Abstract
Microorganisms are critical drivers of biological processes that contribute significantly to plant sustainability and productivity. In recent years, emerging research on plant holobiont theory and microbial invasion ecology has radically transformed how we study plant–microbe interactions. Over the last few years, we have witnessed an accelerating pace of advancements and breadth of questions answered using omic technologies. Herein, we discuss how current state-of-the-art genomics, transcriptomics, proteomics, and metabolomics techniques reliably transcend the task of studying plant–microbe interactions while acknowledging existing limitations impeding our understanding of plant holobionts.
1. Introduction
Technological advancements in high-throughput genome sequencing and mass spectrometry have transformed biological sciences. Today, there is an ever-growing list of revolutionary approaches with the suffix-omics [1] that extends beyond those derived from the central dogma, i.e., genomics, transcriptomics, proteomics, and metabolomics. The application of these technologies in plant sciences has transformed our understanding of plant–microbe interactions [2]. Since the publication of the Arabidopsis thaliana microbiome [3], providing our first detailed look at this complex microbial world, scientists around the world have revealed how plants and microbes have their own sophisticated communication networks and division of labor that is subject to selection in alternative environments [4].
In recent years, it is becoming increasingly apparent that plant phenotypes are a result of the combined expression of the host and associated microbial genomes, leading to the popularization of the ‘holobiont’ theory [5]. As a result, the concept of a plant holobiont, an assemblage of highly cooperative and minimally conflicting plant–microbe interactions, is becoming more frequently used in plant sciences [6]. Today, the field of plant holobiont research invokes the study of interlinkages between plant and individual microbial behavior and evolution to understand how functionally integrated they are or how natural selection operates on them [7]. While the concept of holobionts can seem unnecessarily complex at times, ecological functions provided by microbes are now regarded as an important feature of plant fitness [8]. Thus, unraveling the complexity of holobionts promises to deliver innovations in plant ecosystem productivity for sustainable agriculture [9,10] by rooting out stochasticity and fortifying predictability [11].
The paradigm shift towards the increasingly recognized concept of plant holobionts introduces new questions to be answered [12,13] and, consequently, a new theoretical framework for omic technologies to follow. Conceptually, due to inherently dynamic biotic-abiotic interactions with the environment, the plant holobiont structure and composition will experience adaptive cycles of expansion, consolidation, and resilience [11]. Under such premises, plants and microbes “work together” to continually adapt the plant holobiont to perpetually changing environmental conditions. Efforts to assess the strength of genetic, molecular, and metabolic relationships between plants and microbes across environments would contribute to a more accurate view of the plant holobiont at evolutionary and ecological scales. Therefore, in this perspective, we discuss the promises and challenges of omics technologies in studying plant holobionts and how omics information can be aggregated across these adaptive cycles to effectively understand the long-term fidelity of plant–microbe interactions and discern their connectivity to ecological functions in plant holobionts (Figure 1).
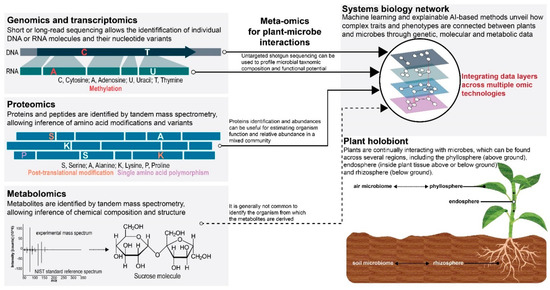
Figure 1.
The application of major omic technologies and their integration provide complementary data necessary for dissecting complex traits and phenotypes associated with plant-microbe interactions. The promises and challenges of each omic technology will be influenced by the genetic, functional, and environmental complexity inherent to the biological system being studied.
2. Advancements in Omics Are Key to Defining Plant Holobionts
Major scientific breakthroughs in the study of plant–microbe interactions are driven by technological advancements that facilitate cost-efficient, high-throughput analysis of DNA, RNA, proteins, and metabolites. In the past decade, high-throughput sequencing technologies, e.g., Illumina (https://www.illumina.com/, accessed on 15 September 2022) Pacific Biosciences (https://www.pacb.com/, accessed on 15 September 2022) and Oxford Nanopore Technologies (https://nanoporetech.com/, accessed on 15 September 2022), have fostered rapid progress in the field of plant–microbe research by delivering insights into relevant genetic and genomic expression signatures. Innovations in tandem mass spectrometry [14] have provided access to how those genomic signatures are translated to proteins and their subsequent metabolic products. Today, as a result of these advances, we are now able to answer questions at astonishing levels of mechanistic detail.
Moving forward, our understanding of the plant holobiont will require host-centered omic strategies paired with commonly used microbial-focused techniques, such as amplicon sequencing and meta-omics [15]. Over the past decade, microbiome sequencing and analysis has improved our understanding of the structure and diversity of the microbial world that grows in (endosphere) and on above-(phyllosphere) and below-ground (rhizosphere) plant tissues. Early efforts that used standardized protocols for 16S ribosomal RNA (rRNA) sequencing expanded our worldly understanding of microbial diversity [16,17], its extent and limit, and how a plant host and/or environment selects for specific taxonomic and phylogenetic composition [18]. Similar to 16S rRNA sequencing, DNA barcoding of Internal Transcribed Spacer (ITS) region of the nuclear DNA has been a key molecular method for our understanding of fungal diversity [19,20,21,22].
