Abstract
The macroalgae surface allows specific bacterial communities to colonize, resulting in complex biological interactions. In recent years, several researchers have studied the diversity and function of the epiphytic bacteria associated with algal host, but largely these interactions remain underexplored. In the present study we analysed the cultivable diversity and polymer degradation potential of epiphytic bacteria associated with five different marine macroalgae (Sargassum, Ulva, Padina, Dictyota and Pterocladia sp.) sampled from the central west coast of India. Out of the total 360 strains isolated, purified and preserved, about 238 strains were identified through 16S rRNA gene sequence analysis and processed for polymer (cellulose, pectin, xylan and starch) degrading activities. Phylogeny placed the strains within the classes Actinobacteria, Bacilli, Alpha-proteobacteria, and Gamma-proteobacteria and clustered them into 45 genera, wherein Vibrio, Bacillus, Pseudoalteromonas, Alteromonas, Staphylococcus and Kocuria spp. were the most abundant with 20 strains identified as potentially novel taxa within the genera Bacillus, Cellulosimicrobium, Gordonia, Marinomonas, Vibrio, Luteimonas and Pseudoalteromonas. In terms of polymer hydrolysis potential, 61.3% had xylanase activity, while 59.7%, 58.8%, and 52.2% had amylase, cellulase, and pectinase activity, respectively. Overall, 75.6% of the strains degraded more than one polysaccharide, 24% degraded all polymers, while nine strains (3.8%) degraded raw sugarcane bagasse. This study showed great potential for seaweed-associated bacteria in the bio-remediation of agro-waste based raw materials, which can be employed in the form of green technology.
1. Introduction
Marine macroalgae/seaweeds that contribute to approximately half of primary sustainable productivity [1,2,3], inhabit the coastal intertidal regions. The micro-environment on algal surfaces is highly dynamic and complex due to colonization by planktonic microbes, such as bacteria, fungi, and diatoms, among other organisms [3,4,5,6,7]. They have emerged as a rich source of microbial diversity and biologically active secondary metabolites in recent years [8,9]. “Algal-microbes” interactions are facilitated through multiple and complex mechanisms involving novel bioactive compounds [10]. Physicochemical properties, metabolite composition, defense mechanism [11] and attractant patterns [12], among different groups of macroalgae (Phaeophyceae, Chlorophyceae and Rhodophyceae) prompt them to have specific and unique microbial architectures [3]. However, the majority of studies over the last decade have focused on an estimation of microbial diversity associated with macroalgae through metagenomics [6,13,14] along with a handful of culture-based approaches [9,15,16,17,18].
India produces ~350 billion tons of organic waste from agriculture [19] and most of it remains untreated and underutilized, disposed of either by burning, dumping or land filling, which leads to air and soil pollution [20,21]. The bioconversion and biodegradation of lignocellulosic compounds and the mitigation of pollution is currently a huge environmental challenge. Many reports dealing with the hydrolysis of the lignocellulosic waste by bacteria, fungi and yeast have suggested the utilization of polymers as a sole carbon and energy source [22]. The bioremediation of lignocellulosic agricultural waste pollutants with marine microorganisms, i.e., algal-associated bacteria, is feasible as they have an inherent ability to hydrolyze polymers into monomers [23]. This is also evident from several patent applications filed for microbe-derived enzymes in the diverse fields of medical, pharmacological, food and textiles [24,25,26,27]. However very few reports are available for the degradation of raw polymers i.e., sugarcane bagasse (constituent: 32–34% cellulose, 19–22% hemicellulose, 25–32% lignin, 6–12% extractives and 2–6% ash [28]), etc., by marine bacteria [29,30]. In the present study we have reported a broad diversity of cultivable epiphytic bacteria isolated from five different marine macroalgae inhabiting the intertidal zones in geographically distinct locations of the central west coast of India and explored their carbohydrate-active enzymatic (CAZymes) profiles in terms of the degradation of polymers and raw substrates.
2. Materials and Methods
2.1. Collection and Identification of Samples
Macroalgae from the different coastal locations (Table 1 and Figure 1) were collected in Nasco sampling bags (HiMedia®, Maharashtra, India) during low tide and were immersed in 500 mL of seawater. The samples were immediately transported to the laboratory in ice packs. Total DNA from the algae were extracted using modified CTAB protocol mentioned by Doyle and Doyle [31]. For the identification of macroalgae, the amplification of COX3 gene was performed using GAZF2 (5′ CCAACCAYAAAGATATWGGTAC 3′) and GAZR2 (5′ GGATGACCAAARAACCAAAA 3′) primers [32]. Amplified gene fragment (~650 bp) was purified by QiaQuick PCR purification kit (Qiagen, Hilden, Germany) and sequenced using Big-dye termination kit (ABI) according to published methods [33]. Generated raw sequence was viewed in FinchTV software version 1.4.0 for removal of ambiguous bases followed by blast of high-quality sequence in NCBI Blast server. The catalogues available from Sahoo et al., [34] and Dhargalkar et al., [35] were also used for the identification of Ulva and Dictyota sp.

Table 1.
Details of collected macroalgae samples with GPS coordinates.

Figure 1.
Photos of macroalgal samples (in-situ) and their designation code (underlined) from different locations: GAMAL (Green Algae from MALwan), MBA (Malwan Brown Algae), MRA (Malwan Red Algae), LLAB (Leaf Like sample from Anjuna Beach), SAB (Sub-tidal algae from Anjuna Beach), CDRSL (Cabo-De-Rama SampLe), LLKUN (Leaf Like sample from KUNkeshwar).
2.2. Cultivation of Epiphytic Bacteria
About 1 g of macroalgal tissue was weighed and washed with sterile distilled water to remove loosely attached microbes and debris from its surface, and vigorously vortexed with 9 mL of sterile 75% artificial sea water (ASW) to re-suspend the epibiotic bacterial community in the diluent [18]. The original suspension was serially diluted (10−1 to 10−8 times) and 100 μL from each dilution was spread plated on six different microbiological media in duplicates, i.e., soyabean casein digest agar (TSBA; HiMedia®), soyabean casein digest broth diluted 100 times with distilled water and solidified with bacteriological agar (TSBAD; HiMedia®), Zobell marine agar (MA; HiMedia®), reasoner’s 2 agar (R2A; HiMedia®), sea water complex agar medium (SWC; containing 6.05 g/L tris base, 12.35 g/L magnesium sulphate, 0.74 g/L potassium chloride, 0.13 g/L diammonium hydrogen phosphate, 17.50 g/L, sodium chloride, 0.14 g/L calcium chloride dihydrate, 1 g/L peptone, 5 g/L yeast extract, 3 mL/L glycerol, and 20 g/L bacteriological grade agar; HiMedia®), and Vaatanen nine salt solution agar medium (VNSS medium containing 17.60 g/L sodium chloride, 1.47 g/L sodium sulphate, 0.08 g/L sodium bicarbonate, 0.25 g/L potassium chloride, 0.04 g/L potassium bromide, 1.87 g/L magnesium chloride hexahydrate, 0.41 g/L calcium chloride dihydrate, 0.01 g/L strontium chloride hexahydrate, 0.01 g/L boric acid, 1 g/L peptone, 0.50 g/L yeast extract, 0.50 g/L glucose, 0.50 g/L soluble starch, 0.01 g/L ferrous sulphate heptahydrate, 0.01 g/L disodium hydrogen phosphate, and 20 g/L agar). The plates were incubated at 30 °C and the colonies were picked after every 24 h for a period of four-six weeks to take care of slow-growers. The CFUs were estimated for a period of 7 days. Bacterial strains were purified by sub-culturing through streaking on the respective fresh medium and preserved at −80 °C in 2 mL cryoprotectant vials (Tarson, 523053) by using 20% (v/v) glycerol.
2.3. Molecular Characterization of Bacterial Strains
Pure colonies of bacterial strains were subjected for genomic DNA isolation using the manual method [36]. Amplification of 16S rRNA gene using universal bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5’-GGTTACCTTGTTACGACTT-3’) was done according to established protocols [37]. The PCR product was purified using QIAquick® PCR purification kit (Qiagen). The sequencing reaction was setup as follows: template DNA (50 ng), sequencing buffer (ABI 5×; 1.5 µL), primers 533F (5′-GTGCCAGCAGCCGCGGTAA-3′), 926F (5’-AAACTCAAAGGAATTGACGG-3’) 685R (5′-TCTACGCATTTCACCGCTAC-3′) and 1100R (5′-GGGTTGCGCTCGTTG-3′) (2 picomoles) and Terminator ready reaction (TRR) mix (1 µL) for sequencing both DNA strands by dideoxy chain terminator method using the Big dye terminator kit followed by capillary electrophoresis on an ABI 3430 genetic analyzer (Applied Biosystem, Waltham, MA, USA). After quality check of raw sequences (Finch Tv software version 1.4.0), the nearly complete 16S rRNA gene sequences (~1400 bp) were subjected to BLAST analysis on EzBioCloud server [38]. The sequences displaying highest similarities affiliated to valid species names were retrieved from NCBI database. The sequences were aligned and phylogenetic analysis was done using MEGA 7.0 software [39]. The 16S rRNA gene sequence of all strains have been deposited to the NCBI gene bank server.
