 0.80 * | −0.74 * | −0.53 | −0.88 ** | −0.81 * | −0.84 ** |
| c | −0.52 | −0.66 | 0.29 | −0.36 | −0.51 | 0.03 | −0.15 | −0.13 |
| a + b | −0.2 | −0.43 | 0.61 | −0.57 | −0.45 | −0.61 | −0.53 | −0.58 |
| ED | −0.61 | −0.92 ** | 0.77 * | −0.71 * | −0.87 ** | −0.57 | −0.73 * | −0.70 * |
| ADF | ||||||||
| a | −0.34 | −0.78 ** | 0.80 ** | −0.85 ** | −0.58 | −0.71 * | −0.60 | −0.75 * |
| b | −0.20 | −0.69 | 0.922 ** | −0.91 ** | −0.55 | −0.72 * | −0.86 ** | −0.64 |
| c | −0.36 | 0.02 | −0.45 | 0.53 | −0.17 | 0.50 | 0.39 | 0.90 ** |
| a + b | −0.16 | −0.67 | 0.91 ** | −0.92 ** | −0.50 | −0.72 * | −0.83 * | −0.63 |
| ED | −0.78 * | −0.87 ** | 0.73 * | −0.73 * | −0.89 ** | −0.53 | −0.73 * | −0.67 |
| pCAest | ||||||||
| a | −0.40 | −0.64 | 0.67 | −0.83 * | −0.45 | −0.38 | −0.41 | −0.35 |
| b | −0.49 | −0.60 | 0.71 * | −0.72 * | −0.60 | −0.54 | −063 | −0.54 |
| c | 0.04 | −0.19 | 0.09 | 0.04 | −0.18 | −0.10 | −0.23 | −0.16 |
| a + b | −0.55 | −0.64 | 0.63 | −0.73 * | −0.56 | −0.31 | −0.45 | −0.34 |
| ED | −0.58 | −0.83 * | 0.69 | −0.63 | −0.81 | −0.56 | −0.72 * | −0.69 |
| FAest | ||||||||
| a | −0.44 | −0.75 * | 0.92 ** | −0.83 * | −0.77 * | −0.73 * | −0.91 ** | −0.71 * |
| b | −0.69 | −0.60 | 0.56 | −0.63 | −0.64 | 0.16 | −0.45 | −0.19 |
| c | −0.05 | −0.05 | 0.14 | 0.15 | −0.33 | −0.30 | −0.40 | −0.33 |
| a + b | −0.67 | −0.77 * | 0.78 * | −0.76 * | −0.82 * | −0.40 | −0.72 * | −0.43 |
| ED | −0.50 | −0.57 | 0.73 * | −0.58 | −0.75 * | −0.59 | −0.86 ** | −0.59 |
| Item | Phenolic Acid and Lignin Concentrations 1 | |||||||
|---|---|---|---|---|---|---|---|---|
| NDF | ADF | ADL | pCAest | pCAeth | FAest | FAeth | pCA/FA | |
| Chao1 index | 0.55 | 0.76 | 0.49 | 0.12 | 0.66 | −0.88 ** | 0.85 ** | 0.55 |
| Simpson | −0.57 | −0.72 | −0.48 | −0.05 | −0.53 | 0.80 * | −0.85 ** | −0.38 |
| Shannon | 0.59 | 0.72 | 0.48 | 0.04 | 0.39 | −0.76 * | 0.86 ** | 0.29 |
| Ace | 0.54 | 0.74 | 0.51 | 0.19 | 0.7 | −0.87 * | 0.84 ** | 0.6 |
| Item | Phenolic Acid and Lignin Concentrations 1 | |||||||
|---|---|---|---|---|---|---|---|---|
| NDF | ADF | pCAest | pCAeth | FAest | FAeth | pCA/FA | ADL | |
| Phylum level | ||||||||
| Firmicutes | −0.37 * | −0.34 | −0.15 | −0.25 | 0.29 | −0.24 | −0.17 | −0.36 * |
| Actinobacteriota | −0.28 | −0.26 | −0.01 | −0.23 | 0.52 * | −0.73 ** | −0.14 | −0.22 |
| Bacteroidota | 0.55 * | 0.51 * | 0.3 | 0.51 | −0.75 ** | 0.88 ** | 0.48 * | 0.50 * |
| Family level | ||||||||
| Lachnospiraceae | −0.18 | 0.03 | −0.12 | 0.06 | 0.01 | −0.11 | 0.01 | −0.22 |
| Erysipelatoclostridiaceae | −0.55 * | −0.47 * | 0.04 | −0.27 | 0.62 ** | −0.75 ** | −0.13 | −0.43 * |
| Acidaminococcaceae | −0.04 | −0.11 | −0.27 | 0.1 | −0.19 | 0.04 | 0.09 | −0.05 |
| Prevotellaceae | 0.3 | 0.35 | 0.3 | 0.46 * | −0.66 ** | 0.63 ** | 0.57 ** | 0.34 |
| Bifidobacteriaceae | −0.13 | −0.1 | 0.1 | 0 | 0.36 | −0.50 * | 0.04 | 0.01 |
