Genetic Diversity and Geographical Distribution of the Red Tide Species Coscinodiscus granii Revealed Using a High-Resolution Molecular Marker
Abstract
:1. Introduction
2. Materials and Methods
2.1. Strain Isolation, Culturing, and Preservation
2.2. Field Sampling and Initial Preparation
2.3. DNA Preparation and Genome Sequencing
2.4. Filtering and Assembly of Sequencing Data
2.5. Phylogenetic Analysis and Synteny Analysis
2.6. Developing Molecular Markers with High Resolution and High Specificity Based on C. granii mtDNAs
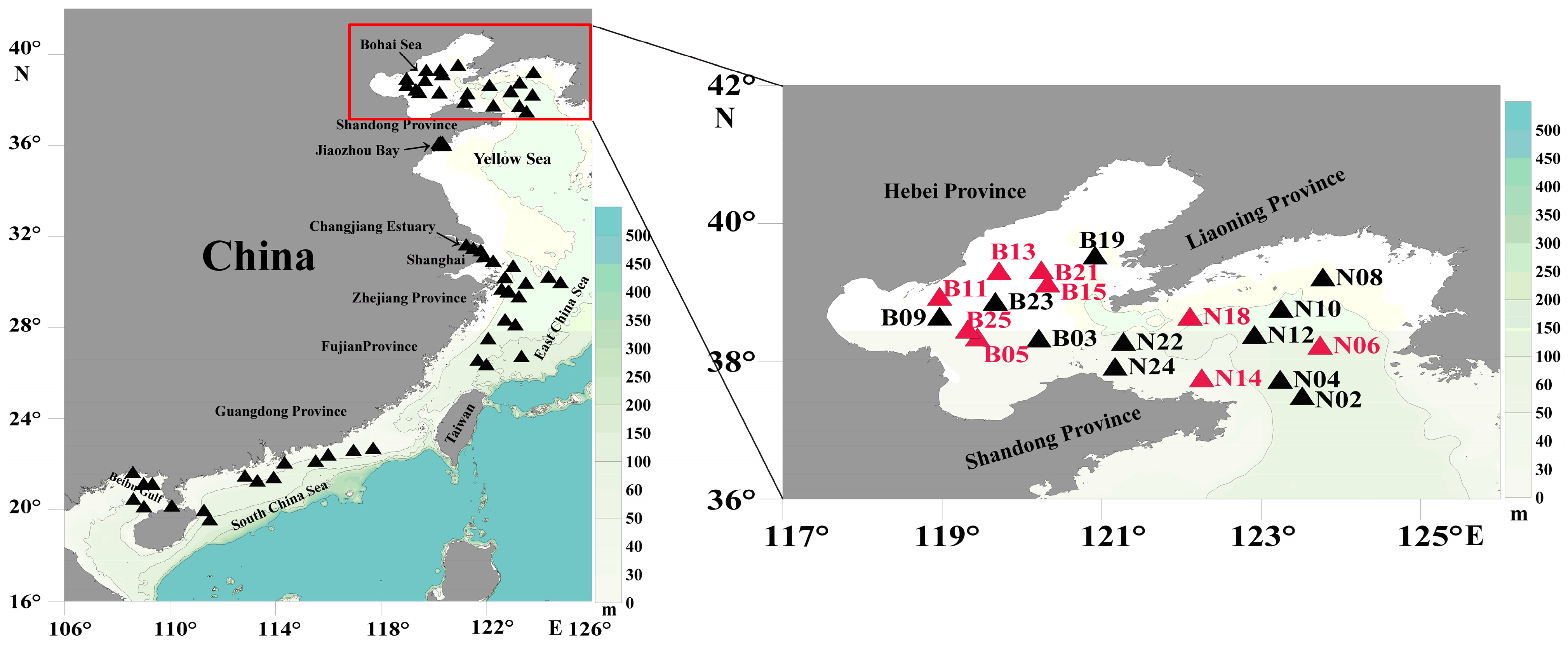
2.7. Genetic Diversity and Biogeographic Distribution Analyses Based on the Molecular Marker
3. Results
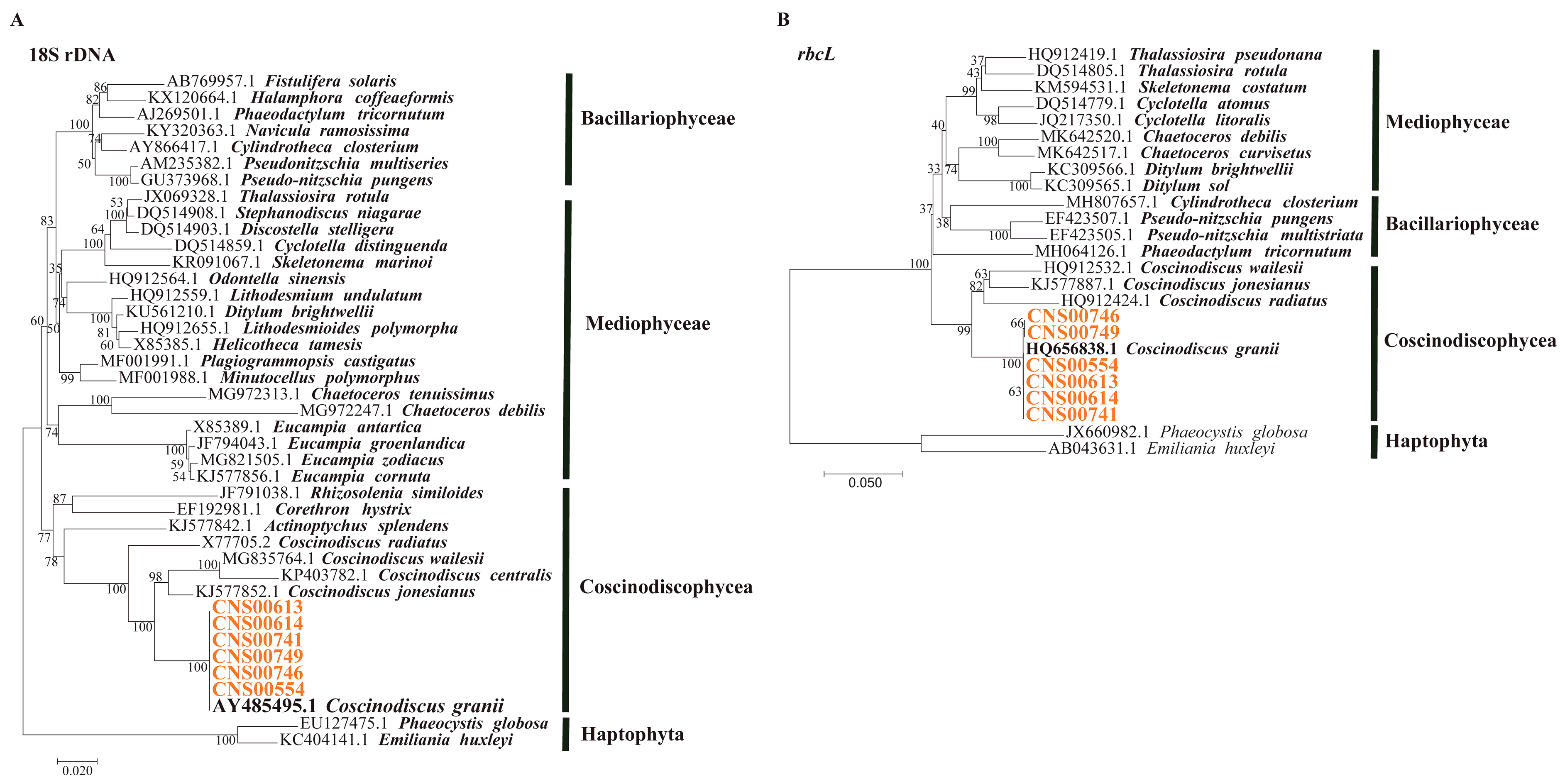
3.1. Morphological and Molecular Identification of C. granii Strains
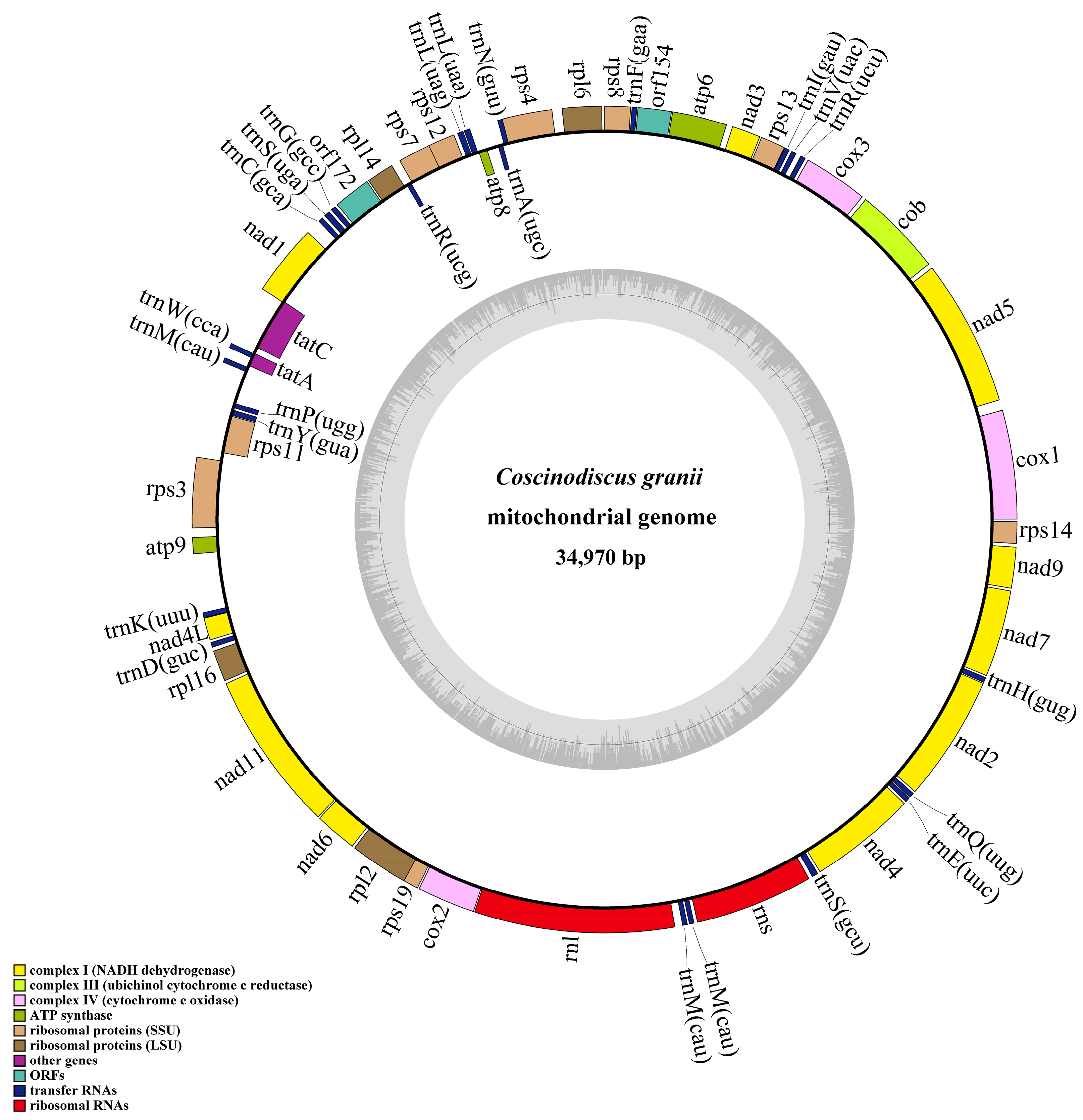
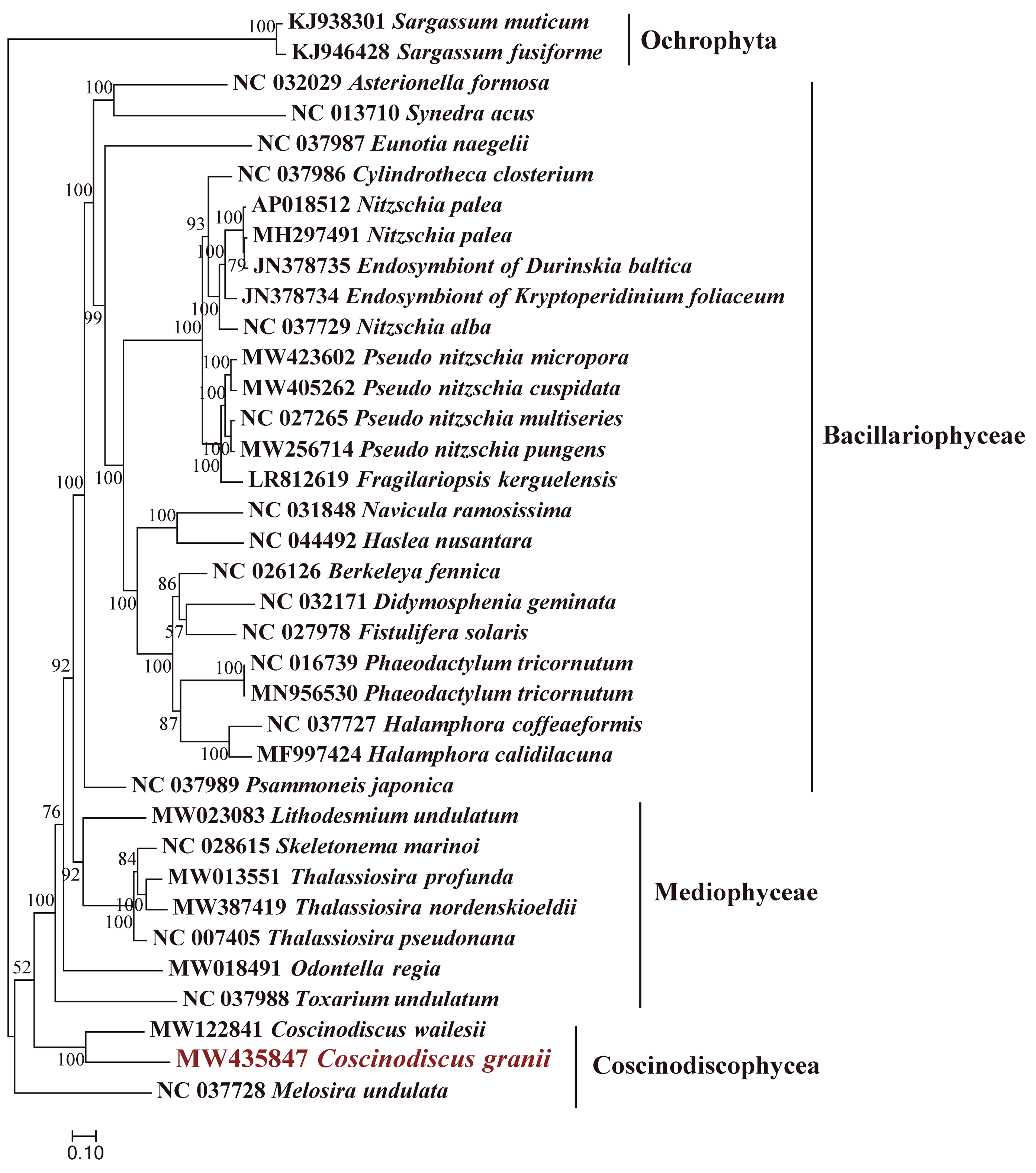
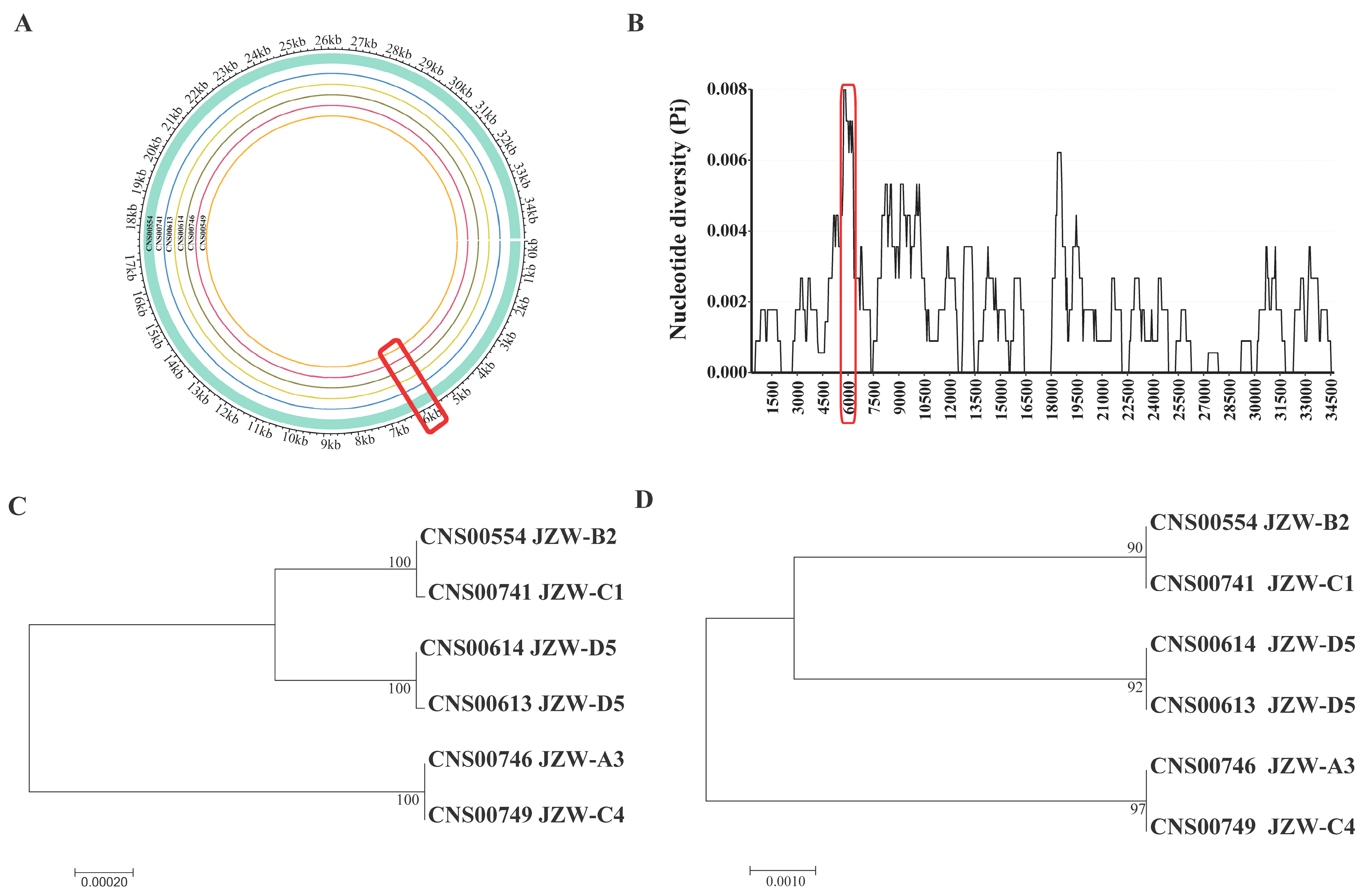
3.2. Phylogenetic Analysis and Synteny Analysis of C. granii mtDNAs
3.3. Defining a Molecular Marker with High Resolution and High Specificity for Distinguishing C. granii Strains
3.4. Probing C. granii Genetic Diversity and Geographical Distribution Using cgmt1
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Malviya, S.; Scalco, E.; Audic, S.; Vincent, F.; Veluchamy, A.; Poulain, J.; Wincker, P.; Iudicone, D.; de Vargas, C.; Bittner, L.; et al. Insights into global diatom distribution and diversity in the world’s ocean. Proc. Natl. Acad. Sci. USA 2016, 113, E1516–E1525. [Google Scholar] [CrossRef] [Green Version]
