Streptococcus salivarius 24SMBc Genome Analysis Reveals New Biosynthetic Gene Clusters Involved in Antimicrobial Effects on Streptococcus pneumoniae and Streptococcus pyogenes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. Whole-Genome Sequencing, De Novo Genome Assembly
2.3. Genomic Data Accession Number
2.4. Genome Annotation
2.5. In Silico Prediction and Characterization of Bacteriocin Biosynthetic Gene Clusters
2.6. Adhesion Assay
2.7. Anti-Adhesion Activity of 24SMBc against S. pneumoniae and S. pyogenes on HEp-2 Cells
2.8. LDH Assay for Cytotoxicity
2.9. Auto- and Co-Aggregation Assay
3. Results
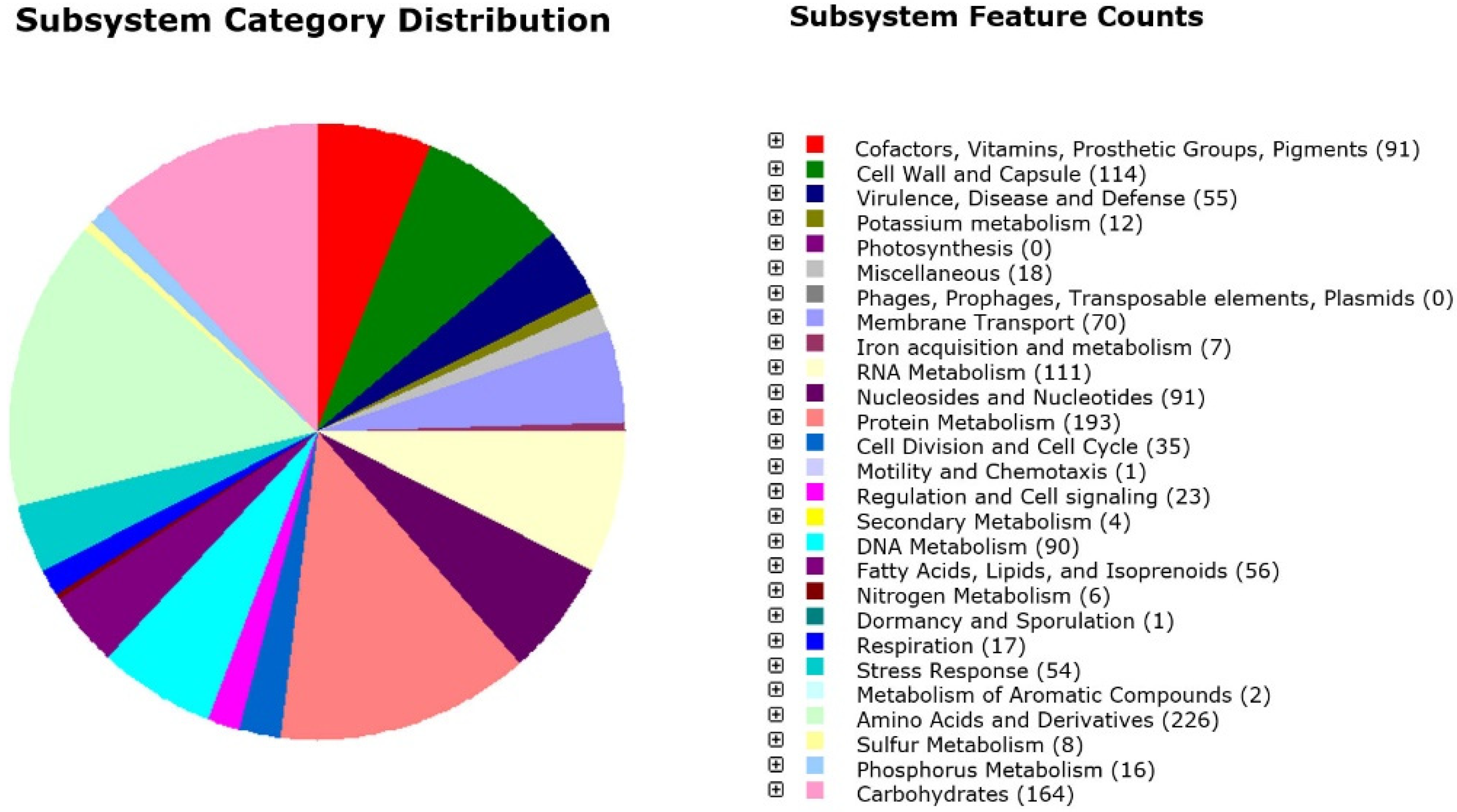
3.1. Genome Annotation
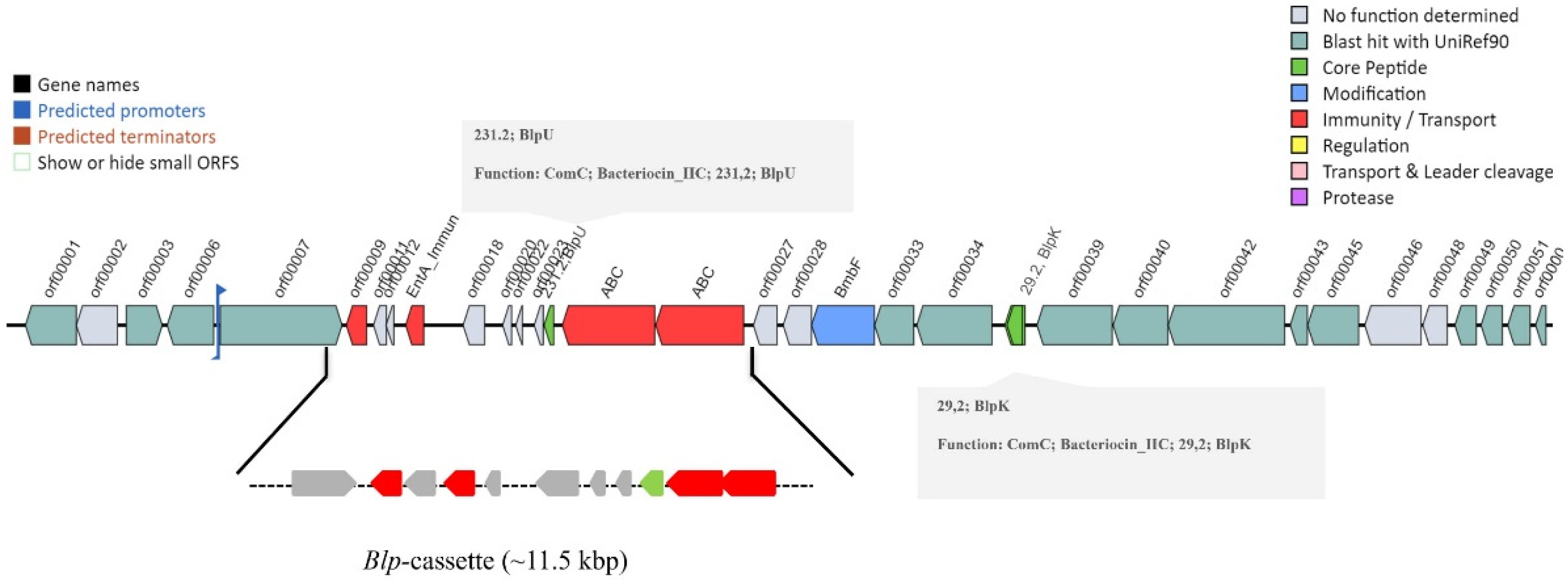
3.2. Microbial Genome Mining for Biosynthetic Gene Clusters
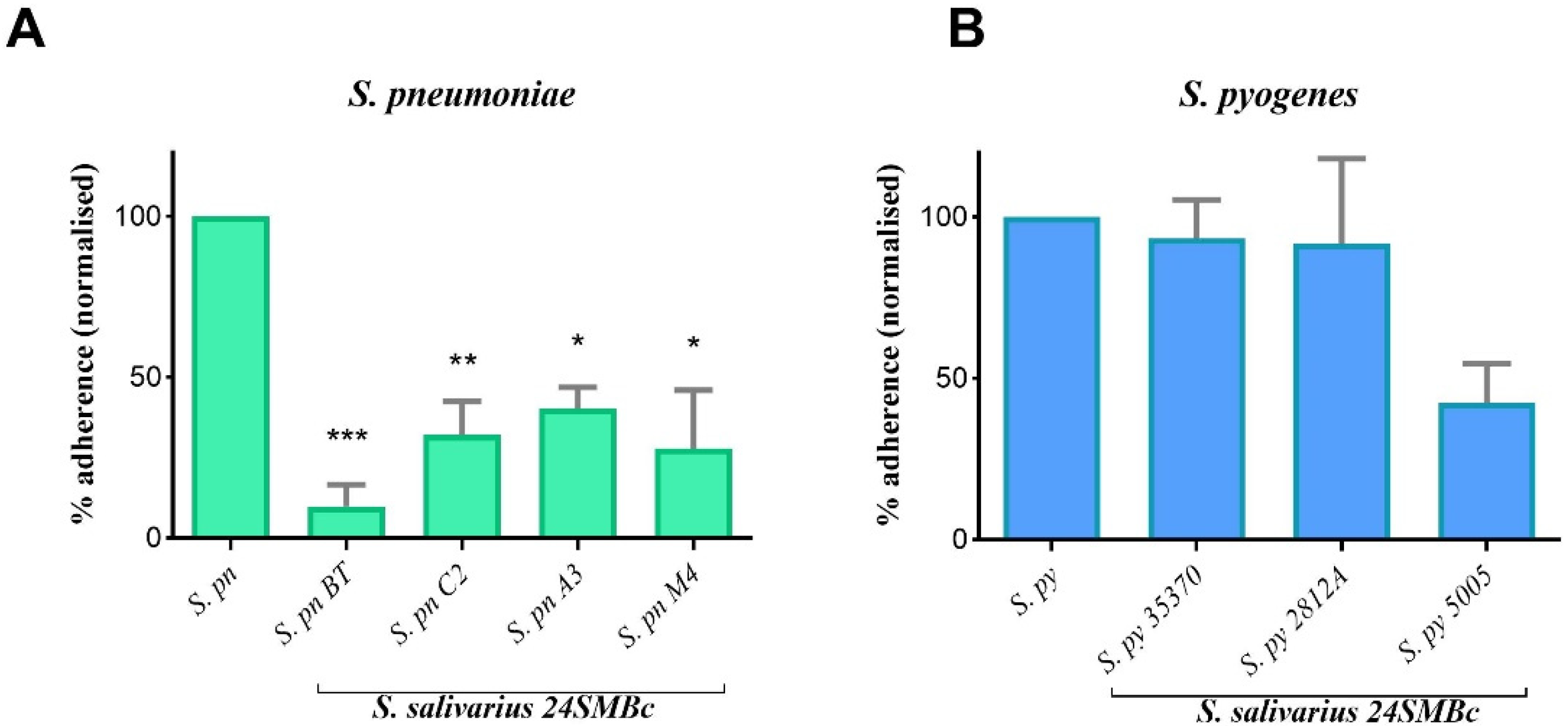
3.3. Antagonistic Activity of S. salivarius 24SMBc against S. pneumoniae and S. pyogenes Adherence to HEp2 Cells
3.4. Cytotoxic Effect of S. salivarius 24SMBc on HEp-2
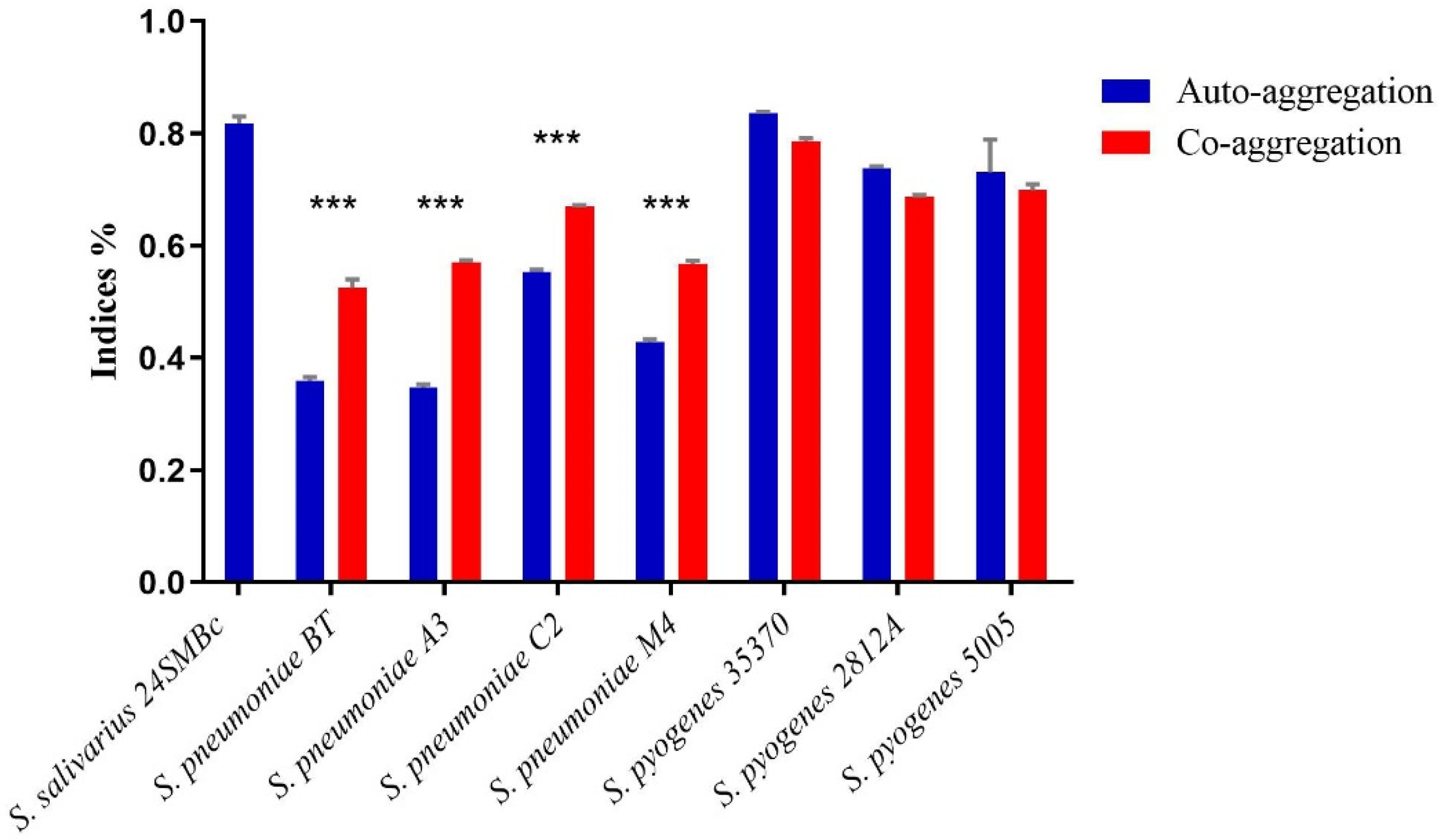
3.5. Auto-Aggregation and Co-Aggregation Ability
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dominguez-Bello, M.G.; Godoy-Vitorino, F.; Knight, R.; Blaser, M.J. Role of the microbiome in human development. Gut 2019, 68, 1108–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Man, W.H.; de Steenhuijsen Piters, W.A.; Bogaert, D. The microbiota of the respiratory tract: Gatekeeper to respiratory health. Nat. Rev. Microbiol. 2017, 15, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.B.; Hussain, M.A.; Nye, R.; Mehta, V.; Mamun, K.T.; Hossain, N. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus 2017, 9, e1403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berg, G.; Rybakova, D.; Fischer, D.; Cernava, T.; Vergès, M.C.; Charles, T.; Chen, X.; Cocolin, L.; Eversole, K.; Corral, G.H.; et al. Microbiome definition re-visited: Old concepts and new challenges. Microbiome 2020, 8, 103. [Google Scholar] [CrossRef]
- Respiratory Tract Infections—Antibiotic Prescribing: Prescribing of Antibiotics for Self-Limiting Respiratory Tract Infections in Adults and Children in Primary Care; National Institute for Health and Clinical Excellence (NICE): London, UK, July 2008.
