Microbial Community in the Permafrost Thaw Gradient in the South of the Vitim Plateau (Buryatia, Russia)
Abstract
1. Introduction
2. Materials and Methods
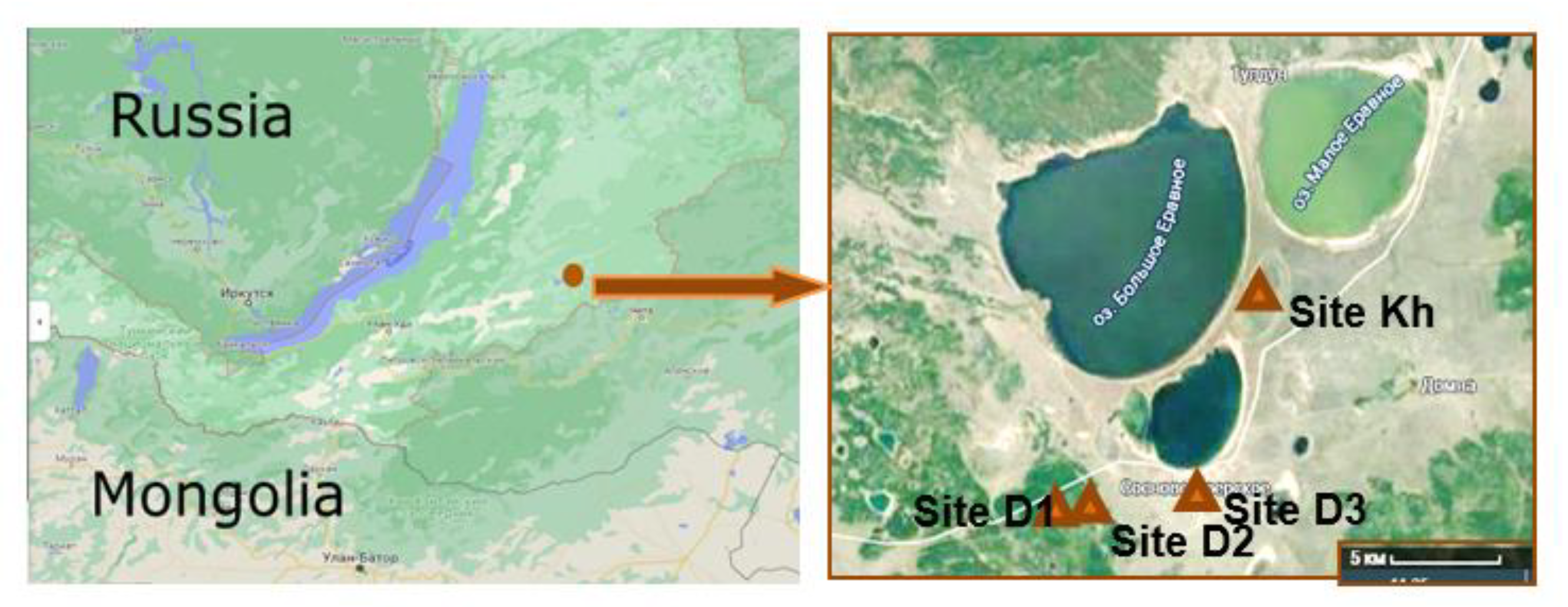
2.1. Soil Sampling and Analysis
2.2. DNA Extraction, Amplification and Sequencing
2.3. Statistical Analysis
3. Results
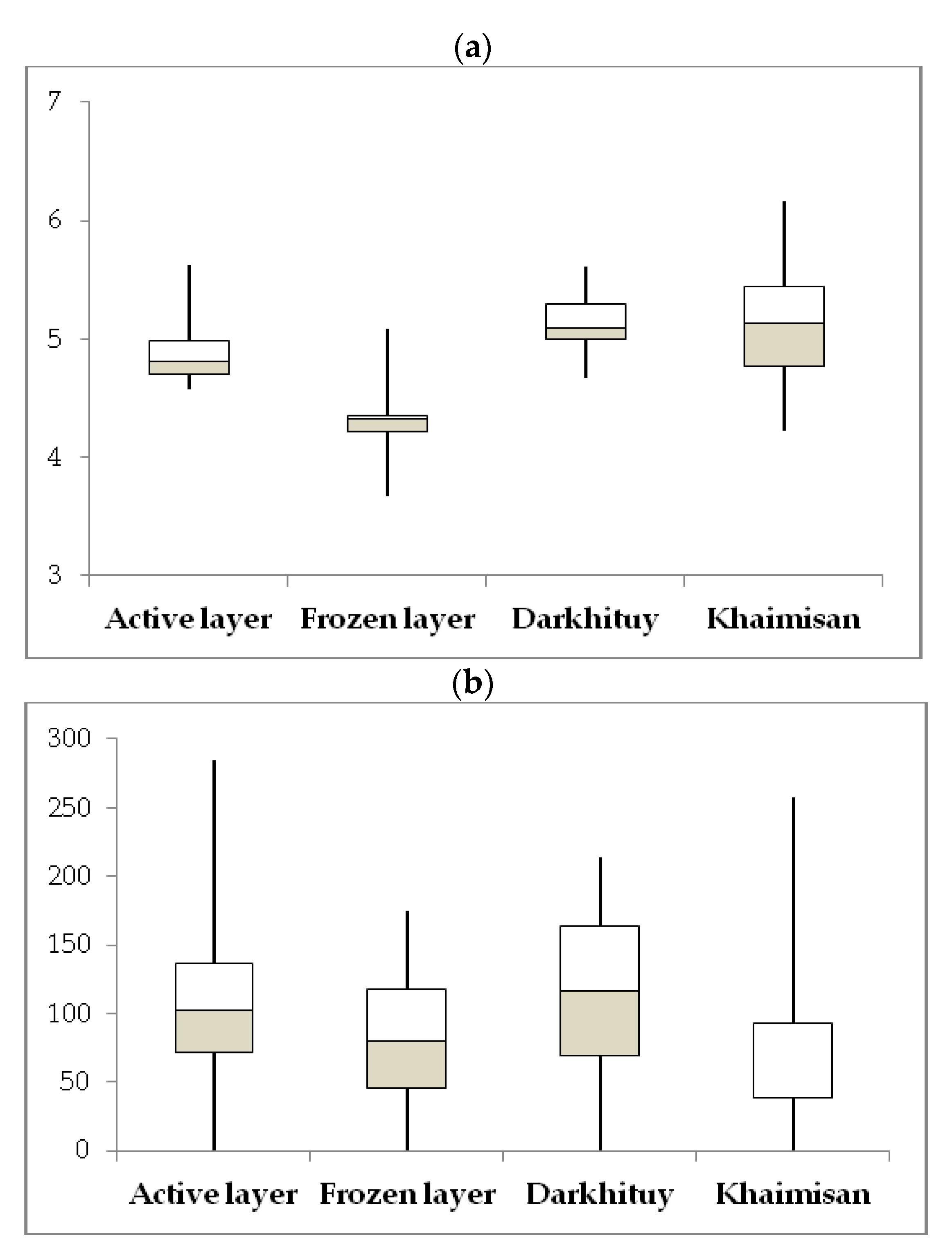
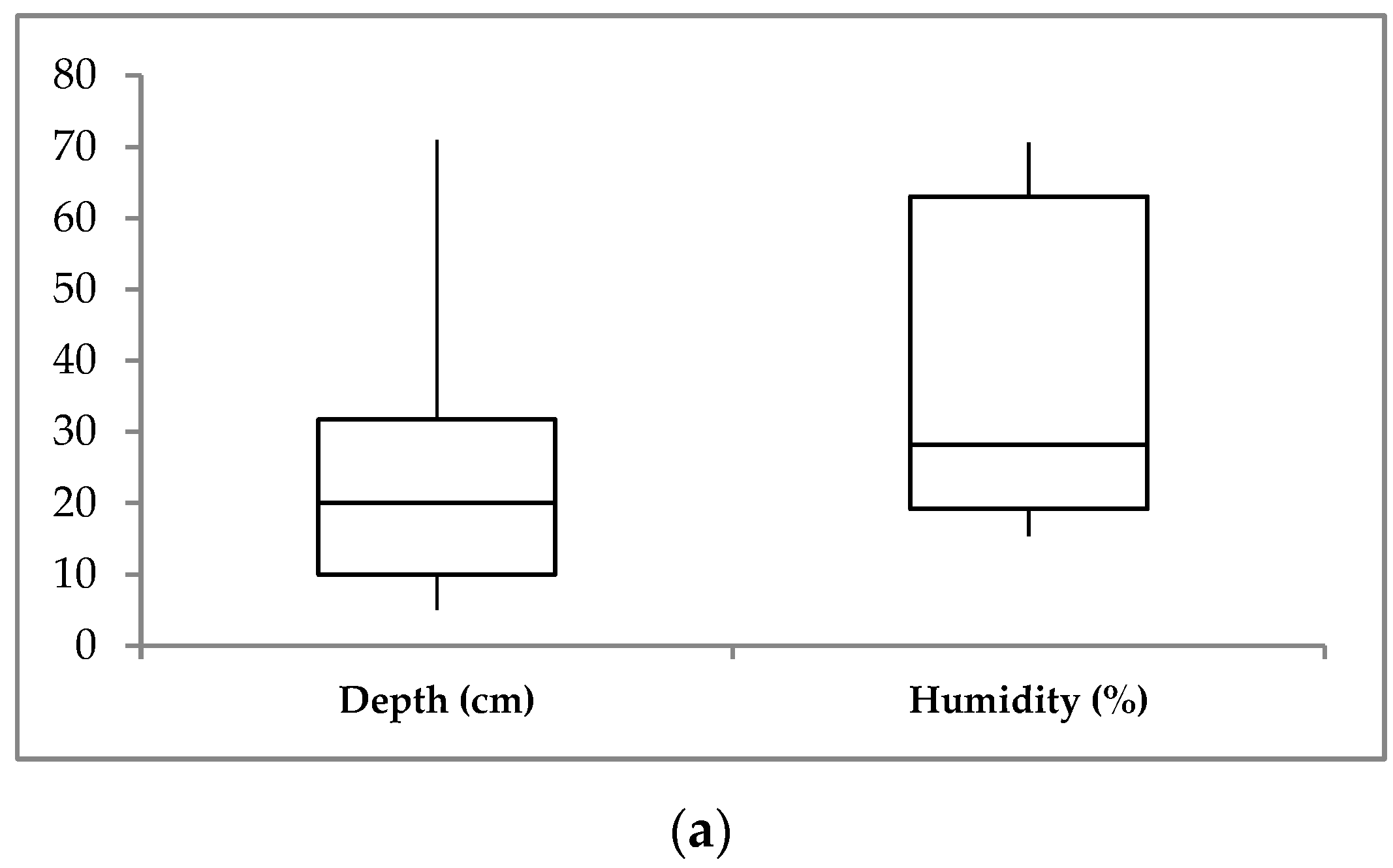
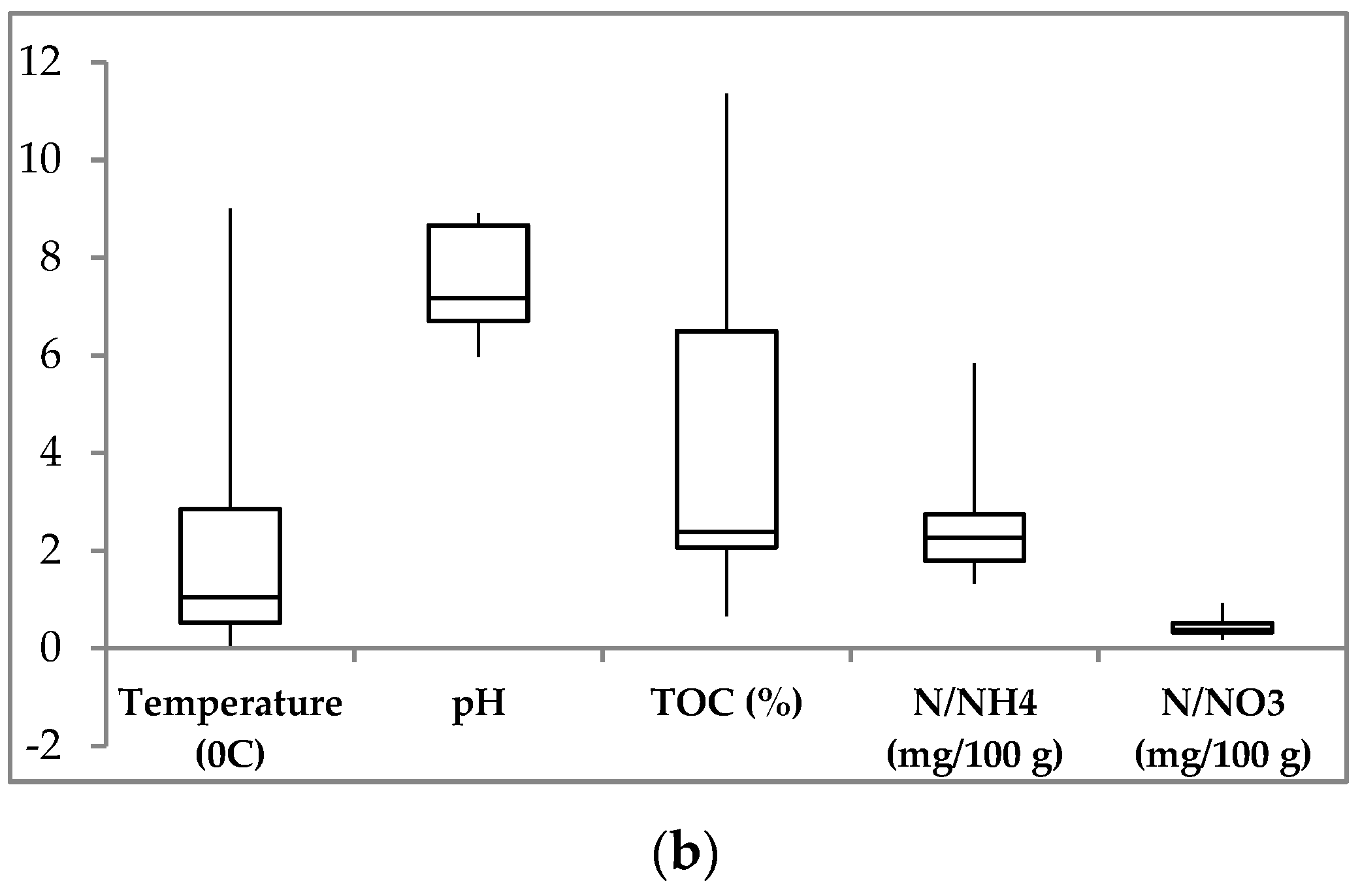
3.1. Soil Environmental Parameters
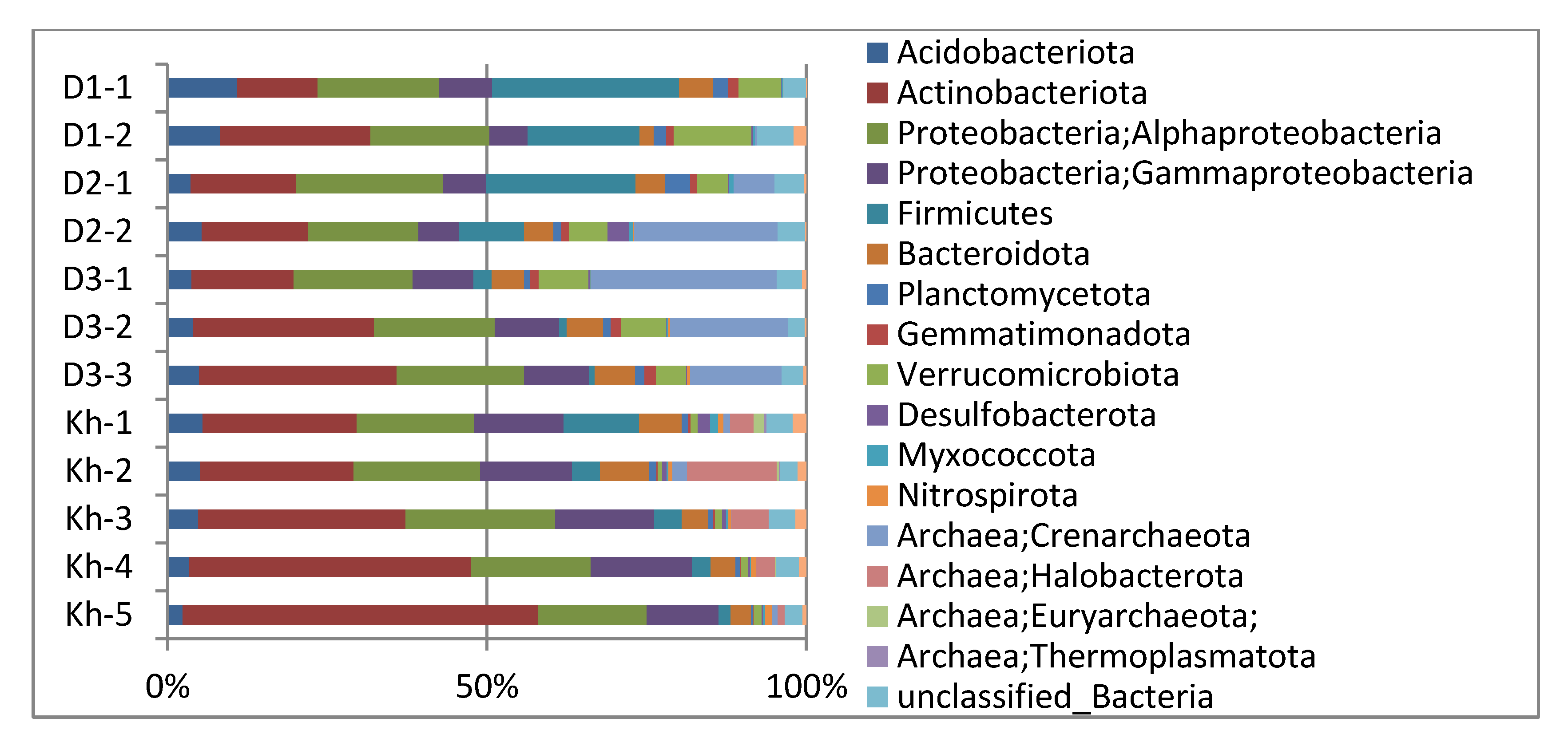
3.2. Soil Microbial Community Structure and Diversity
| Sample | Number of Classified Reads/Share, % | ASVs | Shannon Diversity Indices | Simpson Diversity Indices | Simpson Invers Diversity Indices |
|---|---|---|---|---|---|
| Site D1-1 | 15,023/48.94 | 248 | 4.62 | 0.975 | 40.599 |
| Site D1-2 | 16,288/53.52 | 214 | 4.35 | 0.974 | 38.11 |
| Site D2-1 | 23,363/71.29 | 431 | 5.28 | 0.988 | 86.33 |
| Site D2-2 | 18,774/58.45 | 391 | 5.08 | 0.982 | 56.457 |
| Site D3-1 | 19,000/60.37 | 329 | 4.73 | 0.972 | 35.17 |
| Site D3-2 | 15,939/53.1 | 333 | 4.77 | 0.973 | 37.445 |
| Site D3-3 | 15,829/52.08 | 323 | 4.85 | 0.980 | 49.459 |
| Site Kh-1 | 20,046/59.92 | 533 | 5.62 | 0.990 | 104.01 |
| Site Kh-2 | 19,085/59.4 | 370 | 4.89 | 0.976 | 42.515 |
| Site Kh-3 | 18,920/60.27 | 315 | 4.58 | 0.973 | 37.541 |
| Site Kh-4 | 19,574/54.71 | 316 | 4.22 | 0.943 | 17.667 |
| Site Kh-5 | 18,943/62.37 | 274 | 3.67 | 0.892 | 9.259 |
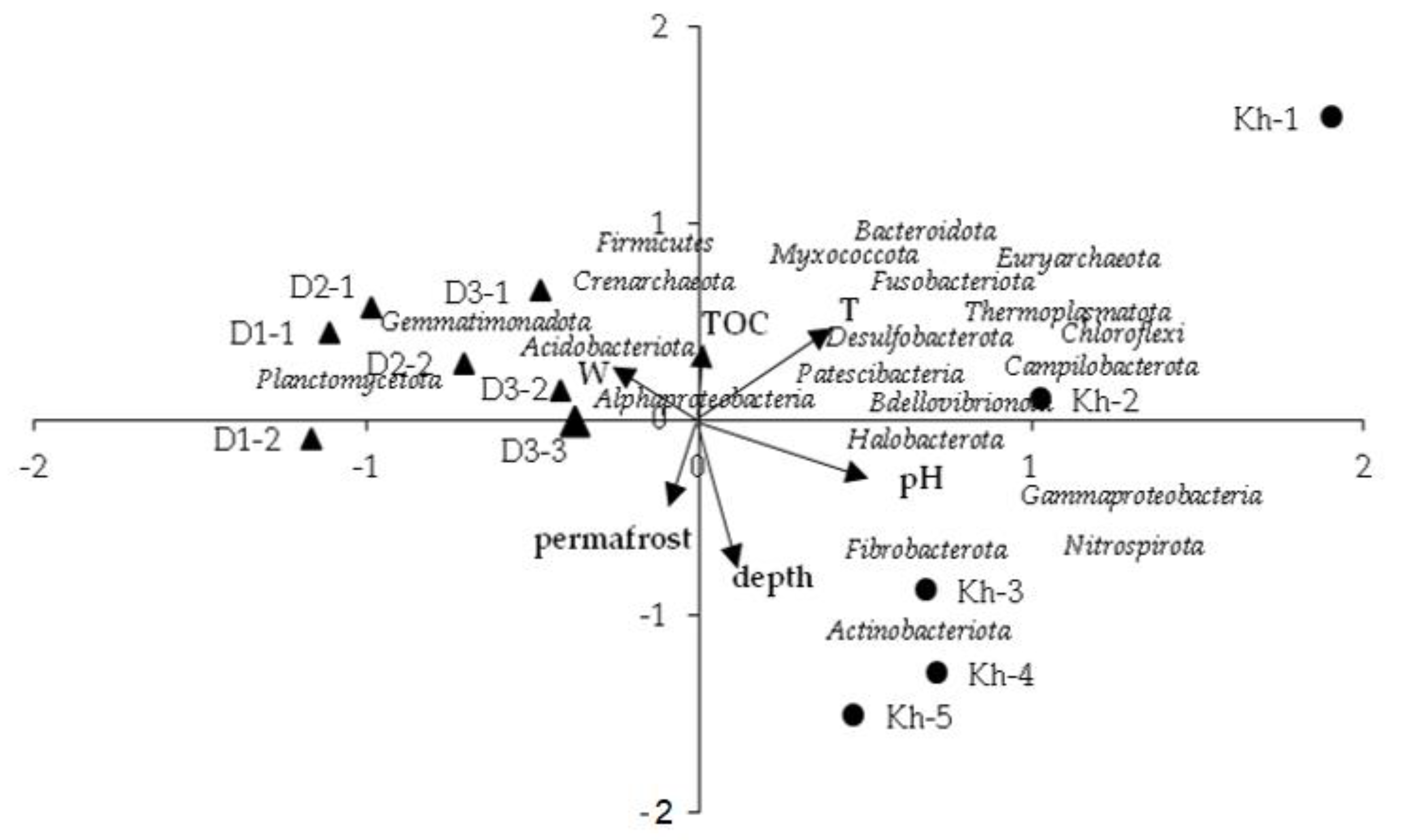
3.3. Relationships between Environmental Variables and Microbial Community Structures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Doherty, S.J.; Barbato, R.A.; Grandy, A.S.; Thomas, W.K.; Ernakovich, J.G. The transition from stochastic to deterministic bacterial community assembly during permafrost thaw succession. Front. Microbiol. 2020, 11, 596589. [Google Scholar] [CrossRef]
- Karelin, D.V.; Zamolodchikov, D.G. Uglerodnyi Obmen V Kriogennykh Ekosistemakh (Carbon Exchange in Cryogenic Ecosystems); Nauka: Moscow, Russia, 2008; p. 342. (In Russian) [Google Scholar]