- Angela, F.; Marianne, J.; Jean-Pierre, B.; Benjamin, B.; Thomas, M. Diatom Molecular Research Comes of Age: Model Species for Studying Phytoplankton Biology and Diversity. Plant Cell 2019, 32, 547–572. [Google Scholar] [CrossRef] [Green Version]
- Mann, D.G.; Vanormelingen, P. An inordinate fondness? The number, distributions, and origins of diatom species. J. Eukaryot. Microbiol. 2013, 60, 414–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, H.; Wang, Y.; Song, H.; Wang, J.; Chen, Y.; Zhao, Y.; Liu, F.; Chen, N. The complete mitochondrial genome and phylogenetic analysis of Coscinodiscus wailesii (Coscinodiscophyceae, Bacillariophyta). Mitochondrial DNA B Res. 2021, 6, 1849–1851. [Google Scholar] [CrossRef] [PubMed]
- Tetsuya, N.; Kazutaka, M.; Satoshi, N. Effects of temperature and salinity on the growth of the giant diatom Coscinodiscus wailesii isolated from Harima-Nada, Seto Inland Sea, Japan. Nippon Suisan Gakkaishi 2000, 66, 993–998. [Google Scholar] [CrossRef] [Green Version]
- Nishikawa, T.; Tarutani, K.; Yamamoto, T. Nitrate and phosphate uptake kinetics of the harmful diatom Coscinodiscus wailesii, a causative organism in the bleaching of aquacultured Porphyra thalli. Harmful Algae 2010, 9, 563–567. [Google Scholar] [CrossRef]
- Laing, I.; Gollasch, S. Coscinodiscus wailesii—A Nuisance Diatom in European Waters. In Invasive Aquatic Species of Europe. Distribution, Impacts and Management; Springer: Amsterdam, The Netherlands, 2002. [Google Scholar]
- Yan, D.; Beardall, J.; Gao, K. Variation in cell size of the diatom Coscinodiscus granii influences photosynthetic performance and growth. Photosynth. Res. 2018, 137, 41–52. [Google Scholar] [CrossRef]