- Barbieri, E.; Donà, D.; Cantarutti, A.; Lundin, R.; Scamarcia, A.; Corrao, G.; Cantarutti, L.; Giaquinto, C. Antibiotic prescriptions in acute otitis media and pharyngitis in Italian pediatric outpatients. Ital. J. Pediatr. 2019, 45, 103. [Google Scholar] [CrossRef] [PubMed]
- Morris, P.S. Upper respiratory tract infections (including otitis media). Pediatr. Clin. N. Am. 2009, 56, 101–117. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, C.; Sarkar, P.; Issa, R.; Haldar, J. Alternatives to Conventional Antibiotics in the Era of Antimicrobial Resistance. Trends Microbiol. 2019, 27, 323–338. [Google Scholar] [CrossRef]
- Ciprandi, G.; La Mantia, I.; Damiani, V.; Passali, D. Local Bacteriotherapy—A promising preventive tool in recurrent respiratory infections. Expert. Rev. Clin. Immunol. 2020, 16, 1047–1052. [Google Scholar] [CrossRef] [PubMed]
- Marchisio, P.; Claut, L.; Rognoni, A.; Esposito, S.; Passali, D.; Bellussi, L.; Drago, L.; Pozzi, G.; Mannelli, S.; Schito, G.; et al. Differences in nasopharyngeal bacterial flora in children with nonsevere recurrent acute otitis media and chronic otitis media with effusion: Implications for management. Pediatr. Infect. Dis. J. 2003, 22, 262–268. [Google Scholar] [CrossRef] [PubMed]
- Wescombe, P.A.; Burton, J.P.; Cadieux, P.A.; Klesse, N.A.; Hyink, O.; Heng, N.C.; Chilcott, C.N.; Reid, G.; Tagg, J.R. Megaplasmids encode differing combinations of lantibiotics in Streptococcus salivarius. Antonie Van Leeuwenhoek 2006, 90, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Marchisio, P.; Santagati, M.; Scillato, M.; Baggi, E.; Fattizzo, M.; Rosazza, C.; Stefani, S.; Esposito, S.; Principi, N. Streptococcus salivarius 24SMB administered by nasal spray for the prevention of acute otitis media in otitis-prone children. Eur. J. Clin. Microbiol. Infect. Dis. Off. Publ. Eur. Soc. Clin. Microbiol. 2015, 34, 2377–2383. [Google Scholar] [CrossRef]
- Cantarutti, A.; Rea, F.; Donà, D.; Cantarutti, L.; Passarella, A.; Scamarcia, A.; Lundin, R.; Damiani, V.; Giaquinto, C.; Corrao, G.; et al. Preventing recurrent acute otitis media with Streptococcus salivarius 24SMB and Streptococcus oralis 89a five months intermittent treatment: An observational prospective cohort study. Int. J. Pediatr. Otorhinolaryngol. 2020, 132, 109921. [Google Scholar] [CrossRef] [PubMed]