- Gyninova, A.; Badmaev, N.; Tsybenov, Y.; Gonchikov, B.; Mangataev, A.; Kulikov, A.; Sympilova, D. Soils of the Darkhitui catena in the southern Vitim Plateau and their micromorphological features. IOP Conf. Ser. Earth Environ. Sci. 2021, 862, 012068. [Google Scholar] [CrossRef]
- Dugarov, V.I.; Kulikov, A.I. Agrophysical Properties of Frozen Soils; Nauka: Novosibirsk, Russia, 1990; p. 254. (In Russian) [Google Scholar]
- Kulikov, A.I.; Dugarov, V.I.; Korsunov, V.M. Frozen Soils: Ecology, Heat Supply and Productivity Forecast; Buryatskoe Knizhnoe Izdatel’stvo: Ulan-Ude, Russia, 1997; p. 313. (In Russian) [Google Scholar]
- Badmaev, N.; Bazarov, A. Monitoring network for atmospheric and soil parameters measurements in permafrost area of Buryatia, Russian Federation. Geosciences 2019, 9, 6. [Google Scholar] [CrossRef]
- Schuur, E.A.G.; McGuire, A.D.; Schädel, C.; Grosse, G.; Harden, J.W.; Hayes, D.J.; Hugelius, G.; Koven, C.D.; Kuhry, P.; Lawrence, D.M.; et al. Climate change and the permafrost carbon feedback. Nature 2015, 520, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Feng, J.; Wang, C.; Lei, J.; Yang, Y.; Yan, Q.; Zhou, X.; Tao, X.; Ning, D.; Yuan, M.M.; Qin, Y.; et al. Warming-induced permafrost thaw exacerbates tundra soil carbon decomposition mediated by microbial community. Microbiome 2020, 8, 3. [Google Scholar] [CrossRef]
- Karelin, D.; Goryachkin, S.; Zazovskaya, E.; Shishkov, V.; Pochikalov, A.; Dolgikh, A.; Sirin, A.; Suvorov, G.; Badmaev, N.; Badmaeva, N.; et al. Greenhouse gas emission from the cold soils of Eurasia in natural settings and under human impact: Controls on spatial variability. Geoderma Reg. 2020, 22, e00290. [Google Scholar] [CrossRef]
- Milkheev, E.Y.; Tsybenov, Y.B. The CO2 emission intensity of wooded steppe soils in the Baikal lake area. Bull 2011, 3, 103–106. (In Russian) [Google Scholar]
- Makushkin, E.O. A comparative assessment of the biological activity of soils in the Selenga river upper delta according to the content of enzymes. Tomsk. State Univ. J. Biol. 2018, 42, 6–23. (In Russian) [Google Scholar] [CrossRef]
- Korsunova, T.D.-T.; Valova, E.E. Biological activity of meadow-chernozem and sod-taiga permafrost soils of the Yeravninsky basin. IOP Conf. Ser. Earth Environ. Sci. 2021, 885, 012065. [Google Scholar] [CrossRef]