- Huang, H.; Song, H.; Zhao, Z.; Liu, F.; Chen, N. Complete mitochondrial genome of Coscinodiscus granii (Coscinodiscophyceae, Bacillariophyta). Mitochondrial DNA Part B 2021, 6, 2332–2334. [Google Scholar] [CrossRef] [PubMed]
- Wasmund, N.; Pollehne, F.; Postel, L.; Siegel, H.B.; Zettler, M.L. Assessment of the biological state of the Baltic Sea in 2007. Proc. Natl. Acad. Sci. USA 2008, 89, 8842–8846. [Google Scholar] [CrossRef] [Green Version]
- Kraberg, A.C.; Baumann, M.; Dürselen, C. Coastal Phytoplankton; Verlag Dr. Friedrich Pfei: München, Germany, 2010. [Google Scholar]
- Zhang, Q.; Yin, C.; Xu, Y.; Shi, H. The phytoplankton community sampled by Nets in the dominant area monitoring red tide in Bohai Bay in summer, 2006. J. Tianjing Univ. Sci. Technol. 2007, 22, 20–23. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, X.; Fang, E.J.; Hou, C.; Chen, W.; Zhang, B. The characteristic of phytoplankton community and its variation at artificial reef area of the coastal sea of Tianjin. Mar. Sci. Bull. 2019, 21, 41–55. [Google Scholar]
- Theriot, E.C.; Ashworth, M.; Ruck, E.; Jansen, N. A preliminary multigene phylogeny of the diatoms. Plant Ecol. Evol. 2010, 143, 278–296. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Zhang, Q.; Chen, Z.; Kong, F.; Wang, J.; Yu, R. Genetic diversity of Phaeocystis globosa strains isolated from the beibu gulf, the south China sea. Oceanol. Limnol. Sin. 2019, 50, 601–610. [Google Scholar]
- Mao, Q.; Yang, S.; Jin, W.; Yan, Z.; Yan, Y.; Jiao, L.; Xu, B.; Jiao, J. Research status and prospects for DNA barcode technology in algal taxonomy. J. Hydroecol. 2020, 41, 9–18. [Google Scholar] [CrossRef]