- Andaloro, C.; Santagati, M.; Stefani, S.; La Mantia, I. Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a oral spray for children with recurrent streptococcal pharyngotonsillitis: A randomized placebo-controlled clinical study. Eur. Arch. Otorhinolaryngol. 2019, 276, 879–887. [Google Scholar] [CrossRef]
- Passali, D.; Passali, G.C.; Vesperini, E.; Cocca, S.; Visconti, I.C.; Ralli, M.; Bellussi, L.M. The efficacy and tolerability of Streptococcus salivarius 24SMB and Streptococcus oralis 89a administered as nasal spray in the treatment of recurrent upper respiratory tract infections in children. Eur. Rev. Med. Pharmacol. Sci. 2019, 23 (Suppl. S1), 67–72. [Google Scholar] [CrossRef]
- Manti, S.; Parisi, G.F.; Papale, M.; Licari, A.; Salpietro, C.; Miraglia Del Giudice, M.; Marseglia, G.L.; Leonardi, S. Bacteriotherapy with Streptococcus salivarius 24SMB and Streptococcus oralis 89a nasal spray for treatment of upper respiratory tract infections in children: A pilot study on short-term efficacy. Ital. J. Pediatr. 2020, 46, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bidossi, A.; De Grandi, R.; Toscano, M.; Bottagisio, M.; De Vecchi, E.; Gelardi, M.; Drago, L. Probiotics Streptococcus salivarius 24SMB and Streptococcus oralis 89a interfere with biofilm formation of pathogens of the upper respiratory tract. BMC Infect Dis. 2018, 18, 653. [Google Scholar] [CrossRef] [PubMed]
- Santagati, M.; Scillato, M.; Patanè, F.; Aiello, C.; Stefani, S. Bacteriocin-producing oral streptococci and inhibition of respiratory pathogens. FEMS Immunol. Med. Microbiol. 2012, 65, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Santagati, M.; Scillato, M.; Stefani, S. Genetic organization of Streptococcus salivarius 24SMBc blp-like bacteriocin locus. Front. Biosci. (Schol. Ed.) 2018, 10, 238–247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stubbendieck, R.M.; Zelasko, S.E.; Safdar, N.; Currie, C.R. Biogeography of Bacterial Communities and Specialized Metabolism in Human Aerodigestive Tract Microbiomes. Microbiol. Spectr. 2021, 9, e0166921. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Kottmann, R.; Yilmaz, P.; Cummings, M.; Biggins, J.B.; Blin, K.; de Bruijn, I.; Chooi, Y.H.; Claesen, J.; Coates, R.C.; et al. Minimum Information about a Biosynthetic Gene cluster. Nat. Chem. Biol. 2015, 11, 625–631. [Google Scholar] [CrossRef]