2.4. Polymer Hydrolysis and Raw Substrate Degradation
The strains were screened for the hydrolysis of polymer substrates (xylan from beechwood, pectin at 5 g/L; starch and cellulose (α-cellulose; crystalline) at 2 g/L; HiMedia®) in their respective growth media (TSBA, TSBAD, MA, SWC, VNSS and R2A) by incubation at 30 °C for 4–7 days followed by visual confirmation of zone clearance. For xylan and pectin, hydrolytic zone was visualized by flooding the plate with 0.1% (w/v) Congo red for 5 min, followed by decanting and washing with 5M NaCl and 1% (v/v) acetic acid. Positive strains for amylase activity (starch hydrolysis) were selected by flooding with gram’s iodine (3 g/L iodine + 2 g/L potassium iodide). Cellulose degradation (through discoloration of medium around growth) was studied in cellulose Congo red agar medium (0.5 g/L monopotassium phosphate, 0.25 g/L magnesium sulphate, 2 g/L cellulose, 0.2 g/L congo red, 2 g/L gelatin, and 15 g/L agar at pH 7.2 ± 0.2 [40,41].
The ability of strains to degrade raw substrates, i.e., sugarcane bagasse (SB) was assayed as described earlier [42]. Briefly, SB was procured from the local market and dried at 65–70 °C for 5 days in a hot air oven followed by grinding in mixer grinder (Philip). About 2% (w/v) of the SB was added in the respective agar media before autoclaving and plates incubated for 4 days were observed for halos after flooding with iodine solution (zone enhancer). For quantitative analyses, broth medium was used for determining the reduced sugar using DNS method [43].
3. Results and Discussion
3.1. Isolation, Identification and Phylogenetic Analysis
For the cultivation of a diverse group of bacterial taxa from all the macroalgal samples, identified as Sargassum polycystum, Padina antilarum, Dictyota sp. (Phaeophyta), Pterocladia musciformis (Rhodophyta), and Ulva sp. (Chlorophyta), a combination of six different media formulations were used which resulted in the isolation of 360 strains based on colony morphology and growth parameters, i.e., the time of the appearance of colonies with the perspective of including slow growers in the collection (Supplementary Table S1 and Figure S1). For the majority of the samples, the highest CFU was reported in MA medium followed by VNSS and SWC, though R2A retrieved the maximum numbers of isolates (91) followed by TSBA (67) and SWC (58) depending on several factors such as the growth of the colonies, cross contamination due to overgrowth/slime production, etc. TSBA100 had minimum CFU for most of the samples with no growth for two of the samples (GAMAL and LLKUN). This might be because of low nutrient and slow growth conditions of bacteria. Among a total of 360, 238 strains (Supplementary Table S2) were prioritized for further identification and enzymatic screening, keeping in perspective the strain details (isolation source, media used and subculture period) and logistics.
BLAST sequence alignment and phylogenetic analysis based on 16S rRNA gene sequences placed them into four different classes in the following descending order of mean abundance: Gamma-proteobacteria (39.5%), Bacilli (37.0%), Actinobacteria (18.0%) and Alpha-Proteobacteria (5.4%) (Figure 2 and Figure 3 and Supplementary Figure S2). The distribution of Gamma-proteobacteria among the samples ranged from 10% (LLAB) to 52% (GAMAL) with majority of the strains belonging to Vibrionaceae, Pseudoalteromonadaceae and Alteromonadaceae (Figure 3B). The Bacilli, Actinobacteria and Alpha-Proteobacteria abundance varied from 14% (MBA)—70% (LLAB), 13.8% (CDRSL)—28.5% (MBA) and 0% (LLKUN)—14.8% (GAMAL), respectively. Overall, the genera Vibrio (20.6%), Bacillus (12.6%), Pseudoalteromonas (7.6%), Alteromonas (6.3%), Staphylococcus (7.6%), Kocuria (4.6%), Micrococcus (2.9%), Streptomyces (2.1%), Shewanella (2.5%), and Microbacterium sp. (2.5%) were cosmopolitan in distribution and abundant in the majority of the samples (at least three), whereas Gordonia (2.1%), Neobacillus (1.3%), Psychrobacter (1.3%) and Enterobacter sp. (1.7%) were moderately abundant in few of the samples (Figure 3C). The rare taxa (<1%) Aeromonas, Brachybacterium, Brevundimonas, Catenococcus, Cellulomonas, Cellulosimicrobium, Cobetia, Domibacillus, Lederbergia, Halobacillus, Fredinandcohnia, Niallia, Metabacillus, Exiguobacterium and Fictibacillus sp., etc., (Figure 3C) were isolated from one or more samples. The genera Vibrio, Bacillus and Micrococcus formed the core and Alteromonas, Gordonia Psychrobacter, Kocuria, Microbacterium, Micrococcus, Pseudoalteromonas, Shewanella, Sphingomonas, Staphylococcus and Streptomyces were shared among two or more host samples (Figure 3C and Figure 4A and Supplementary Table S3). It was interesting to note that Sargassum polycystum sampled from three different locations (MBA, LLAB and LLKUN, Table 1) shared Vibrio, Bacillus, Pseudoalteromonas and Brevibacillus sp. constituting ≥ 50% of the total bacterial taxa thus emphasizing the importance of the phylogenetic identity of the host in selecting epiphytic microbial communities (Figure 4B) [3]. However, no unique bacterial species were observed in the Pterocladia sp. (MRA). Moreover, Padina antillarum (CDRSL) and Dictyota sp. (SAB) (had altogether distinct abundance patterns (i.e., β-diversity) and clustered separately from the other samples (Supplementary Figure S3). All three Malwan samples (MBA, MRA, GAMAL) clustered together in proximity to the Kunkeshwar sample (LLKUN) suggesting that geography along with host phylogeny influences the community composition. Considering host phylotype at the phylum level, the genera Exiguobacterium and Sanguibacter were unique to Chlorophyta (GAMAL) whereas Aeromonas, Alteromonas, Brevundimonas, Psychrobacter, Catenococcus, Cellulomonas, Gordonia, Gracibaccilus, Halobacillus, Klebsiella, Citrobacter, Cobetia, Photobacterium, Paracoccus, Fictibacillus, Domibacillus, etc. were unique to the Phaeophyta algal phyla (SAB, CDRSL, LLKUN, LLAB, MBA). The phyla Phaeophyta and Rhodophyta (MRA) shared Brachybacterium and Microbacterium while Shewanella was shared by Rhodophyta and Chlorophyta. Similarly, Streptomyces and Sphingomonas genera were shared by Phaeophyta and Chlorophyta algae (Supplementary Figure S4).
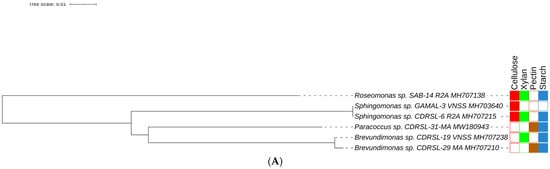
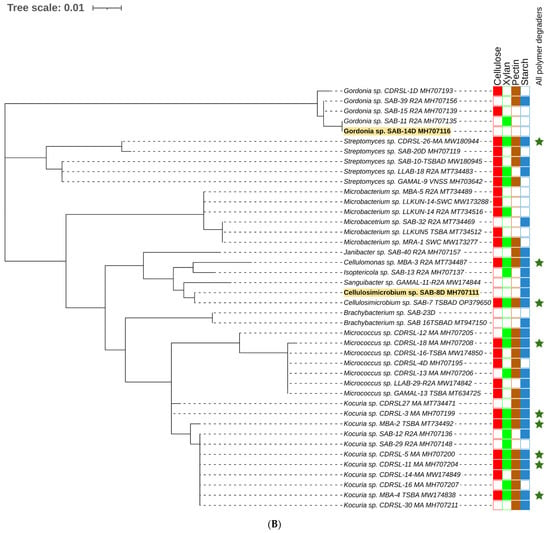
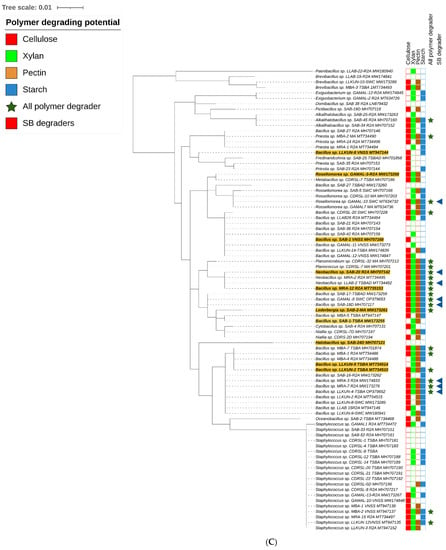
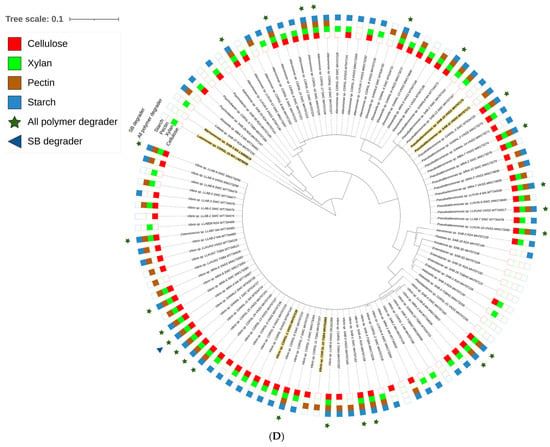
Figure 2.