- Chen, Y.; Xu, Q.; Gibson, K.; Chen, N. Metabarcoding dissection of harmful algal bloom species in the East China Sea off Southern Zhejiang Province in late spring. Mar. Pollut. Bull. 2021, 169, 112586. [Google Scholar] [CrossRef]
- Song, H.; Chen, Y.; Gibson, K.; Liu, S.; Yu, Z.; Chen, N. High genetic diversity of the harmful algal bloom species Phaeocystis globosa revealed using the molecular marker COX1. Harmful Algae 2021, 107, 102065. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, Y.; Wang, J.; Liu, F.; Chen, N. Mitochondrial genome of the harmful algal bloom species Odontella regia (Mediophyceae, Bacillariophyta). J. Appl. Phycol. 2021, 33, 855–868. [Google Scholar] [CrossRef]
- Plese, B.; Kenny, N.J.; Rossi, M.E.; Cardenas, P.; Schuster, A.; Taboada, S.; Koutsouveli, V.; Riesgo, A. Mitochondrial evolution in the Demospongiae (Porifera): Phylogeny, divergence time, and genome biology. Mol. Phylogenet. Evol. 2021, 155, 107011. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Wang, Y.; Liu, K.; Liu, F.; Chen, N. Development of a high-resolution molecular marker for tracking Pseudo-nitzschia pungens genetic diversity through comparative analysis of mitochondrial genomes. J. Appl. Phycol. 2021, 33, 2283–2298. [Google Scholar] [CrossRef]
- Zhang, M.; Cui, Z.; Liu, F.; Chen, N. Definition of a High-Resolution Molecular Marker for Tracking the Genetic Diversity of the Harmful Algal Species Eucampia zodiacus Through Comparative Analysis of Mitochondrial Genomes. Front. Microbiol. 2021, 12, 631144. [Google Scholar] [CrossRef] [PubMed]