- Tracanna, V.; de Jong, A.; Medema, M.H.; Kuipers, O.P. Mining prokaryotes for antimicrobial compounds: From diversity to function. FEMS Microbiol. Rev. 2017, 41, 417–429. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santagati, M.; Spanu, T.; Scillato, M.; Santangelo, R.; Cavallaro, F.; Arena, V.; Castiglione, G.; Falcone, M.; Venditti, M.; Stefani, S. Rapidly fatal hemorrhagic pneumonia and group A Streptococcus serotype M1. Emerg. Infect. Dis. 2014, 20, 98–101. [Google Scholar] [CrossRef]
- Iannelli, F.; Santagati, M.; Santoro, F.; Oggioni, M.R.; Stefani, S.; Pozzi, G. Nucleotide sequence of conjugative prophage Φ1207.3 (formerly Tn1207.3) carrying the mef(A)/msr(D) genes for efflux resistance to macrolides in Streptococcus pyogenes. Front. Microbiol. 2014, 5, 687. [Google Scholar] [CrossRef] [Green Version]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. MiSeq: A Next Generation Sequencing Platform for Genomic Analysis. Methods Mol. Biol. 2018, 1706, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [Green Version]
- Lagesen, K.; Hallin, P.; Rødland, E.A.; Staerfeldt, H.H.; Rognes, T.; Ussery, D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007, 35, 3100–3108. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997, 25, 955–964. [Google Scholar] [CrossRef] [PubMed]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef]
- Sachdeva, G.; Kumar, K.; Jain, P.; Ramachandran, S. SPAAN: A software program for prediction of adhesins and adhesin-like proteins using neural networks. Bioinformatics 2005, 21, 483–491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cosentino, S.; Voldby Larsen, M.; Møller Aarestrup, F.; Lund, O. PathogenFinder—Distinguishing friend from foe using bacterial whole genome sequence data. PLoS ONE 2013, 8, e77302. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Kapil, R.; Dhakan, D.B.; Sharma, V.K. MP3: A software tool for the prediction of pathogenic proteins in genomic and metagenomic data. PLoS ONE 2014, 9, e93907. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- van Heel, A.J.; de Jong, A.; Song, C.; Viel, J.H.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Scillato, M.; Spitale, A.; Mongelli, G.; Privitera, G.F.; Mangano, K.; Cianci, A.; Stefani, S.; Santagati, M. Antimicrobial properties of Lactobacillus cell-free supernatants against multidrug-resistant urogenital pathogens. Microbiologyopen 2021, 10, e1173. [Google Scholar] [CrossRef]