Maximum likelihood method based phylogenetic sub tree of cultured strains (n = 238) belonging to (A) Alpha-proteobacteria (B) Actinobacteria, (C) Bacilli group and (D) Gamma-proteobacteria were inferred using Mega 7 software [39] considering a total of 1514 positions in final datasets with general time reversal (GTR+G+I) method. Branch length is observed as 1.81442122 and analysis involved 239 nucleotide sequences. The tree contains datasets with polymer degrading potentials represented outside taxon name in coloured blocks. Filled and blank blocks represent positive and negative activity respectively against tested substrates by the respective taxa. Strains degrading all polymers ( ) and raw sugarcane bagasse (
) and raw sugarcane bagasse ( ) are appropriately highlighted. Novel strains at genus and species level (published and putative) are highlighted with bold and yellow background. Gaps and ambiguous bases were removed from the final data sets. The final tree was visualized and edited in iTOL server.
) are appropriately highlighted. Novel strains at genus and species level (published and putative) are highlighted with bold and yellow background. Gaps and ambiguous bases were removed from the final data sets. The final tree was visualized and edited in iTOL server.
 ) and raw sugarcane bagasse (
) and raw sugarcane bagasse ( ) are appropriately highlighted. Novel strains at genus and species level (published and putative) are highlighted with bold and yellow background. Gaps and ambiguous bases were removed from the final data sets. The final tree was visualized and edited in iTOL server.
) are appropriately highlighted. Novel strains at genus and species level (published and putative) are highlighted with bold and yellow background. Gaps and ambiguous bases were removed from the final data sets. The final tree was visualized and edited in iTOL server.
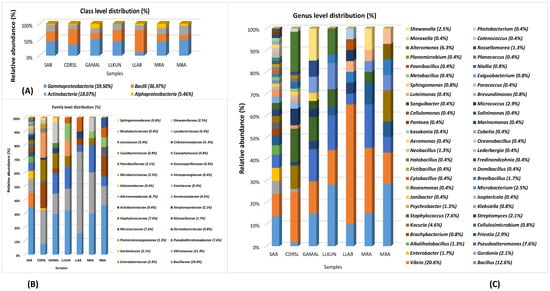
Figure 3.
Stacked bar plots of (A) Class, (B) Family and (C) genus level distribution of microbial taxa. The percent value mentioned along with the genera name signifies abundance (mean %) among all the samples.
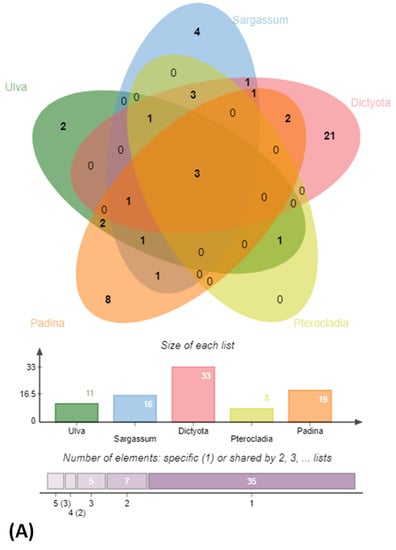
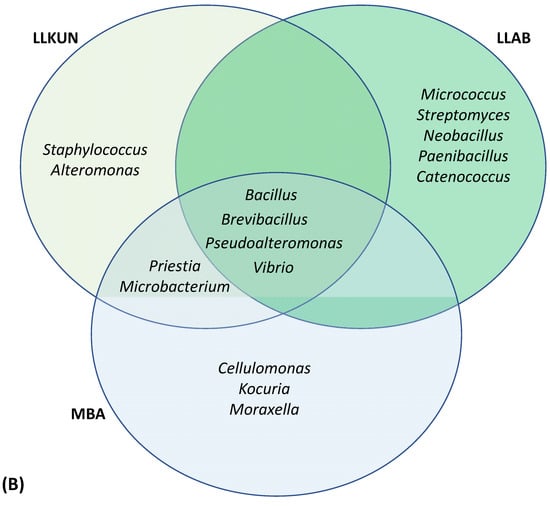
Figure 4.
Venn diagram highlighting (A) distribution of bacterial general among algal samples with barplots mentioning the total number of unique genera in each sample and (B) Identity of unique and shared bacterial genera in Sargassum polycystum sampled from 3 different locations.
Geography, sampling sites and host organism shape the abundance and occurrence of epiphytic bacterial communities [3,13,18,44,45,46,47,48,49,50,51,52]. Several culture-based studies have highlighted that the members of Proteobacteria (Vibrio; class: Gamma-proteobacteria), Actinomyctota (Micrococcus; class: Actinobacteria) and Bacilliota (Bacillus and Staphylococcus) are shared between some groups of algae (Rhodophyta, Phaeophyta and Chlorophyta) [16,53,54]. Among these group of macroalage, Vibrio is abundant in the marine ecosystem and has important role in terms of organic matter mineralization [9], pathogenesis of marine organisms [55] and the protection of macroalgae from antifouling [56,57]. Furthermore, Bacillus and Micrococcus are known to have growth and morphogenetic effects [7] along with antibacterial activity, respectively [58]. Considering geography and climate distribution, the genera Alteromonas, Bacillus, Cobetia, Labrenzia, Microbacterium, Micrococcus, Pseudoalteromonas, Shewanella, Vibrio and Arthrobacter are cosmopolitan both in the tropical and temperate environments in all the major groups of algae (red, green and brown seaweeds) (Supplementary Table S4) [9,16,52,59,60,61,62]. The genera Pseudoalteromonas and Alteromonas are involved in nutrient cycling processes through their ability to produce polysaccharide degrading enzymes [5,63]. Several Pseudoalteromonas and Alteromonas strains display algicidal activities and play an important role in protecting shellfish farms from toxic dinoflagellate blooms [9,64,65]. Pseudoalteromonas and Shewanella sp. are involved in antibacterial processes, including antifouling, and either stimulate or inhibit the settlement of zoopsores of Ulva sp. through the production of quorum sensing metabolites thus protecting the alga from pathogens, herbivores and fouling organisms and thereby making them an important part of the epiphytic community [5,66,67,68,69]. In fact, Pseudoalteromonas tunicata is a model organism for antifouling and displays activities against algal spores, larvae, diatoms, bacteria, fungi, protists and nematodes [63,70,71,72]. However, contrary to several previous reports, we could not identify any member of the phylum Bacteriodetes. One of the reasons could be increasing the sequencing depth to cover more strains and collection of fresh algal samples attached to the intertidal rocks, unlike Barbato et al., [61] and Ihua et al., [54] who had used decaying algae as the starting material, different culture conditions and algal species which targeted the isolation of algal polysaccharide degrading bacteria. Interestingly, culture independent analyses of the same samples (unpublished study) identified the classes Gamma-proteobacteria, Bacteroidetes, Alpha-Proteobacteria, Beta-Proteobacteria, Bacilli and Actinobacteria with the families Pseudoalteromonadaceae, Vibrionaceae, Flavobacteriaceae and Bacillaceae as the core community, as supported in other metabarcoding surveys [4,63,64].
Analyses of the culturable diversity identified a total of 20 strains as putative novel taxa at genus and species level indicating that marine macroalgae harbour a vast reservoir of unexplored bacterial diversity. Also as compared to terrestrial isolates, the marine counterparts represent a potential pool of novel metabolic capacities which are yet to be fully explored and exploited for industrial applications [73]. The criteria for selection of novel taxa were based on the assumption that strains sharing low 16S rRNA gene sequence identities (≤98.7%) with valid species names in the EzTaxon server. are potential candidates for novel taxa description based on correlation plot analyses between 16S rRNA gene sequence similarities and corresponding overall genome relatedness indices of GGDC and ANI [74,75]. These potential novel strains were affiliated to the phyla Firmicutes (11), Proteobacteria (7) and Actinobacteria (2) belonging to the families Bacillaceae, Oceanospirulaceae, Microbacteriaceae, Gordoniaceae, Pseudoalteromonadaceae, Micrococcaceae, Vibrionaceae and Lysobacteriaceae. Few strains have already been described as valid species names (SAB 38T; Domibacillus epiphyticus sp. nov., SAB 3T; Marinomonas epiphytica sp. nov., and CDRSL-15T; Luteimonas padinae sp. nov. [37,76,77] and the remaining are undergoing polyphasic taxonomic characterization including phylogenomic analyses to ascertain their exact taxonomic status (Supplementary Table S5 and Figure S2).