- Makarova, A.P.; Naprasnikova, E.V. Microbiological and biochemical characteristics of gley cryogenic soils of Northern Transbaikalia. Bull. Irkutsk. State Univ. Series Biol. Ecol. 2011, 4, 25–32. (In Russian) [Google Scholar]
- Gyninova, A.; Badmaev, N.; Andreeva, D.; Zech, W.; Syrenzhapova, A. On frozen chernozem genesis and evolution at lacustrine plains in the southern Vitim plateau (Eastern Siberia, Russia). IOP Conf. Ser. Earth Environ. Sci. 2021, 908, 012033. [Google Scholar] [CrossRef]
- Mineev, V.G.; Sychev, V.G.; Amelyanchik, O.A.; Bolysheva, T.N.; Gomonova, N.F.; Durynina, E.P.; Egorov, B.C.; Egorova, E.V.; Edemskaya, N.L.; Karpova, E.A.; et al. Workshop on Agrochemistry, 2nd ed.; Publishing House: Moscow, Russia, 2001; p. 689. [Google Scholar]
- Bates, S.T.; Berg-Lyons, J.G.; Caporaso, W.A.; Walters, W.A.; Knight, R.; Fierer, N. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2010, 5, 908–917. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- McMurdie, P.J.; Holmes, S.P. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Zuur, A.F.; Ieno, E.N.; Smith, G.M. Analyzing Ecological Data; Springer: New York, NY, USA, 2007; p. 672. [Google Scholar]
- ter Braak, C.J.F.; Verdonschot, P.F.M. Canonical correspondence analysis and related multivariate methods in aquatic ecology. Aquat. Sci. 1995, 57, 255–289. [Google Scholar] [CrossRef]
- Steven, B.; Pollard, W.H.; Greer, C.W.; Whyte, L.G. Microbial diversity and activity through a permafrost/ground ice core profile from the Canadian high Arctic. Environ. Microbiol. 2008, 10, 3388–3403. [Google Scholar] [CrossRef]
- Chen, Y.L.; Deng, Y.; Ding, J.Z.; Hu, H.W.; Xu, T.L.; Li, F. Distinct microbial communities in the active and permafrost layers on the Tibetan Plateau. Mol. Ecol. 2017, 26, 6608–6620. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, Q.; Tian, T.; Li, D.; Cheng, G.; Mu, J.; Wu, Q.; Niu, F.; Stegen, J.C.; An, L. Relative roles of deterministic and stochastic processes in driving the vertical distribution of bacterial communities in a permafrost core from the Qinghai-Tibet Plateau, China. PLoS ONE 2015, 10, e0145747. [Google Scholar] [CrossRef]