- Dunne, E.M.; Toh, Z.Q.; John, M.; Manning, J.; Satzke, C.; Licciardi, P. Investigating the effects of probiotics on pneumococcal colonization using an in vitro adherence assay. J. Vis. Exp. 2014, 28, 51069. [Google Scholar] [CrossRef] [Green Version]
- Kumar, P.; Nagarajan, A.; Uchil, P.D. Analysis of Cell Viability by the Lactate Dehydrogenase Assay. Cold Spring Harb. Protoc. 2018, 2018, pdb-prot095497. [Google Scholar] [CrossRef]
- Chaffanel, F.; Charron-Bourgoin, F.; Soligot, C.; Kebouchi, M.; Bertin, S.; Payot, S.; Le Roux, Y.; Leblond-Bourget, N. Surface proteins involved in the adhesion of Streptococcus salivarius to human intestinal epithelial cells. Appl. Microbiol. Biotechnol. 2018, 102, 2851–2865. [Google Scholar] [CrossRef] [Green Version]
- Dawid, S.; Roche, A.M.; Weiser, J.N. The blp bacteriocins of Streptococcus pneumoniae mediate intraspecies competition both in vitro and in vivo. Infect Immun. 2007, 75, 443–451. [Google Scholar] [CrossRef]
- Kawasaki, T.; Hayashi, Y.; Kuzuyama, T.; Furihata, K.; Itoh, N.; Seto, H.; Dairi, T. Biosynthesis of a natural polyketide-isoprenoid hybrid compound, furaquinocin A: Identification and heterologous expression of the gene cluster. J. Bacteriol. 2006, 188, 1236–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shimizu, Y.; Ogata, H.; Goto, S. Type III Polyketide Synthases: Functional Classification and Phylogenomics. Chembiochem 2017, 18, 50–65. [Google Scholar] [CrossRef] [PubMed]
- Calderone, C.T. Isoprenoid-like alkylations in polyketide biosynthesis. Nat. Prod. Rep. 2008, 25, 845–853. [Google Scholar] [CrossRef]
- Löfling, J.; Vimberg, V.; Battig, P.; Henriques-Normark, B. Cellular interactions by LPxTG-anchored pneumococcal adhesins and their streptococcal homologues. Cell Microbiol. 2011, 13, 186–197. [Google Scholar] [CrossRef] [PubMed]
- Nobbs, A.H.; Lamont, R.J.; Jenkinson, H.F. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 2009, 73, 407–450. [Google Scholar] [CrossRef] [Green Version]