3.2. Analysis of Polymer Hydrolysis Potential
Algal-associated microbial communities are crucial for algal biomass degradation and mineralization [78,79] and can bio-remediate algal waste material [79]. In further pursuance with an intent to utilize the macroalgal-associated bacterial communities, screening for polymer hydrolysis was undertaken and, among a total of 238 strains, the majority (89%) were positive for hydrolysis of at least a single polymer with the highest activity for xylanase (61.3%) followed by amylase (59.7%) and cellulase (58.8%) (Figure 2 and Figure 5A, B). Approximately 24% of isolates were able to degrade all the screened substrates and among those belonging to Alteromonas, Pseudoalteromonas, Psychrobacter, Shewanella and Vibrio sp. (Gamma-proteobacteria); Brevundimonas, Paracoccus, Roseomonas and Sphingomonas sp. (Alpha-Proteobacteria), Bacillus and related genera, Staphylococcus, Alkalihalobacillus sp. (Firmicutes); Cellulomonas, Cellulosimicrobium, Isoptericola, Kocuria, Micrococcus and Streptomyces sp. (Actinobacteria) could degrade at least two polymers (Figure 2A–D, Supplementary Figure S5 and Table S2). The members of genera Alkalihalobacillus (1), Alteromonas (4), Bacillus (11), Cellulomonas (1), Cellulosimicrobium (1), Catenococcus (1), Kocuria (5), Lederbergia (1), Micrococcus (1), Neobacillus (3), Planococcus (1), Planomicrobium (1), Priestia (1), Pseudoalteromonas (6), Rossellomorea (1), Shewanella (1), Staphylococcus (2), Streptomyces (1), and Vibrio (13) sp. were the most potent degraders and the majority of the strains hydrolyzed all the polymers while Cobetia sp. And Luteimonas sp. Could degrade only xylan. However, polysaccharide degradation potential varied among both strain levels (Supplementary Table S2). For example, SAB 34 R2A, SAB 25 R2A and SAB 45 R2A isolates identified as Alkalihalobacillus algicola from Dictyota sp. were differentially capable of degrading 3, 1 and 4 polymers respectively and, similarly, strains CDRSL 8 VNSS, CDRSL 8 MA, CDRSL 16 SWC, and LLKUN 7 VNSS identified as Alteromonas macleodii isolated from Padina sp. and Sargassum polycystum, showed varied hydrolytic potential (Supplementary Table S2) suggesting impact of the isolation source for the induction of enzymatic activity. The highest frequency (n = 58) of polymer degraders were observed in the CDRSL samples, followed by SAB (n = 51), and GAMAL (n = 27). Several researchers have isolated bacteria belonging to the phyla Bacillaeota, Bacteriodetes, Sphingobacteria and Actinomyctota from live and decaying macroalgae and screened for the degradation of algal biomass and associated polysaccharides [18,61,80]. Barbato et al., [61] isolated and tested 634 bacterial isolates from decaying Rhodophyta and Phaeophyta for algal polysaccharide degrading activity wherein approximately 65% strains were capable of degrading at least one algal polysaccharide. Furthermore, Ihua et al., [54] reported that among 800 isolates, 7% of bacteria had polysaccharidase producing activity (cellulase, lichenase and pectinase) and Rodrigues et al., [18] reported 71% bacteria with ulvan lyase, 11.5% carbohydrate sulfatase, 32.3% cellulase and 29% with glucosidase activity. Martin et al., [10] identified association of the genera Maribacter, Algibacter, Celluophaga, Pseudoalteromonas, Vibrio, Cobetia, Shewanella, Marinomonas and Paraglaceciola sp. of the classes Flavobacteria and Gamma-proteobacteria with Ascophylum nodosum along with the degradation of marine polymeric carbon associated with macro- and microphytes. Furthermore, some other studies [13,15,16,17,52,53,54,81,82] have shown that the phyla Proteobacteria (genera Vibrio, Alteromonas Pseudoalteromonas), Bacillaeota (Bacillus, Alkalihalobacillus), Actinobacteria (Streptomyces) and Bacteroidetes (Algibacter, Zobelia, Maribacter) predominate the marine environment and can degrade agar, alginate, xylan, carrageenan, cellulose and chitin (Supplementary Table S4). Congruent to the above reports, our study retrieved a similar pattern in bacterial diversity profiles with the genera Bacillus and Vibrio sp. as the predominant taxa, except Bacteroidetes, which was entirely absent despite using media conditions (MA and seawater agar) suitable for their isolation. This could be attributed to the fact that reports highlighting their abundance used degrading algal biomass as the starting material (substrates) for isolation, therefore, the sequencing depth (total strains identified) was insufficient to cover the range of strain diversity and culture conditions relative to previous culturable diversity reports [17,75,76]. Interestingly, compared with earlier findings, the frequency of polysaccharide degraders was much higher in our work (Supplementary Figure S6). One of the aims of culture-dependent studies has been to focus on the ability of the isolated marine bacterial communities associated with sediment, water, molluscs, sponge and seaweed, etc., to bio-remediate algal waste material [17,61,79,83]. Imran et al., [82] reported that the multiple polysaccharide (cellulose, chitin, fucoidan, pectin, laminarin, pullulan, xylan, agar, alginate and starch) degraders (Microbulbifer and Sacchrophagus) were capable of decomposing red seaweed thallus. Although most of the reports targeted algal specific sulphated polymers, such as alginate, agar and carrageenan, we screened for hydrolysis of non-sulphated plant polymers (cellulose, starch, pectin and xylan) for testing our hypothesis that algal-associated bacterial communities can be exploited for the bioremediation of agricultural wastes because of similarities in structure of the two types of polymers. Incidentally, we did recover several strains with multiple polysaccharides degrading activities (56% of the total strains screened, Figure 2 and Supplementary Table S2).
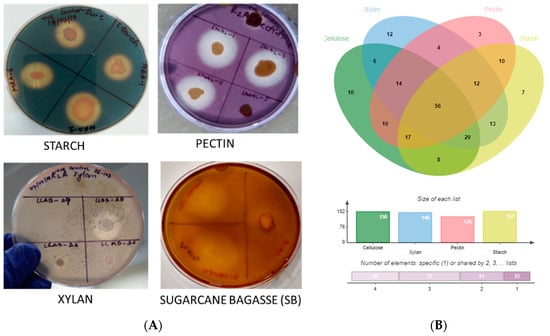
Figure 5.
(A) Zone of clearance due to hydrolysis of polymer substrates, (B) Venn diagram revealing polymer degradation patterns.
The above strains belonged to the genera Alteromonas, Bacillus, Catenococcus, Cellulomonas, Cellulosimicrobium, Kocuria, Priestia, Lederbergia, Micrococcus, Planomicrobium, Pseudoalteromonas, Shewanella, Staphylococcus, Streptomyces and Vibrio sp. capable of hydrolyzing all the four tested polysaccharides (cellulosic and hemi-cellulosic; Figure 2 and Supplementary Table S2). In the next phase, they were checked for the degradation of complex lignocellulosic plant polymer substrate, i.e., sugarcane bagasses with an aim towards bio-remediation of the agriculture waste material in plate assays. Out of the total strains tested (n = 56), nine showed an ability for the degradation of raw sugarcane bagasse without any pre-treatment (Figure 5A and Supplementary Table S6). Further quantitation experiments suggested that the strain GAMAL 10 SWC (Rosellomorea marisflavi), GAMAL 2 SWC (Vibrio owensii) and LLAB 2 TSBAD (Neobacillus derentis) produced ~0.2–0.3 g/L of reducing sugars in the medium (Figure 6) after 24 hrs of incubation. Slow hydrolysis was observed for strains SAB 18 TSBAD (Bacillus infantis) and SAB 20 R2A (Neobacillus cucumis), almost close to the negative control (SAOS 207 TSBAD; Novosphingobium arabidopsis). Recently, Gebbie et al., [84] had explored the microbial communities (fungal, bacterial and yeast) of the stored bagasse piles using mixed cultures (Bacillus, Burkholderia and Talaromyces sp.) and metabarcoding techniques and shown the abundance of bacteria [(Proteobacteria (24%), Actinobacteria (18%), Firmicutes (18), Acidobacteria (12%), Verrucomicrobia (6%) and Bacteroidetes (4%)] and fungi [(Ascomycota (87%), Basidiomycota (11%), Zygomycota (small fraction)] along with their polymer hydrolysing enzymes (cellulose, xylan, laccase and peroxidase). However, several studies related to the hydrolysis of plant-based polysaccharides, such as xylan, cellulose, starch, pectin and others using acid, organo-solvent, hydrothermal-acid-alkaline-enzymatic [85,86,87] and bacterial taxa isolated from agricultural waste landfills and other habitats have been reported, wherein pre-treated substrate were efficiently hydrolysed by enzymatic activity [88,89,90,91]. Similarly, Maeda et al., [30] and Wobiwo et al., [42] demonstrated that the hydrolysis of raw and hydrothermally pre-treated sugarcane bagasse and banana bulb could be achieved by a commercially available enzymatic cocktail (Multifect®) of fungal isolates: Penicillum funiculosum, Trichoderma harzianum and Saccharomyces cerevisiae. Pre-treatment of recalcitrant starchy lignocellulose leads to the opening of the plant cell wall structure that facilitates the enzymatic action more efficiently [92]. Several patents relating to the bacterial degradation of agricultural residue (lignocellulosic waste material) has been granted [26,27], however, in our study we have not used any pre-treatment, except sterilization of the bagasse powder in the autoclave (that might loosen the bagasse). Kunamneni et al., [93] had reported 70–80% of treated maize sugar reduction within 24 hrs using enzymatic hydrolysis, and pre-treated the substrate at 80–105 °C. Similarly, other reports have highlighted celluase, xylanase and raw biomass degrading enzymes acting on sugarcane bagasse wherein 1–5 U/g of enzyme activity per min have been achieved [94,95,96,97]. However, unlike Kunamneni et al., (70–80%) [93], we were able to achieve only 1% of reduced sugar in 24 h. This could be due to the utilization of untreated bagasse substrates that are directly recalcitrant to bacterial activity [92,98].