- Hu, W.; Zhang, Q.; Tian, T.; Li, D.; Cheng, G.; Mu, J.; Wu, Q.; Niu, F.; An, L.; Feng, H. Characterization of the prokaryotic diversity through a stratigraphic permafrost core profile from the Qinghai-Tibet Plateau. Extremophiles 2016, 20, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Mackelprang, R.; Waldrop, M.P.; DeAngelis, K.M.; David, M.M.; Chavarria, K.L.; Blazewicz, S.J.; Rubin, E.M.; Jansson, J.K. Metagenomic analysis of a permafrost microbial community reveals a rapid response to thaw. Nature 2011, 480, 368–371. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Hu, Y.; Chen, B.; Zhang, Y.; Thiele, J.; Shi, R.; Liu, M.; Bu, R. Soil pH and plant diversity shape soil bacterial community structure in the active layer across the latitudinal gradients in continuous permafrost region of Northeastern China. Sci. Rep. 2018, 8, 5619. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; Hu, Y.; Bu, R. Vertical distribution patterns and drivers of soil bacterial communities across the continuous permafrost region of northeastern China. Ecol. Processes 2022, 11, 6. [Google Scholar] [CrossRef]
- Yergeau, E.; Hogues, H.; Whyte, L.G.; Greer, C.W. The functional potential of high Arctic permafrost revealed by metagenomic sequencing, qPCR and microarray analyses. ISME J. 2010, 4, 1206–1214. [Google Scholar] [CrossRef] [PubMed]
- Wilhelm, R.C.; Niederberger, T.D.; Greer, C.; Whyte, L.G. Microbial diversity of active layer and permafrost in an acidic wetland from the Canadian High Arctic. Can. J. Microbiol. 2011, 57, 303–315. [Google Scholar] [CrossRef] [PubMed]
- Goordial, J.; Davila, A.; Greer, C.; Cannam, R.; DiRuggiero, J.; McKay, C.; Whyte, L. Comparative activity and functional ecology of permafrost soils and lithic niches in a hyper-arid polar desert. Environ. Microbiol. 2017, 19, 443–458. [Google Scholar] [CrossRef]
- Blanco, Y.; Prieto-Ballesteros, O.; Gómez, M.J.; Moreno-Paz, M.; García-Villadangos, M.; Rodríguez-Manfredi, J.A.; Cruz-Gil, P.; Sánchez-Román, M.; Rivas, L.A.; Parro, V. Prokaryotic communities and operating metabolisms in the surface and the permafrost of Deception Island (Antarctica). Environ. Microbiol. 2012, 14, 2495–2510. [Google Scholar] [CrossRef]