- Couvigny, B.; Lapaque, N.; Rigottier-Gois, L.; Guillot, A.; Chat, S.; Meylheuc, T.; Kulakauskas, S.; Rohde, M.; Mistou, M.Y.; Renault, P.; et al. Three glycosylated serine-rich repeat proteins play a pivotal role in adhesion and colonization of the pioneer commensal bacterium, Streptococcus salivarius. Environ. Microbiol. 2017, 19, 3579–3594. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foster, T.J. The MSCRAMM Family of Cell-Wall-Anchored Surface Proteins of Gram-Positive Cocci. Trends Microbiol. 2019, 27, 927–941. [Google Scholar] [CrossRef]
- Zhu, M.; Ajdić, D.; Liu, Y.; Lynch, D.; Merritt, J.; Banas, J.A. Role of the Streptococcus mutans irvA gene in GbpC-independent, dextran-dependent aggregation and biofilm formation. Appl. Environ. Microbiol. 2009, 75, 7037–7043. [Google Scholar] [CrossRef] [Green Version]
- Couvigny, B.; Kulakauskas, S.; Pons, N.; Quinquis, B.; Abraham, A.L.; Meylheuc, T.; Delorme, C.; Renault, P.; Briandet, R.; Lapaque, N.; et al. Identification of New Factors Modulating Adhesion Abilities of the Pioneer Commensal Bacterium Streptococcus salivarius. Front. Microbiol. 2018, 9, 273. [Google Scholar] [CrossRef] [Green Version]
- Monteagudo-Mera, A.; Rastall, R.A.; Gibson, G.R.; Charalampopoulos, D.; Chatzifragkou, A. Adhesion mechanisms mediated by probiotics and prebiotics and their potential impact on human health. Appl. Microbiol. Biotechnol. 2019, 103, 6463–6472. [Google Scholar] [CrossRef]

| Genomic Features | |
|---|---|
| Size (bp) | 2,131,204 |
| G + C content (%) | 39.85 |
| N50 length (bp) | 171,173 |
| L50 | 5 |
| No. of contigs | 32 |
| Number of Subsystems | 217 |
| CDS | 1.933 |
| rRNA | 6 |
| tRNA | 44 |
| Bacteriocin | Property | Amino Acid Sequences | CDS | Genome Locus Tag |
|---|---|---|---|---|
| blpU-like | Class IIc bacteriocin with double-glycine leader peptide (PF10439) | MTTQTMNNFETLDLEALANVEGGGWVKCYAGTIGSALVGSAGGPVGYWGGALVGYATFC | Contig_24_87554_87375 | LFGOLEEL_01643 |
| blpK-like | Class IIc bacteriocin with double-glycine leader peptide (PF10439) | MTTQIINNFNSLNSEDLSIIEGGGVIGCVAGTAGSAGLGFLTGTSVGTVTFPIVGTVSGGAFGA | Contig_24_96179_95985 | LFGOLEEL_01651 |
| bact_IIb_cerein family | Class IIb bacteriocin, lactobin A/cerein 7B family (TIGR03949) | MTNKERNTSDLASVTGGGWKTNLAVGGLCLASGPIGTMICLGAYNGYMDSAR | Contig_26_37678_37836 | LFGOLEEL_01810 |