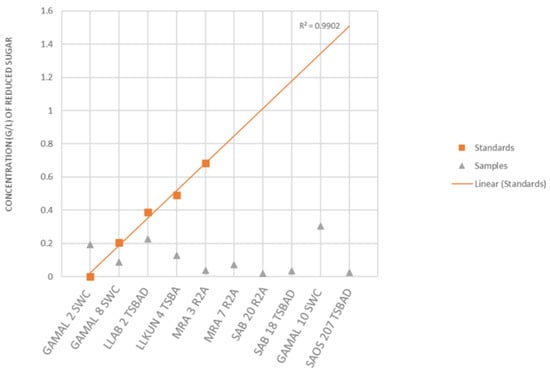
Figure 6.
Determination of reducing sugar concentration by DNS method.
4. Conclusions
In the present studythe macroalgal samples were examined for their bacterial diversity, as well as their polymer degrading potential in the context of their application related to agriculture wastes bioremediation. The highest average CFU and isolates were recovered on MA and R2A medium, respectively. Among all the macroalgal samples, the maximum and minimum diversity with respect to diversity richness and evenness was observed in Dictyota sp. and Sargassum polycystum respectively. The most frequently isolated bacteria (≥5) belonged to Vibrio, Bacillus, Pseudoalteromonas, Staphylococcus, Alteromonas, Kocuria, Micrococcus, Shewanella, Microbacterium, Streptomyces, Brevibacillus and Gordonia sp. in descending order of abundance. The genera Bacillus and Vibrio were shared among all the algal species whereas Brevibacillus and Pseudoalteromonas sp. were cosmopolitan for S. polycsytum (constituting ≥ 50% of the total community) collected from different coastal locations. The genera Micrococcus and Catenococcus were specific to S. polysystum collected from Anjuna beach (LLAB) while Gracibacillus, Alteromonas and Microbacteriun were specific to Kunkeshwar (LLKUN) and Kocuria, Enhydrobacter and Cellulomonas were uniquely observed in S. polysystum collected from the Malwan region (MBA) emphasizing that host specificity and biogeographic conditions play an important role in the selection of microbial community. About 20 novel taxa were identified in our study with three strains published as novel species, i.e., Marinomonas epiphytica sp. nov. (SAB 3T), Luteimonas padinae sp. nov. (CDRSL 15T) and Domibacillus epiphyticus sp. nov. (SAB 38T) while others await description at novel species and genus level distributed in the classes Gamma-proteobacteria (Vibrio sp., Pseudoalteromonas sp.), Bacilli (Bacillus sp.) and Actinobacteria (Microbacterium and Gordonia sp.).
Out of the total strains screened, 54% (n = 129) were positive for degrading at least three substrates, 24% (n = 56) degraded all the four polymers, and the majority (89%; n = 212) degraded at least one polymer and species in the genera Alteromonas (4), Cellulomonas (1), Cellulosimicrobium (1), Catenococcus (1), Pseudoalteromonas (6), Shewanella (1), Vibrio (13) (phylum Proteobacteria) Alkalihalobacillus (1), Bacillus (11), Lederbergia (1), Neobacillus (3), Planococcus (1), Planomicrobium (1), Priestia (1), Rossellomorea (1), Staphylococcus (2) [phylum Firmicutes], Kocuria (5), Micrococcus (1), Streptomyces (1) [phylum Actinobacteria] were the most potent degraders. In terms of degradation efficiency and scope, the genera Vibrio and Bacillus seemed best since they could hydrolyze several substrates. However, actinobacterial strains isolated in a lower frequency appeared to contain high potential, since the majority of the strains were able to degrade at least two polymers. From our preliminary screening procedures, we identified nine strains with the capability for degrading raw sugarcane bagasse (without any pre-treatment) in both plate assays and broth medium. The majority of the strains were identified as Bacillus sp. and related genera, phylogenetically clustering in three different lineages. Strains GAMAL-10 SWC (Rossellomorea marisflavi), LLKUN-4 TSBA (Bacillus pseudomycoides), LLAB-2 TSBAD (Neobacillus derentis) and GAMAL-8 SWC (Bacillus infentis) were able to maximally produce reduced sugar from raw sugarcane bagasse (0.1–0.3 g/L) in 24 h. Since these are marine strains with the ability to grow optimally at 35 ppt salinity, desiccation and moderately high temperatures, therefore high potential exists for the application of their enzyme systems as a bio-remediation option for agro-wastes in the form of green technology after process optimization. Further work is required in terms of pre-treatment and the formulation of microbial consortia/strain optimization.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10122513/s1, Supplementary Figure S1: parallel wise horizontal bar plot showing the number of isolates preserved from six different microbiological media. Supplementary Figure S2: Phylogenetic analysis of selected cultured isolates. The dataset contains sequence of representative isolates only (multiple strains having similar taxonomic identity had been removed). Strains with a lower 16S rRNA gene sequence similarity was marked in white background. Maximum likelihood method-based tree was inferred using Mega7 software (Kumar et al., 2016) with kimura-2 parameter. There was total 1486 positions in the final datasets. Gaps and ambiguous bases were removed from the final datasets. Supplementary Figure S3: representation of abundance and clustering pattern of bacterial taxa isolated among different samples based on PCA analysis. Supplementary Figure S4: shared and unique taxa among three different algal phyla. Supplementary Figure S5: a 3D bar plot showing occurrences of multi-polysaccharides degrading strains. Supplementary Figure S6: extent of polysaccharide degraders recovered in different study. Supplementary Figure S7: diversity indices showing bacterial taxa abundance, richness and evenness among the samples. Supplementary Table S1: CFU counts and total number of bacterial isolates preserved from intertidal macroalgal samples on different media. Supplementary Table S2: list of total identified strains and their polymer degrading potentials (sample wise) with NCBI accession number. Supplementary Table S3: sample wise distribution profile of unique and shared taxa. Supplementary Table S4: showing comparative analysis of the present study with existing study. Supplementary Table S5: list of potential novel isolates. Supplementary Table S6: raw sugarcane bagasse degradation potential of strains from macroalgae.
Author Contributions
A.V. and S.K. collected the samples. P.K., A.V., S.S.S. and A.K.O. were involved in strain identification and screening process. Sugarcane bagasse hydrolysis was assayed by P.K. Result interpretation and draft preparation was done by P.K. and A.V. Writing review and editing was performed by P.K., A.V., S.S.S. and S.K. Funding, project administration, and supervision was taken care by S.K. All authors have read and agreed to the published version of the manuscript.
Funding
The current study was financially supported by the Council of Scientific and Industrial Research (CSIR; grant no. BSC0402) and the Department of Biotechnology (DBT), Government of India (grant no. BT/PR7368/INF/22/177/2012) and the jointly supported program ‘Expansion and modernization of Microbial Type Culture Collection and Gene Bank (MTCC)’. P.K. is a recipient of DBT fellowship. A.V. is a recipient of a CSIR fellowship. S.S. is a recipient of CSIR fellowship. A.K.O. had worked in the DST funded GAP0128 project “Study of Diversity of marine macroalgae associated epiphytic bacteria along the Central West Coast of India”.
Data Availability Statement
16S rRNA gene sequence could be availed from NCBI server with the help of accession number mentioned in Supplementary Table S2. Other data can be found in the Supplementary File attached.