- Goessling, J.W.; Paulo, C.; Michael, K. Photo-Protection in the Centric Diatom Coscinodiscus granii is Not Controlled by Chloroplast High-Light Avoidance Movement. Front. Mar. Sci. 2016, 2, 115. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Song, H.; Liu, F.; Li, Z.; Xu, Q.; Chen, Y.; Yu, Z.; Chen, N. Development of a high-resolution molecular marker for tracking Phaeocystis globosa genetic diversity through comparative analysis of chloroplast genomes. Harmful Algae 2020, 99, 101911. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kajitani, R.; Yoshimura, D.; Okuno, M.; Minakuchi, Y.; Kagoshima, H.; Fujiyama, A.; Kubokawa, K.; Kohara, Y.; Toyoda, A.; Itoh, T. Platanus-allee is a de novo haplotype assembler enabling a comprehensive access to divergent heterozygous regions. Nat. Commun. 2019, 10, 1702. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jackman, S.D.; Vandervalk, B.P.; Mohamadi, H.; Chu, J.; Yeo, S.; Hammond, S.A.; Jahesh, G.; Khan, H.; Coombe, L.; Warren, R.L.; et al. ABySS 2.0: Resource-efficient assembly of large genomes using a Bloom filter. Genome Res. 2017, 27, 768–777. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, T.; Yamada, K.D.; Tomii, K.; Katoh, K. Parallelization of MAFFT for large-scale multiple sequence alignments. Bioinformatics 2018, 34, 2490–2492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capella-Gutierrez, S.; Silla-Martinez, J.M.; Gabaldon, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Chen, Y.; Wang, Y.; Liu, K.; Xu, Q.; Li, Y.; Chen, N. Comparative analysis of Pseudo-nitzschia chloroplast genomes revealed extensive inverted region variation and Pseudo-nitzschia speciation. Front. Mar. Sci. 2022, 9, 784579. [Google Scholar] [CrossRef]