- Rivkina, E.; Petrovskaya, L.; Vishnivetskaya, T.; Krivushin, K.; Shmakova, L.; Tutukina, M.; Meyers, A.; Kondrashov, F. Metagenomic analyses of the late Pleistocene permafrost-Additional tools for reconstruction of environmental conditions. Biogeosciences 2016, 13, 2207–2219. [Google Scholar] [CrossRef]
- Deng, J.; Gu, Y.; Zhang, J.; Xue, K.; Qin, Y.J.; Yuan, M.T.; Yin, H.; He, Z.; Wu, L.; Schuur, E.A.G.; et al. Shifts of tundra bacterial and archaeal communities along a permafrost thaw gradient in Alaska. Mol. Ecol. 2015, 24, 222–234. [Google Scholar] [CrossRef]
- Aksenov, A.S.; Shirokova, L.S.; Kisil, O.Y.; Kolesova, S.N.; Lim, A.G.; Kuzmina, D.; Pouillé, S.; Alexis, M.A.; Castrec-Rouelle, M.; Loiko, S.V.; et al. Bacterial Number and Genetic Diversity in a Permafrost Peatland (Western Siberia): Testing a Link with Organic Matter Quality and Elementary Composition of a Peat Soil Profile. Diversity 2021, 13, 328. [Google Scholar] [CrossRef]
- Wagner, D.; Kobabe, S.; Pfeiffer, E.-M.; Hubberten, H.-W. Microbial controls on methane fluxes from a polygonal tundra of the Lena Delta, Siberia. Permafr. Periglac Process 2003, 14, 173–185. [Google Scholar] [CrossRef]
- Gittel, A.; Bárta, J.; Kohoutová, I.; Mikutta, R.; Owens, S.; Gilbert, J.; Schnecker, J.; Wild, B.; Hannisdal, B.; Maerz, J.; et al. Distinct microbial communities associated with buried soils in the Siberian tundra. ISME J. 2014, 8, 841–853. [Google Scholar] [CrossRef] [PubMed]
- Ganzert, L.; Bajerski, F.; Wagner, D. Bacterial community composition and diversity of five different permafrost-affected soils of Northeast Greenland. FEMS Microbiol. Ecol. 2014, 89, 426–441. [Google Scholar] [CrossRef]
- Tytgat, B.; Verleyen, E.; Sweetlove, M.; D’hondt, S.; Clercx, P.; Ranst, V.E.; Peeters, K.; Roberts, S.; Namsaraev, Z.; Wilmotte, A.; et al. Bacterial community composition in relation to bedrock type and macrobiota in soils from the Sør Rondane Mountains, East Antarctica. FEMS Microbiol. Ecol. 2016, 92, fiw126. [Google Scholar] [CrossRef]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microb. 2009, 75, 5111–5120. [Google Scholar] [CrossRef]