Acknowledgments
This manuscript is prepared under CSIR-IMTECH communication number 094/2020. We would like to thank Deepak Bhatt for sequencing facility.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Roy, S.; Salvi, H.; Brahmbhatt, B.; Vaghela, N.; Das, L.; Pathak, B. Diversity and Distribution of Seaweeds in Selected Reefs and Island in Gulf of Kachchh. Seaweed Res. Utiln. 2015, 37, 12–19. [Google Scholar]
- Tait, L.W.; Schiel, D.R. Dynamics of productivity in naturally structured macroalgal assemblages: Importance of canopy structure on light-use efficiency. Mar. Ecol. Prog. Ser. 2011, 421, 97–107. [Google Scholar] [CrossRef][Green Version]
- Lachnit, T.; Blümel, M.; Imhoff, J.F.; Wahl, M. Specific epibacterial communities on macroalgae: Phylogeny matters more than habitat. Aquat. Biol. 2009, 5, 181–186. [Google Scholar] [CrossRef]
- Lachnit, T.; Meske, D.; Wahl, M.; Harder, T.; Schmitz, R. Epibacterial community patterns on marine macroalgae are host-specific but temporally variable. Environ. Microbiol. 2010, 13, 655–665. [Google Scholar] [CrossRef]
- Goecke, F.; Labes, A.; Wiese, J.; Imhoff, J.F. Chemical interactions between marine macroalgae and bacteria. Mar. Ecol. Prog. Ser. 2010, 409, 267–299. [Google Scholar] [CrossRef]
- Burke, C.; Thomas, T.; Lewis, M.; Steinberg, P.; Kjelleberg, S. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 2010, 5, 590–600. [Google Scholar] [CrossRef]
- Burke, C.; Steinberg, P.; Rusch, D.; Kjelleberg, S.; Thomas, T. Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad. Sci. USA 2011, 108, 14288–14293. [Google Scholar] [CrossRef]
- Leiva, S.; Alvarado, P.; Huang, Y.; Wang, J.; Garrido, I. Diversity of pigmented Gram-positive bacteria associated with marine macroalgae from Antarctica. FEMS Microbiol. Lett. 2015, 362. [Google Scholar] [CrossRef]
- Albakosh, M.A.; Naidoo, R.K.; Kirby, B.; Bauer, R. Identification of epiphytic bacterial communities associated with the brown alga Splachnidium rugosum. J. Appl. Phycol. 2015, 28, 1891–1901. [Google Scholar] [CrossRef]
- Martin, M.; Portetelle, D.; Michel, G.; Vandenbol, M. Microorganisms living on macroalgae: Diversity, interactions, and biotechnological applications. Appl. Microbiol. Biotechnol. 2014, 98, 2917–2935. [Google Scholar] [CrossRef]
- Potin, P.; Bouarab, K.; Salaün, J.; Pohnert, G.; Kloareg, B. Biotic interactions of marine algae. Curr. Opin. Plant Biol. 2002, 5, 308–317. [Google Scholar] [CrossRef] [PubMed]
- Pasmore, M.; Costerton, J.W. Biofilms, bacterial signaling, and their ties to marine biology. J. Ind. Microbiol. Biotechnol. 2003, 30, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Florez, J.Z.; Camus, C.; Hengst, M.B.; Buschmann, A.H. A Functional Perspective Analysis of Macroalgae and Epiphytic Bacterial Community Interaction. Front. Microbiol. 2017, 8, 2561. [Google Scholar] [CrossRef]
- Selvarajan, R.; Sibanda, T.; Venkatachalam, S.; Ogola, H.J.O.; Obieze, C.C.; Msagati, T. Distribution, Interaction and Functional Profiles of Epiphytic Bacterial Communities from the Rocky Intertidal Seaweeds, South Africa. Sci. Rep. 2019, 9, 19835. [Google Scholar] [CrossRef] [PubMed]
- Abdullah, N.S.; Naim, M.A.; Mohd-Nor, N.; Abidin, Z.A.Z. Diversity of cultivable bacteria by strategic enrichment isolated from farmed edible red seaweed, Gracilaria sp. J. CleanWAS 2020, 4, 17–20. [Google Scholar] [CrossRef]
- Hinojosa, V.S.; Asenjo, J.; Leiva, S. Agarolytic culturable bacteria associated with three antarctic subtidal macroalgae. World J. Microbiol. Biotechnol. 2018, 34, 73. [Google Scholar] [CrossRef]
- Naik, M.M.; Naik, D.; Charya, L.; Mujawar, S.Y.; Vaingankar, D.C. Application of Marine Bacteria Associated with Seaweed, Ulva lactuca, for Degradation of Algal Waste. Proc. Natl. Acad. Sci. India Sect. B Boil. Sci. 2018, 89, 1153–1160. [Google Scholar] [CrossRef]
- Odaneth, A.A. Diversity of Ulvan and Cellulose Depolymerizing Bacteria Associated with the Green Macroalgae ulva Spp. J. Appl. Biotechnol. Bioeng. 2017, 2, 136–142. [Google Scholar] [CrossRef][Green Version]
- Pappu, A.; Saxena, M.; Asolekar, S.R. Solid wastes generation in India and their recycling potential in building materials. Build. Environ. 2007, 42, 2311–2320. [Google Scholar] [CrossRef]
- Bernard, S.; Kazmin, A. Dirty Air: How India Became the Most Polluted Country on Earth. Financ. Times 2018, 12. [Google Scholar]
- Iqbal, N.; Agrawal, A.; Dubey, S.; Kumar, J. Role of Decomposers in Agricultural Waste Management. In Biotechnological Applications of Biomass; IntechOpen: London, UK, 2020. [Google Scholar] [CrossRef]
- Kumar, A.; Chandra, R. Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon 2020, 6, e03170. [Google Scholar] [CrossRef] [PubMed]
- Nayak, S.; Mukherjee, A.K. Management of Agricultural Wastes Using Microbial Agents. Waste Manag. Chall. Threat. Oppor. 2015, 65–91. [Google Scholar]
- Santanna, L.; Maria, M.; Pereira, N.; Bitancur, G.; Jaime, V.; Bevilaqua, J. Process for the Fermentative Production of Ethanol by Pichia Stipitis from the Hemicellulose Hydrolysate of Sugarcane Bagasse. WO Patent WO 2009/004273 Al, 8 January 2009. [Google Scholar]
- Rarbach, M.; Dragovic, Z. Efficient Lignocellulose Hydrolysis with Integrated Enzyme Production. EP Patent EP 2 471 940 A1, 2007. [Google Scholar]
- Dottori, F.A.; Ashley, R.; Benson, C.; Ca, B.; Benech, R. Bagasse Fractionation for Cellulosc Ethanoland Chemical Production. U.S. Patent 9,187,862 B2, 17 November 2015. [Google Scholar]
- Delmas, M.; Benjelloum, B.; Mlayah, B.B. Process for Producing Bioethanol by Enzymatic Hydrolysis of Cellulose. U.S. Patent 9,518,274B2, 13 December 2016. [Google Scholar] [CrossRef]
- Sakdaronnarong, C.; Jonglertjunya, W. Rice straw and sugarcane bagasse degradation mimicking lignocellulose decay in nature: An alternative approach to biorefinery. ScienceAsia 2012, 38, 364–372. [Google Scholar] [CrossRef]
- Bian, J.; Peng, P.; Peng, F.; Xiao, X.; Xu, F.; Sun, R.-C. Microwave-assisted acid hydrolysis to produce xylooligosaccharides from sugarcane bagasse hemicelluloses. Food Chem. 2014, 156, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Maeda, R.N.; Serpa, V.I.; Rocha, V.A.L.; Mesquita, R.A.A.; De Santa Anna, L.M.M.S.; Castro, A.M.; Driemeier, C.E.; Pereira, N.; Polikarpov, I. Enzymatic hydrolysis of pretreated sugar cane bagasse using Penicillium funiculosum and Trichoderma harzianum cellulases. Process. Biochem. 2011, 46, 1196–1201. [Google Scholar] [CrossRef]
- Doyle, J.; Doyle, J. A Rapid DNA Isolation Procedure for Small Quantities of Fresh Leaf Tissue. Phytochem. Bull. 1987, 19, 11–15. [Google Scholar]
- Saunders, G.W.; McDevit, D.C. Methods for DNA Barcoding Photosynthetic Protists Emphasizing the Macroalgae and Diatoms. Methods Mol. Biol. 2012, 858, 207–222. [Google Scholar] [CrossRef]
- Verma, A.; Sundharam, S.S.; Pal, Y.; Bisht, B.; Yadav, P.; Krishnamurthi, S. Yangia mangrovi sp. nov., a novel member of the Roseobacter clade isolated from mangrove soil and emended description of Yangia pacifica Dai et al. 2006. Int. J. Syst. Evol. Microbiol. 2021, 71, 005021. [Google Scholar] [CrossRef]
- Sahoo, D.; Debasish, N. Seaweeds of Indian Coast; APH Pub. Corp.: Delhi, India, 2001. [Google Scholar]
- Dhargalkar, V.; Aquaculture, X.V. Undefined Southern Ocean Seaweeds: A Resource for Exploration in Food and Drugs; Elsevier: Amsterdam, The Netherlands, 2009. [Google Scholar]