- Liu, K.; Liu, S.; Chen, Y.; Liu, F.; Chen, N. Complete mitochondrial genome of Thalassiosira profunda (Mediophyceae, Bacillariophyta). Mitochondrial DNA Part B 2021, 6, 1560–1562. [Google Scholar] [CrossRef] [PubMed]
- Koboldt, D.C.; Zhang, Q.; Larson, D.E.; Shen, D.; McLellan, M.D.; Lin, L.; Miller, C.A.; Mardis, E.R.; Ding, L.; Wilson, R.K. VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 2022, 22, 568–576. [Google Scholar] [CrossRef] [Green Version]
- Gu, Z.; Gu, L.; Eils, R.; Schlesner, M.; Brors, B. circlize Implements and enhances circular visualization in R. Bioinformatics 2014, 30, 2811–2812. [Google Scholar] [CrossRef]
- Evans, K.M.; Wortley, A.H.; Mann, D.G. An assessment of potential diatom “barcode” genes (cox1, rbcL, 18S and ITS rDNA) and their effectiveness in determining relationships in Sellaphora (Bacillariophyta). Protist 2007, 158, 349–364. [Google Scholar] [CrossRef]
- Secq, O.L.; Green, B.R. Complex repeat structures and novel features in the mitochondrial genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana. Gene 2011, 476, 20–26. [Google Scholar] [CrossRef]
- Kamikawa, R.; Azuma, T.; Ishii, K.I.; Matsuno, Y.; Miyashita, H. Diversity of Organellar Genomes in Non-photosynthetic Diatoms. Protist 2018, 169, 351–361. [Google Scholar] [CrossRef]