- Chong, C.W.; Pearce, D.; Convey, P.; Tan, G.Y.A.; Wong, R.C.S.; Tan, I.K.R. High levels of spatial heterogeneity in the biodiversity of soil prokaryotes on Signy Island, Antarctica. Soil Biol. Biochem. 2010, 42, 601–610. [Google Scholar] [CrossRef]
- Yang, S.; Wen, X.; Jin, H.; Wu, Q. Pyrosequencing investigation into the bacterial community in permafrost soils along the China-Russia Crude Oil Pipeline (CRCOP). PLoS ONE 2012, 7, e52730. [Google Scholar] [CrossRef]
- Griffiths, R.I.; Thomson, B.C.; James, P.; Bell, T.; Bailey, M.; Whiteley, A.C. The bacterial biogeography of British soils. Environ. Microbiol. 2011, 13, 1642–1654. [Google Scholar] [CrossRef]
- Chu, H.; Fierer, N.; Lauber, C.L.; Caporaso, J.G.; Knight, R.; Grogan, P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol. 2010, 12, 2998–3006. [Google Scholar] [CrossRef]
- Wallenstein, M.D.; McMahon, S.; Schimel, J. Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol. Ecol. 2007, 59, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Zinger, L.; Lejon, D.P.H.; Baptist, F.; Bouasria, A.; Aubert, S.; Geremia, R.A.; Choler, P. Contrasting diversity patterns of crenarchaeal, bacterial and fungal soil communities in an alpine landscape. PLoS ONE 2011, 6, e19950. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Green, S.; Tfaily, M.M.; Prakash, O.; Konstantinidis, K.T.; Corbett, J.E.; Chanton, J.P.; Cooper, W.T.; Kostka, J.E. Microbial community structure and activity linked to contrasting biogeochemical gradients in bog and fen environments of the Glacial Lake Agassiz Peatland. Appl. Environ. Microb. 2012, 78, 7023–7031. [Google Scholar] [CrossRef] [PubMed]
- Rousk, J.; Bååth, E.; Brookes, P.; Lauber, C.L.; Lozupone, C.; Caporaso, J.G.; Knight, R.; Fierer, N. Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 2010, 4, 1340–1351. [Google Scholar] [CrossRef]
- Hu, H.-W.; Zhang, L.-M.; Dai, Y.; Di, H.-J.; He, J.-Z. pH-dependent distribution of soil ammonia oxidizers across a large geographical scale as revealed by high-throughput pyrosequencing. J. Soil Sediment 2013, 13, 1439–1449. [Google Scholar] [CrossRef]
- Ivanova, A.A.; Zhelezova, A.D.; Chernov, T.I.; Dedysh, S.N. Linking ecology and systematics of acidobacteria: Distinct habitat preferences of the Acidobacteriia and Blastocatellia in tundra soils. PLoS ONE 2020, 15, e0230157. [Google Scholar] [CrossRef]