| bact_IIb_cerein family | Class IIb bacteriocin, lactobin A/cerein 7B family (TIGR03949) | MTKTINNRKHMTTQELEAVSGGVVPWAAISVGMAAAKLTYDLSYAAGKSFYNLTH | Contig_26_37857_38024 | LFGOLEEL_01811 |
| Genome Locus Tag | Protein/ Domain | Property | Putative Function |
|---|---|---|---|
| LFGOLEEL_00404 | GbpC superfamily | Streptococcus glucan-binding protein C (PF08363) | Adhesion, dextran-induced aggregation |
| Streccoc_I_II | Antigen I/II family LPXTG-anchored adhesin (NF033804) | Adhesion, aggregation | |
| LFGOLEEL_00415 | GBS Bsp-like | GBS Bsp-like repeat (PF08481) | Adhesion |
| SH3_5 | SH3 domain-containing protein (PF08460) | Adhesion, aggregation | |
| LFGOLEEL_00538 | Collagen_binding domain | Collagen_bind (PF05737) | Adhesion, colonization |
| FctA | FctA family (PF12892) | Adhesion, colonization | |
| LFGOLEEL_00802 | Agg_substance superfamily | LPXTG-anchored aggregation substance (NF033875) | Aggregation |
| LFGOLEEL_00836 | CshA_NR2 | Surface adhesin CshA non-repetitive domain 2 (PF18651) | Adhesion, aggregation |
| CshA_repeat | Surface adhesin CshA repetitive domain (PF19076) | Adhesion | |
| CshA_fibril_rpt | CshA-type fibril repeat (TIGR04225) | Adhesion | |
| SSSPR-51 | SSSPR-51 domain (PF18877) | Adhesion | |
| LFGOLEEL_00906 | Streccoc_I_II | Antigen I/II family LPXTG-anchored adhesion (NF033804) | Adhesion, aggregation |
| LPXTG anchor | LPXTG-motif cell wall anchor domain (TIGR01167) | Adhesion | |
| LFGOLEEL_00907 | GbpC superfamily | Streptococcus glucan-binding protein C (PF08363) | Adhesion, dextran-induced aggregation |
| ProTailRpt superfamily | Proline-rich tail region repeat (TIGR04307) | Adhesion | |
| LFGOLEEL_01308 | MucBP_2 | Mucin binding domain (PF17965) | Mucin adhesion |
| Mub_B2 | Mub B2-like domain (PF17966) | Mucin adhesion | |
| LFGOLEEL_01314 | MucBP_2 | Mucin binding domain (PF17965) | Mucin adhesion |
| Mub_B2 | Mub B2-like domain (PF17966) | Mucin adhesion | |
| LFGOLEEL_01317 | MSCRAMM_ClfA | MSCRAMM family adhesin clumping factor ClfA (NF033609) | Adhesion |
| LFGOLEEL_01318 | MSCRAMM_SdrD | MSCRAMM family adhesin SdrD (NF012181) | Adhesion |
| aRib | Atypical Rib domain (PF18938) | Adhesion | |
| LFGOLEEL_01320 | MSCRAMM_SdrC | MSCRAMM family adhesin SdrC (NF000535) | Adhesion, aggregation |
| MSCRAMM_SdrD | MSCRAMM family adhesin SdrD (NF012181) | Adhesion | |