- Wynn, E.L.; Christensen, A.C. Repeats of Unusual Size in Plant Mitochondrial Genomes: Identification, Incidence and Evolution. G3 2019, 9, 549–559. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Liu, S.; Huang, T.; Chen, N. Construction and comparative analysis of mitochondrial genome in the brown tide forming alga Aureococcus anophagefferens (Pelagophyceae, Ochrophyta). J. Appl. Phycol. 2020, 32, 441–450. [Google Scholar] [CrossRef]
- Pogoda, C.S.; Keepers, K.G.; Hamsher, S.E.; Stepanek, J.G.; Kane, N.C.; Kociolek, J.P. Comparative analysis of the mitochondrial genomes of six newly sequenced diatoms reveals group II introns in the barcoding region of cox1. Mitochondrial DNA Part A 2019, 30, 43–51. [Google Scholar] [CrossRef]
- Ravin, N.V.; Galachyants, Y.P.; Mardanov, A.V.; Beletsky, A.V.; Petrova, D.P.; Sherbakova, T.A.; Zakharova, Y.R.; Likhoshway, Y.V.; Skryabin, K.G.; Grachev, M.A. Complete sequence of the mitochondrial genome of a diatom alga Synedraacus and comparative analysis of diatom mitochondrial genomes. Curr. Genet. 2010, 56, 215–223. [Google Scholar] [CrossRef]
- Lei, B.; Li, S.; Liu, G.; Chen, Z.; Su, A.; Li, P.; Li, Z.; Hua, J. Evolution of mitochondrial gene content: Loss of genes, tRNAs and introns between Gossypium harknessii and other plants. Plant Syst. Evol. 2013, 299, 1889–1897. [Google Scholar] [CrossRef]
- Chen, S.; Wei, D.; Shao, R.; Shi, J.; Dou, W.; Wang, J. Evolution of multipartite mitochondrial genomes in the booklice of the genus Liposcelis (Psocoptera). BMC Genom. 2014, 15, 861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kikuchi, T.; Afrin, T.; Yoshida, M. Complete mitochondrial genomes of four entomopathogenic nematode species of the genus Steinernema. Parasit. Vectors 2016, 9, 430. [Google Scholar] [CrossRef] [PubMed]