- Zhalnina, K.; Dörr de Quadros, P.; Camargo, F.A.O.; Triplet, E.W. Drivers of archaeal ammonia-oxidizing communities in soil. Front. Microbiol. 2012, 3, 210. [Google Scholar] [CrossRef]
- Stopnisek, N.; Gubry Rangin, C.; Hofferle, S. Thaumarchaeal ammonia oxidation in an acidic forest peat soil is not influenced by ammonium amendment. Appl. Environ. Microbiol. 2010, 76, 7626–7634. [Google Scholar] [CrossRef]
- Kerou, M.; Offre, P.; Valledor, L.; Abby, S.S.; Melchera, M.; Naglera, M.; Weckwerthb, W.; Schlepera, C. Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc. Natl. Acad. Sci. USA 2016, 113, E7937–E7946. [Google Scholar] [CrossRef]

| Darkhituy | Khaimisan | |||
|---|---|---|---|---|
| Site D1 | Site D2 | Site D3 | Site Kh | |
| Coordinates * | 52°31.222′ N, 111°26.346′ E | 52°31.506′ N, 111°28.226′ E | 52°030.905′ N, 111°032.770′ E | 52°035.677′ N, 111°035.611′ E |
| Parent rock (according to genesis) | Eluvium; eluvio- deluvium | deluvium; deluvium carbonate | deluvium carbonate, deluvium, lacustrine deposits | lacustrine deposits |
| Vegetation | larch forest with Rhododendron dauricum, Vaccinium vitisidaea, Pirola rotundifolia, Calamagrostis epigeios | larch-birch forest with Carex pediformis, Sanguisorba officinalis, Calamagrostis epigeios, Achillea and parches of moss | meadow steppe with Carex duriuscula, Sanguisorba officinalis, Artemisia mongolica | herb-sedge community |
| a.s.l., m * | 1052 | 965 | 957 | 944 |
| Position in landscape (facies) | eluvial, transeluvial | transeluvial; transaccumula- tive | Transaccumula-tive; accumulative | accumulative |
| Thawing depth, cm (for the season) | 270–300 | 230–250 | 275–290 | 170–200 |
| WRB | Skeletic Cambisols (Gelic) | Cambic Calcic Cryosols (Siltic, Humic) | Eutric Chernozems Calcaric | Calcaric Humic Fluvisols Limnic Turbic |
| Site | Sample | Layer, cm | Color | T, °C | W, % | pH | TOC, % | N-NH4, mg/100 g | N-NO3, mg/100 g |
|---|---|---|---|---|---|---|---|---|---|
| D1 | D1-1 | 5–15 | brown | 1.6 ÷ 0.1 | 65.86 | 5.97 | 1.59 | 28.0 | 6.0 |
| D1-2 | 17–25 | dark brown | −0.5 | 62.56 | 6.1 | 0.81 | 22.3 | 1.8 | |