- Pitcher, D.G.; Saunders, N.A.; Owen, R.J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett. Appl. Microbiol. 1989, 8, 151–156. [Google Scholar] [CrossRef]
- Ojha, A.K.; Verma, A.; Pal, Y.; Bhatt, D.; Mayilraj, S.; Krishnamurthi, S. Marinomonas epiphytica sp. nov., isolated from a marine intertidal macroalga. Int. J. Syst. Evol. Microbiol. 2017, 67, 2746–2751. [Google Scholar] [CrossRef]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Hendricks, C.W.; Doyle, J.D.; Hugley, B. A new solid medium for enumerating cellulose-utilizing bacteria in soil. Appl. Environ. Microbiol. 1995, 61, 2016–2019. [Google Scholar] [CrossRef]
- Catherine, G.V. Isolation of Cellulose Degrading Bacteria and Yeasts From Pineapple Waste. Int. J. Curr. Res. Rev. 2012, 4, 7. [Google Scholar]
- Awedem Wobiwo, F.; Chaturvedi, T.; Boda, M.; Fokou, E.; Emaga, T.H.; Cybulska, I.; Deleu, M.; Gerin, P.A.; Thomsen, M.H. Bioethanol Potential of Raw and Hydrothermally Pretreated Banana Bulbs Biomass in Simultaneous Saccharification and Fermentation Process with Saccharomyces Cerevisiae. Biomass Convers. Biorefinery 2019, 9, 553–563. [Google Scholar] [CrossRef]
- Garriga, M.; Almaraz, M.; Marchiaro, A. Determination of Reducing Sugars in Extracts of Undaria Pinnatifida (Harvey) Algae by UV-Visible Spectrophotometry (DNS Method) Determinación de Azúcares Reduc-tores En Extractos de Alga Undaria Pinnatifida (Harvey) Por Espectrofotometriía UV-Visible (Meétodo DNS). Actas Ing. 2017, 3, 173–179. [Google Scholar]
- Lyngwi, N.A.; Koijam, K.; Sharma, D.; Joshi, S.R. Cultivable bacterial diversity along the altitudinal zonation and vegetation range of tropical Eastern Himalaya. Rev. Biol. Trop. 2013, 61, 467–490. [Google Scholar] [CrossRef]
- Keller, A.G.; Apprill, A.; Lebaron, P.; Robbins, J.; Romano, T.A.; Overton, E.; Rong, Y.; Yuan, R.; Pollara, S.; Whalen, E.K. Characterizing the culturable surface microbiomes of diverse marine animals. FEMS Microbiol. Ecol. 2021, 97, fiab040. [Google Scholar] [CrossRef]
- Jan, B.; Reshi, Z.A.; Mohiddin, F.A. Site and Organ-Specific Culture-Dependent Endophytic Diversity of Crocus sativus L. (Saffron) in Kashmir Himalaya, India. Microb. Ecol. 2021, 83, 989–1006. [Google Scholar] [CrossRef]
- Aires, T.; Serrão, E.A.; Engelen, A.H. Host and Environmental Specificity in Bacterial Communities Associated to Two Highly Invasive Marine Species (Genus Asparagopsis). Front. Microbiol. 2016, 7, 559. [Google Scholar] [CrossRef]
- Tang, J.-C.; Taniguchi, H.; Chu, H.; Zhou, Q.; Nagata, S. Isolation and characterization of alginate-degrading bacteria for disposal of seaweed wastes. Lett. Appl. Microbiol. 2009, 48, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Kizhakkekalam, V.K.; Chakraborty, K. Pharmacological properties of marine macroalgae-associated heterotrophic bacteria. Arch. Microbiol. 2018, 201, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Kai, W.; Peisheng, Y.; Rui, M.; Wenwen, J.; Zongze, S. Diversity of culturable bacteria in deep-sea water from the South Atlantic Ocean. Bioengineered 2017, 8, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Beleneva, I.A.; Zhukova, N.V. Bacterial communities of some brown and red algae from Peter the Great Bay, the Sea of Japan. Microbiology 2006, 75, 348–357. [Google Scholar] [CrossRef]
- Menezes, C.B.; Bonugli-Santos, R.C.; Miqueletto, P.B.; Passarini, M.R.; Silva, C.H.; Justo, M.R.; Leal, R.R.; Fantinatti-Garboggini, F.; Oliveira, V.M.; Berlinck, R.G.; et al. Microbial diversity associated with algae, ascidians and sponges from the north coast of São Paulo state, Brazil. Microbiol. Res. 2010, 165, 466–482. [Google Scholar] [CrossRef] [PubMed]
- Alvarado, R.; Leiva, S. Agar-degrading bacteria isolated from Antarctic macroalgae. Folia Microbiol. 2017, 62, 409–416. [Google Scholar] [CrossRef] [PubMed]
- Ihua, M.W.; Guihéneuf, F.; Mohammed, H.; Margassery, L.M.; Jackson, S.A.; Stengel, D.B.; Clarke, D.J.; Dobson, A.D.W. Microbial Population Changes in Decaying Ascophyllum nodosum Result in Macroalgal-Polysaccharide-Degrading Bacteria with Potential Applicability in Enzyme-Assisted Extraction Technologies. Mar. Drugs 2019, 17, 200. [Google Scholar] [CrossRef]
- Reen, F.J.; Almagro-Moreno, S.; Ussery, D.; Boyd, E.F. The genomic code: Inferring Vibrionaceae niche specialization. Nat. Rev. Genet. 2006, 4, 697–704. [Google Scholar] [CrossRef]
- Dobretsov, S.; Abed, R.M.; Teplitski, M. Mini-review: Inhibition of biofouling by marine microorganisms. Biofouling 2013, 29, 423–441. [Google Scholar] [CrossRef]
- Dobretsov, S.; Dahms, H.-U.; Qian, P.-Y. Inhibition of biofouling by marine microorganisms and their metabolites. Biofouling 2006, 22, 43–54. [Google Scholar] [CrossRef]
- Sinimol, S.; Sarika, A.R.; Nair, A.J. Diversity and antagonistic potential of marine microbes collected from south-west coast of India. 3 Biotech 2015, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Vieira, C.; Engelen, A.; Guentas, L.; Aires, T.; Houlbreque, F.; Gaubert, J.; Serrao, E.; De Clerck, O.; Payri, C. Species Specificity of Bacteria Associated to the Brown Seaweeds Lobophora (Dictyotales, Phaeophyceae) and Their Potential for Induction of Rapid Coral Bleaching in Acropora muricata. Front. Microbiol. 2016, 7, 316. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Chen, L.; Zhang, Z.; Wang, X.; Qin, S.; Yan, P. Screening of alginate lyase-excreting microorganisms from the surface of brown algae. AMB Express 2017, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Barbato, M.; Vacchini, V.; Engelen, A.H.; Patania, G.; Mapelli, F.; Borin, S.; Crotti, E. What lies on macroalgal surface: Diversity of polysaccharide degraders in culturable epiphytic bacteria. AMB Express 2022, 12, 98. [Google Scholar] [CrossRef]
- Hollants, J.; Leliaert, F.; De Clerck, O.; Willems, A. What We Can Learn from Sushi: A Review on Sea-weed-Bacterial Associations. FEMS Microbiol. Ecol. 2013, 83, 1–16. [Google Scholar] [CrossRef]
- Bowman, J.P. Bioactive Compound Synthetic Capacity and Ecological Significance of Marine Bacterial Genus. Pseudoalteromonas 2007, 5, 220–241. [Google Scholar] [CrossRef]
- Skerratt, J.H.; Bowman, J.; Hallegraeff, G.; James, S.; Nichols, P.D. Algicidal bacteria associated with blooms of a toxic dinoflagellate in a temperate Australian estuary. Mar. Ecol. Prog. Ser. 2002, 244, 1–15. [Google Scholar] [CrossRef]
- Lee, B.-K.; Katano, T.; Kitamura, S.-I.; Oh, M.-J.; Han, M.-S. Monitoring of Algicidal Bacterium, Alteromonas sp. Strain A14 in Its Application to Natural Cochlodinium Polykrikoides Blooming Seawater Using Fluorescence in Situ Hybridization. J. Microbiol. 2008, 46, 274–282. [Google Scholar] [CrossRef]
- Egan, S.; James, S.; Holmström, C.; Kjelleberg, S. Inhibition of Algal Spore Germination by the Marine Bacterium Pseudoalteromonas Tunicata. FEMS Microbiol. Ecol. 2001, 35, 67–73. [Google Scholar] [CrossRef]
- Dobretsov, S.V.; Qian, P.-Y. Effect of Bacteria Associated with the Green Alga Ulva reticulata on Marine Micro- and Macrofouling. Biofouling 2002, 18, 217–228. [Google Scholar] [CrossRef]
- Patel, P.; Callow, M.E.; Joint, I.; Callow, J.A. Specificity in the settlement-modifying response of bacterial biofilms towards zoospores of the marine alga Enteromorpha. Environ. Microbiol. 2003, 5, 338–349. [Google Scholar] [CrossRef] [PubMed]