| Atopobiaceae | −0.52 * | −0.46 * | −0.02 | −0.27 | 0.58 ** | −0.66 ** | −0.2 | −0.43 * |
| Eubacterium_coprostanoligenes_group | −0.26 | −0.04 | 0.35 | −0.14 | 0.17 | −0.01 | −0.02 | −0.24 |
| Christensenellaceae | 0.18 | 0.08 | −0.1 | −0.27 | −0.05 | 0.3 | −0.32 | 0.05 |
| Anaerovoracaceae | −0.31 | −0.15 | 0.06 | −0.28 | 0.15 | −0.08 | −0.12 | −0.33 |
| Oscillospiraceae | 0.25 | 0.19 | −0.02 | −0.12 | −0.2 | 0.45 * | −0.19 | 0.13 |
| Muribaculaceae | 0.47 * | 0.39 * | 0.01 | 0.16 | −0.50 * | 0.65 ** | 0.08 | 0.37 * |
| Ruminococcaceae | 0.3 | 0.15 | −0.2 | −0.16 | −0.26 | 0.41 * | −0.18 | 0.2 |
| Genus level | ||||||||
| Erysipelotrichaceae_UCG−002 | −0.57 * | −0.48 * | 0.03 | −0.27 | 0.62 ** | −0.74 ** | −0.13 | −0.45 * |
| Lachnospiraceae_NK3A20_group | −0.14 | −0.02 | 0.01 | −0.15 | 0.03 | 0.11 | −0.15 | −0.21 |
| Succiniclasticum | 0.06 | −0.07 | −0.19 | 0.13 | −0.19 | 0.07 | 0.11 | 0.07 |
| Prevotella | 0.35 | 0.40 * | 0.38 * | 0.50 * | −0.65 ** | 0.63 ** | 0.60 ** | 0.42 * |
| Olsenella | −0.46 * | −0.49 * | −0.07 | −0.28 | 0.60 ** | −0.68 ** | −0.23 | −0.37 * |
| Ruminococcus_gauvreauii_group | −0.35 | −0.22 | 0.03 | −0.03 | 0.35 | −0.51 * | 0.02 | −0.27 |
| Acetitomaculum | −0.66 ** | −0.40 * | −0.1 | −0.2 | 0.52 * | −0.59 ** | −0.22 | −0.64 ** |
| norank_f_Eubacterium_coprostanoligenes_group | −0.23 | −0.01 | 0.33 | −0.09 | 0.11 | 0.03 | 0.02 | −0.22 |
| Christensenellaceae_R−7_group | 0.16 | 0.07 | −0.14 | −0.29 | −0.03 | 0.26 | −0.34 | 0.03 |
| Bifidobacterium | −0.06 | −0.21 | −0.04 | −0.19 | 0.42 * | −0.50 * | −0.15 | 0.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-L.; Wang, W.-K.; Wu, Q.-C.; Zhang, F.; Li, W.-J.; Li, S.-L.; Wang, W.; Cao, Z.-J.; Yang, H.-J. In Situ Rumen Degradation Characteristics and Bacterial Colonization of Corn Silages Differing in Ferulic and p-Coumaric Acid Contents. Microorganisms 2022, 10, 2269. https://doi.org/10.3390/microorganisms10112269
Wang Y-L, Wang W-K, Wu Q-C, Zhang F, Li W-J, Li S-L, Wang W, Cao Z-J, Yang H-J. In Situ Rumen Degradation Characteristics and Bacterial Colonization of Corn Silages Differing in Ferulic and p-Coumaric Acid Contents. Microorganisms. 2022; 10(11):2269. https://doi.org/10.3390/microorganisms10112269
Chicago/Turabian StyleWang, Yan-Lu, Wei-Kang Wang, Qi-Chao Wu, Fan Zhang, Wen-Juan Li, Sheng-Li Li, Wei Wang, Zhi-Jun Cao, and Hong-Jian Yang. 2022. "In Situ Rumen Degradation Characteristics and Bacterial Colonization of Corn Silages Differing in Ferulic and p-Coumaric Acid Contents" Microorganisms 10, no. 11: 2269. https://doi.org/10.3390/microorganisms10112269
APA StyleWang, Y.-L., Wang, W.-K., Wu, Q.-C., Zhang, F., Li, W.-J., Li, S.-L., Wang, W., Cao, Z.-J., & Yang, H.-J. (2022). In Situ Rumen Degradation Characteristics and Bacterial Colonization of Corn Silages Differing in Ferulic and p-Coumaric Acid Contents. Microorganisms, 10(11), 2269. https://doi.org/10.3390/microorganisms10112269