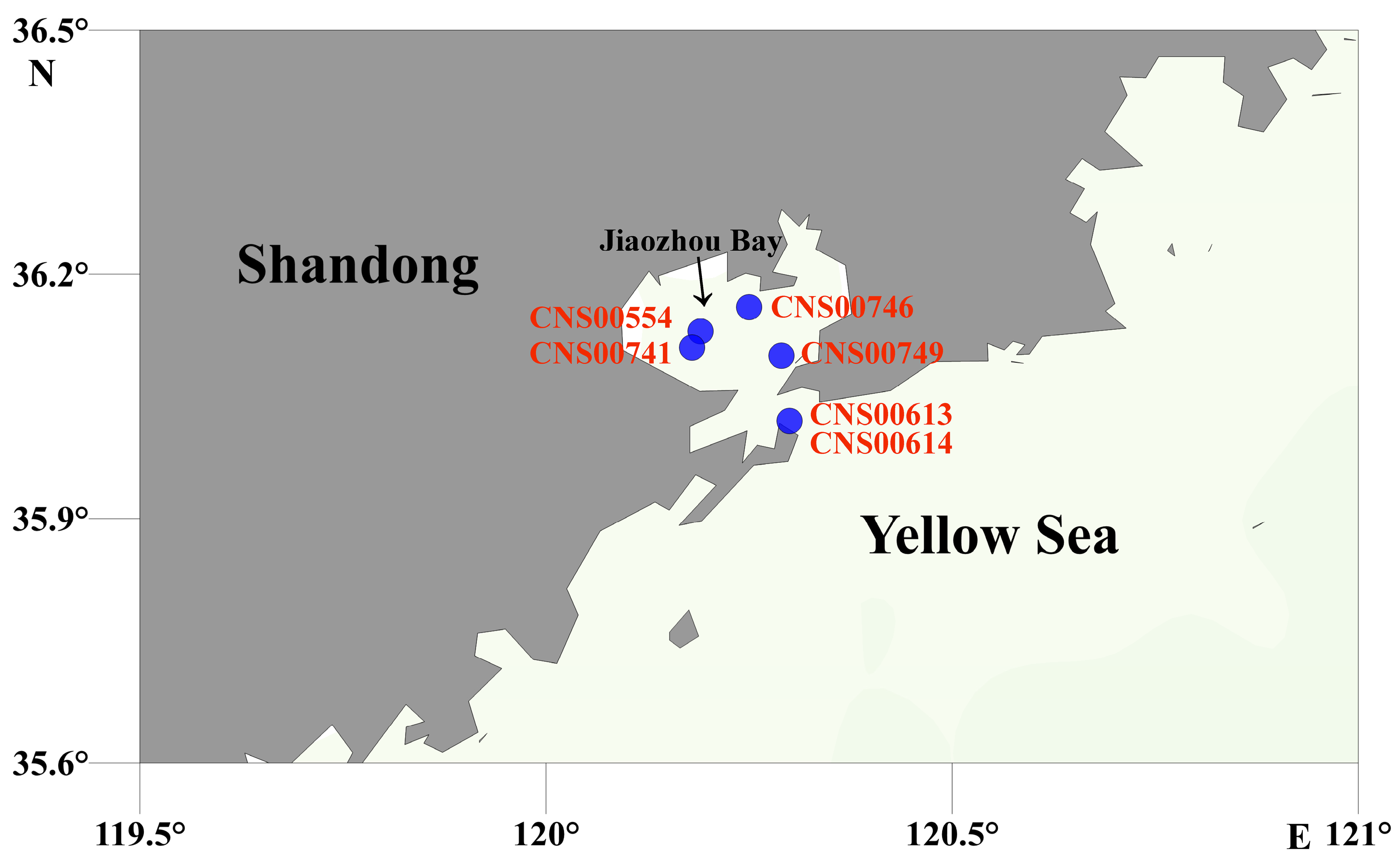

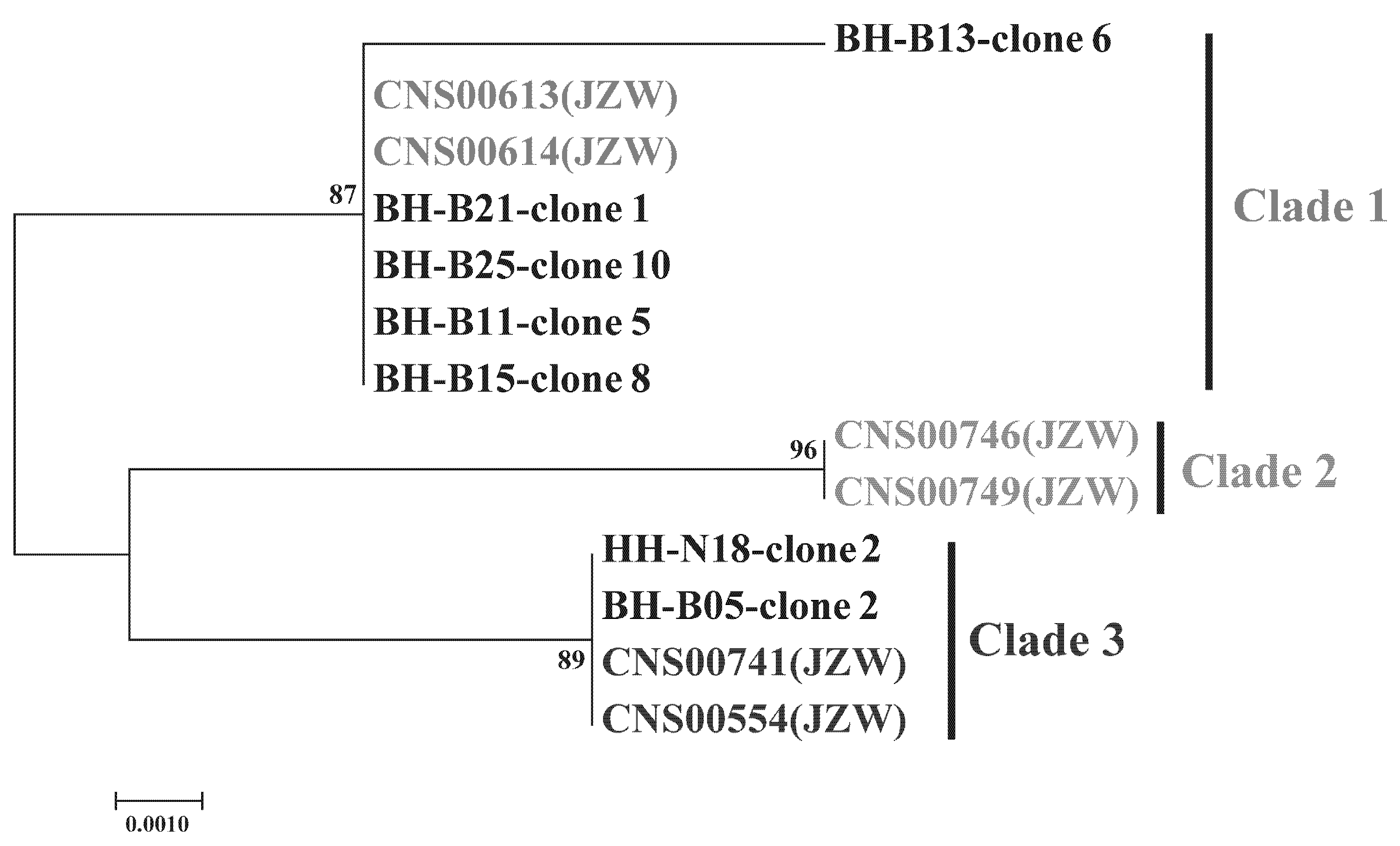
| Strain Number | Stations | Sampling Time | Longitude (°E) | Latitude (°N) |
|---|---|---|---|---|
| CNS00554 | JZW-B2 | 2020.08 | 120.19 | 36.13 |
| CNS00741 | JZW-C1 | 2020.08 | 120.18 | 36.11 |
| CNS00613 | JZW-D5 | 2020.09 | 120.30 | 36.02 |
| CNS00614 | JZW-D5 | 2020.09 | 120.30 | 36.02 |
| CNS00746 | JZW-A3 | 2020.11 | 120.25 | 36.16 |
| CNS00749 | JZW-A4 | 2020.11 | 120.29 | 36.10 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, H.; Xu, Q.; Song, H.; Chen, N. Genetic Diversity and Geographical Distribution of the Red Tide Species Coscinodiscus granii Revealed Using a High-Resolution Molecular Marker. Microorganisms 2022, 10, 2028. https://doi.org/10.3390/microorganisms10102028
Huang H, Xu Q, Song H, Chen N. Genetic Diversity and Geographical Distribution of the Red Tide Species Coscinodiscus granii Revealed Using a High-Resolution Molecular Marker. Microorganisms. 2022; 10(10):2028. https://doi.org/10.3390/microorganisms10102028
Chicago/Turabian StyleHuang, Hailong, Qing Xu, Huiyin Song, and Nansheng Chen. 2022. "Genetic Diversity and Geographical Distribution of the Red Tide Species Coscinodiscus granii Revealed Using a High-Resolution Molecular Marker" Microorganisms 10, no. 10: 2028. https://doi.org/10.3390/microorganisms10102028
APA StyleHuang, H., Xu, Q., Song, H., & Chen, N. (2022). Genetic Diversity and Geographical Distribution of the Red Tide Species Coscinodiscus granii Revealed Using a High-Resolution Molecular Marker. Microorganisms, 10(10), 2028. https://doi.org/10.3390/microorganisms10102028