| MSCRAMM_ClfB | MSCRAMM family adhesin clumping factor ClfB (NF033845) | Adhesion, colonization | |
| LFGOLEEL_01321 | Agg_substance super family | LPXTG-anchored aggregation substance (NF033875) | Aggregation |
| LFGOLEEL_01338 | MucBP_2 | Mucin binding domain (PF17965) | Mucin adhesion |
| Adhesin_LEA super family | LEA family epithelial adhesin N-terminal domain (NF033647) | Adhesion | |
| LFGOLEEL_01344 | AgI_II_C2 | Cell surface antigen I/II C2 terminal domain (PF17998) | Adhesion, aggregation |
| Agg_substance super family | LPXTG-anchored aggregation substance (NF033875)) | Aggregation | |
| FctA | Spy0128-like isopeptide containing domain (PF12892) | Adhesion | |
| LFGOLEEL_01933 | DUF4097 superfamily | Putative adhesin (cl23960) | Adhesion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vertillo Aluisio, G.; Spitale, A.; Bonifacio, L.; Privitera, G.F.; Stivala, A.; Stefani, S.; Santagati, M. Streptococcus salivarius 24SMBc Genome Analysis Reveals New Biosynthetic Gene Clusters Involved in Antimicrobial Effects on Streptococcus pneumoniae and Streptococcus pyogenes. Microorganisms 2022, 10, 2042. https://doi.org/10.3390/microorganisms10102042
Vertillo Aluisio G, Spitale A, Bonifacio L, Privitera GF, Stivala A, Stefani S, Santagati M. Streptococcus salivarius 24SMBc Genome Analysis Reveals New Biosynthetic Gene Clusters Involved in Antimicrobial Effects on Streptococcus pneumoniae and Streptococcus pyogenes. Microorganisms. 2022; 10(10):2042. https://doi.org/10.3390/microorganisms10102042
Chicago/Turabian StyleVertillo Aluisio, Gaia, Ambra Spitale, Luca Bonifacio, Grete Francesca Privitera, Aldo Stivala, Stefania Stefani, and Maria Santagati. 2022. "Streptococcus salivarius 24SMBc Genome Analysis Reveals New Biosynthetic Gene Clusters Involved in Antimicrobial Effects on Streptococcus pneumoniae and Streptococcus pyogenes" Microorganisms 10, no. 10: 2042. https://doi.org/10.3390/microorganisms10102042
APA StyleVertillo Aluisio, G., Spitale, A., Bonifacio, L., Privitera, G. F., Stivala, A., Stefani, S., & Santagati, M. (2022). Streptococcus salivarius 24SMBc Genome Analysis Reveals New Biosynthetic Gene Clusters Involved in Antimicrobial Effects on Streptococcus pneumoniae and Streptococcus pyogenes. Microorganisms, 10(10), 2042. https://doi.org/10.3390/microorganisms10102042