- Kanagasabhapathy, M.; Sasaki, H.; Haldar, S.; Yamasaki, S.; Nagata, S. Antibacterial activities of marine epibiotic bacteria isolated from brown algae of Japan. Ann. Microbiol. 2006, 56, 167–173. [Google Scholar] [CrossRef]
- Egan, S.; Thomas, T.; Kjelleberg, S. Unlocking the diversity and biotechnological potential of marine surface associated microbial communities. Curr. Opin. Microbiol. 2008, 11, 219–225. [Google Scholar] [CrossRef] [PubMed]
- Matz, C.; Webb, J.; Schupp, P.J.; Phang, S.Y.; Penesyan, A.; Egan, S.; Steinberg, P.; Kjelleberg, S. Marine Biofilm Bacteria Evade Eukaryotic Predation by Targeted Chemical Defense. PLoS ONE 2008, 3, e2744. [Google Scholar] [CrossRef] [PubMed]
- Ballestriero, F.; Thomas, T.; Burke, C.; Egan, S.; Kjelleberg, S. Identification of Compounds with Bioactivity against the Nematode Caenorhabditis elegans by a Screen Based on the Functional Genomics of the Marine Bacterium Pseudoalteromonas tunicata D2. Appl. Environ. Microbiol. 2010, 76, 5710–5717. [Google Scholar] [CrossRef]
- Prakash, O.; Nimonkar, Y.; Shouche, Y.S. Practice and prospects of microbial preservation. FEMS Microbiol. Lett. 2012, 339, 1–9. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Chun, J.; Oren, A.; Ventosa, A.; Christensen, H.; Arahal, D.R.; Da Costa, M.S.; Rooney, A.P.; Yi, H.; Xu, X.-W.; De Meyer, S.; et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018, 68, 461–466. [Google Scholar] [CrossRef]
- Verma, A.; Ojha, A.K.; Dastager, S.G.; Natarajan, R.; Mayilraj, S.; Krishnamurthi, S. Domibacillus mangrovi sp. nov. and Domibacillus epiphyticus sp. nov., isolated from marine habitats of the central west coast of India. Int. J. Syst. Evol. Microbiol. 2017, 67, 3063–3070. [Google Scholar] [CrossRef]
- Verma, A.; Ojha, A.K.; Kumari, P.; Sundharam, S.S.; Mayilraj, S.; Krishnamurthi, S.; Mual, P. Luteimonas padinae sp. nov., an epiphytic bacterium isolated from an intertidal macroalga. Int. J. Syst. Evol. Microbiol. 2016, 66, 5444–5451. [Google Scholar] [CrossRef]
- Brunet, M.; Le Duff, N.; Barbeyron, T.; Thomas, F. Consuming fresh macroalgae induces specific catabolic pathways, stress reactions and Type IX secretion in marine flavobacterial pioneer degraders. ISME J. 2022, 16, 2027–2039. [Google Scholar] [CrossRef] [PubMed]
- Muñoz, C.; Hidalgo, C.; Zapata, M.; Jeison, D.; Riquelme, C.; Rivas, M. Use of Cellulolytic Marine Bacteria for Enzymatic Pretreatment in Microalgal Biogas Production. Appl. Environ. Microbiol. 2014, 80, 4199–4206. [Google Scholar] [CrossRef] [PubMed]
- Naik, M.M.; Dubey, S.K. Marine Pollution and Microbial Remediation; Springer: Berlin/Heidelberg, Germany, 2017. [Google Scholar] [CrossRef]
- Martin, M.; Barbeyron, T.; Martin, R.; Portetelle, D.; Michel, G.; Vandenbol, M. The Cultivable Surface Microbiota of the Brown Alga Ascophyllum nodosum is Enriched in Macroalgal-Polysaccharide-Degrading Bacteria. Front. Microbiol. 2015, 6, 1487. [Google Scholar] [CrossRef] [PubMed]
- Imran; Poduval, P.B.; Ghadi, S.C. Bacterial Degradation of Algal Polysaccharides in Marine Ecosystem. In Marine Pollution and Microbial Remediation; Springer: Singapore, 2016; pp. 189–203. ISBN 9789811010446. [Google Scholar]
- Comba-González, N.B.; Ruiz-Toquica, J.S.; Lopez-Kleine, L.; Montoya-Castano, D. Epiphytic Bacteria of Macroalgae of the Genus Ulva and their Potential in Producing Enzymes Having Biotechnological Interest. J. Mar. Biol. Oceanogr. 2016, 5, 2–9. [Google Scholar] [CrossRef]
- Gebbie, L.; Dam, T.T.; Ainscough, R.; Palfreyman, R.; Cao, L.; Harrison, M.; O’Hara, I.; Speight, R. A snapshot of microbial diversity and function in an undisturbed sugarcane bagasse pile. BMC Biotechnol. 2020, 20, 12. [Google Scholar] [CrossRef]
- Laopaiboon, P.; Thani, A.; Leelavatcharamas, V.; Laopaiboon, L. Acid hydrolysis of sugarcane bagasse for lactic acid production. Bioresour. Technol. 2010, 101, 1036–1043. [Google Scholar] [CrossRef]
- Mesa, L.; González, E.; Cara, C.; Castro, E.; Mussatto, S. The effect of organosolv pretreatment variables on enzymatic hydrolysis of sugarcane bagasse. Chem. Eng. J. 2011, 168, 1157–1162. [Google Scholar] [CrossRef]
- Guilherme, A.A.; Dantas, P.V.F.; Santos, E.; Fernandes, F.; Macedo, G.R. Evaluation of composition, characterization and enzymatic hydrolysis of pretreated sugar cane bagasse. Braz. J. Chem. Eng. 2015, 32, 23–33. [Google Scholar] [CrossRef]
- Gaur, R.; Tiwari, S.; Pathak, P. Sugarcane Baggase Agro-waste Material Used for Renewable Cellulase Production from Streptococcus and Bacillus sp. Res. J. Microbiol. 2017, 12, 255–265. [Google Scholar] [CrossRef][Green Version]
- Nargotra, P.; Vaid, S.; Bajaj, B.K. Cellulase Production from Bacillus subtilis SV1 and Its Application Potential for Saccharification of Ionic Liquid Pretreated Pine Needle Biomass under One Pot Consolidated Bioprocess. Fermentation 2016, 2, 19. [Google Scholar] [CrossRef]
- Talia, P.; Sede, S.M.; Campos, E.; Rorig, M.; Principi, D.; Tosto, D.; Hopp, H.E.; Grasso, D.; Cataldi, A. Biodiversity characterization of cellulolytic bacteria present on native Chaco soil by comparison of ribosomal RNA genes. Res. Microbiol. 2012, 163, 221–232. [Google Scholar] [CrossRef] [PubMed]
- Bharathiraja, S.; Suriya, J.; Krishnan, M.; Manivasagan, P.; Kim, S.-K. Production of Enzymes from Agricultural Wastes and Their Potential Industrial Applications. In Advances in Food and Nutrition Research; Elsevier BV: Amsterdam, The Netherlands, 2017; Volume 80, pp. 125–148. [Google Scholar]
- Taheri, M.E.; Salimi, E.; Saragas, K.; Novakovic, J.; Barampouti, E.M.; Mai, S.; Malamis, D.; Moustakas, K.; Loizidou, M. Effect of pretreatment techniques on enzymatic hydrolysis of food waste. Biomass-Convers. Biorefinery 2020, 11, 219–226. [Google Scholar] [CrossRef]
- Kunamneni, A.; Singh, S. Response surface optimization of enzymatic hydrolysis of maize starch for higher glucose production. Biochem. Eng. J. 2005, 27, 179–190. [Google Scholar] [CrossRef]
- Bohra, V.; Dafale, N.A.; Hathi, Z.; Purohit, H.J. Genomic annotation and validation of bacterial consortium NDMC-1 for enhanced degradation of sugarcane bagasse. Ann. Microbiol. 2019, 69, 695–711. [Google Scholar] [CrossRef]
- Di Marco, E.; Soraire, P.M.; Romero, C.M.; Villegas, L.B.; Martínez, M.A. Raw sugarcane bagasse as carbon source for xylanase production by Paenibacillus species: A potential degrader of agricultural wastes. Environ. Sci. Pollut. Res. 2017, 24, 19057–19067. [Google Scholar] [CrossRef] [PubMed]
- Delabona, P.D.S.; Pirota, R.D.B.; Codima, C.A.; Tremacoldi, C.R.; Rodrigues, A.; Farinas, C.S. Using Amazon forest fungi and agricultural residues as a strategy to produce cellulolytic enzymes. Biomass-Bioenergy 2012, 37, 243–250. [Google Scholar] [CrossRef]
- Dos Santos, B.S.L.; Gomes, A.F.S.; Franciscon, E.G.; Oliveira, J.; Baffi, M.A. Thermotolerant and mesophylic fungi from sugarcane bagasse and their prospection for biomass-degrading enzyme production. Braz. J. Microbiol. 2015, 46, 903–910. [Google Scholar] [CrossRef]
- Popper, Z.A.; Michel, G.; Hervé, C.; Domozych, D.S.; Willats, W.G.; Tuohy, M.G.; Kloareg, B.; Stengel, D.B. Evolution and Diversity of Plant Cell Walls: From Algae to Flowering Plants. Annu. Rev. Plant Biol. 2011, 62, 567–590. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).