| D2 | D2-1 | 5–15 | almost black | 2.0 ÷ 0.2 | 64.43 | 6.16 | 7.38 | 31.0 | 9.2 |
| D2-2 | 27–37 | dark brown | −0.4 | 70.61 | 7.16 | 0.66 | 17.7 | 3.6 | |
| D3 | D3-1 | 5–10 | black | 4.0 | 18.35 | 6.88 | 11.36 | 23.0 | 4.8 |
| D3-2 | 10–20 | yellow brown | 2.1 | 19.05 | 7.05 | 8.58 | 18.7 | 3.2 | |
| D3-3 | 20–30 | light brown | 0.9 | 19.23 | 7.2 | 4.9 | 13.3 | 3.9 | |
| Kh | Kh-1 | 0–10 | black | 9.6 ÷ 7.5 | 41.97 | 8.91 | 6.2 | 16.7 | 3.7 |
| Kh-2 | 10–30 | black brown | 4.1 ÷ 2.1 | 23.6 | 8.9 | 2.3 | 58.3 | 5.3 | |
| Kh-3 | 30–50 | black brown | 1.9 ÷ 1.3 | 15.36 | 8.62 | 2.47 | n.d. | n.d. | |
| Kh-4 | 50–70 | grey-yellow | 0.2 | 32.72 | 8.62 | 2.3 | 25.7 | 2.8 | |
| Kh-5 | 68–75 | grey-yellow | −0.2 | 22.12 | 8.8 | 2.23 | n.d. | n.d. |
| Parameter | Abbreviated in Figure 6 | PC1 (29.8%) | PC2 (13.9%) | PC3 (11.7%) |
|---|---|---|---|---|
| Depth | depth | −0.72 | ||
| Soil temperature | T | 0.48 | ||
| Humidity | W | −0.41 | 0.61 | |
| pH | pH | 0.57 | ||
| Total organic carbon | TOC | −0.54 | ||
| Permafrost | permafrost | −0.43 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zaitseva, S.; Badmaev, N.; Kozyreva, L.; Dambaev, V.; Barkhutova, D. Microbial Community in the Permafrost Thaw Gradient in the South of the Vitim Plateau (Buryatia, Russia). Microorganisms 2022, 10, 2202. https://doi.org/10.3390/microorganisms10112202
Zaitseva S, Badmaev N, Kozyreva L, Dambaev V, Barkhutova D. Microbial Community in the Permafrost Thaw Gradient in the South of the Vitim Plateau (Buryatia, Russia). Microorganisms. 2022; 10(11):2202. https://doi.org/10.3390/microorganisms10112202
Chicago/Turabian StyleZaitseva, Svetlana, Nimazhap Badmaev, Lyudmila Kozyreva, Vyacheslav Dambaev, and Darima Barkhutova. 2022. "Microbial Community in the Permafrost Thaw Gradient in the South of the Vitim Plateau (Buryatia, Russia)" Microorganisms 10, no. 11: 2202. https://doi.org/10.3390/microorganisms10112202
APA StyleZaitseva, S., Badmaev, N., Kozyreva, L., Dambaev, V., & Barkhutova, D. (2022). Microbial Community in the Permafrost Thaw Gradient in the South of the Vitim Plateau (Buryatia, Russia). Microorganisms, 10(11), 2202. https://doi.org/10.3390/microorganisms10112202