Today, next-generation sequencing technologies are increasingly used in attempts to identify key or “core” microbiome members that consistently engage with plants directly; a key ecological parameter in holobiont theory. The most actively applied approach to define a core microbiome prioritizes membership by taxonomic rank, which is determined by a member’s occupancy and abundance across longitudinal studies [23]. In the simplest way, taxa with relatively high abundance or observed more frequently can be interpreted as core taxa, though conditionally rare species can also play important roles [24]. While high-throughput sequencing using marker genes (e.g., 16S rRNA, ITS or 18S rRNA gene) is being performed at unprecedented spatial and temporal levels [25], these methods lack functional information. Therefore, in addition to taxonomic approaches, it is important to integrate or, at the very least, follow up with functional data (metagenomics [26], metatranscriptomics [27], metaproteomics [28], and metabolomics [29,30,31]) because we are learning that microbiomes having different species can still encode similar functions [32]. It is expected that comparative functional metagenomics combined with other downstream meta-omic methods represents a critical step to our discoveries of interacting mechanisms between plants and microbes that explain consistently defined core microbiome taxa or function [33,34].
As discussed later in this perspective, integrating multi-omics data is inherently difficult because genome expression, transcription, translation, and metabolism all operate on different timescales [35]. As the field of plant sciences trends towards ever-larger data sets with multiple omic layers, state-of-the-art approaches will employ machine-learning and explainable artificial intelligence approaches that serve as a means to classify and interpret key relationships across a multitude of variables (e.g., plant host genotype, microbiome composition and function, environment, time, space, etc.) [36].
3. From Genes to Ecosystems: Studying Plant–Microbe Interactions across the Complexity Landscape
Incorporating natural genetic, environmental, and functional complexity into a single experimental framework represents a key challenge for all omic technologies—sparse data combined with methodological limitations can lead to insufficient information or, even worse, misleading biological inferences [37]. Hence, identifying the genetic, molecular, and metabolic factors underpinning emergent plant microbiome-associated phenotypes in the environment is recognized as a daunting task. To address this, plant–microbe interaction studies adopt experimental frameworks that either seek to control or embrace natural complexity (Figure 2) [38,39].
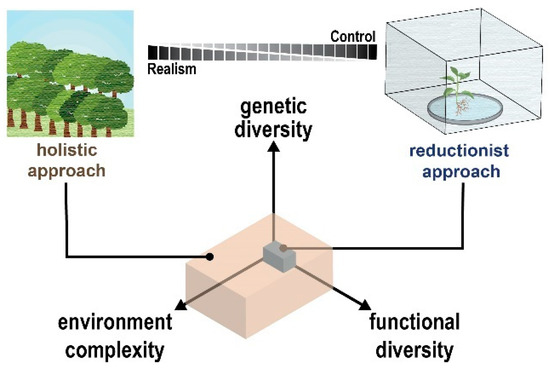
Figure 2.
Dimensions of complexity and diversity in plant–microbe interactions. Studying plant–microbe interactions in both reduced and highly complex biological systems is necessary to obtain a complete understanding of these complex relationships. As complexity increases, the completeness and reliability of omic technologies will be challenged.
Using a reductionist approach, relatively low levels of genotypic and functional diversity are studied in habitats operating with a highly controlled environment. Under this framework, simplicity and experimental control offers greater interpretability. Today, a major advantage of using a reductionist approach is the ability to develop and use engineered plant–microbe habitats, for example, EcoFab devices [40], to control complexity while seamlessly integrating with omic measurements. On the one hand, the reductionist strategy offers an opportunity to define plant holobionts with exquisite mechanistic detail, providing genetic and/or molecular explanations for plant–microbe interactions. On the other hand, this strategy is not poised for studying higher-order ecological processes and their importance.
Moving to the other side of the complexity-control spectrum, a holistic approach captures ecological interactions with the plant holobiont. Using this experimental framework, large-scale surveys incorporating marker gene or metagenomic sequencing are used to study plant–microbe interactions across complex natural environments. This approach offers an opportunity to disentangle the relative influence of genotypic, environmental, and functional variables and the ecological importance between these variables [41]. Except for a few exemplary examples [42,43], holistic approaches are often without omic information due to cost and labor.
Arguably the next step forward in plant–microbe interaction studies is to integrate reductionist and holistic approaches [44]. Unfortunately, integrating these approaches is not likely achievable for a single research group. Instead, integration takes place across large, interdisciplinary research projects such as several notable efforts supported by the Department of Energy (https://genomicscience.energy.gov/sfas/, accessed on 15 September 2022) and the Earth Microbiome Project (https://earthmicrobiome.org/, accessed on 15 September 2022) [17]. When complete integration is not feasible, it is recommended that careful experimental design be implemented to bridge the knowledge gaps between the reductionist-holistic divide by working inward from both sides of the complexity-control spectrum [38]. In the sections below, we highlight state-of-the-art examples of omics technologies being applied across the complexity-control spectrum and acknowledge key challenges that must be addressed.
3.1. Recent Advancements and Current Impediments for Genomics, Transcriptomics, Proteomics, and Metabolomics for Studying the Plant Holobiont
As discussed in the section above, designing an experiment that generates accurate and meaningful results that can be built upon is a challenging task when studying plant holobionts because there are many confounding factors that warrant careful attention. Coordination between research objectives and technology approaches is essential to deploy the appropriate measurements to acquire the desired information. This section aims to highlight the current state-of-the-art research employing omic technologies to better understand plant–microbe interactions as well as notable impediments to their application in complex biological systems.
3.1.1. Genomic and Transcriptomics
In the past decade, considerable efforts have been made using next-generation genomics and transcriptomics to understand how plant genomes influence the presence and function of bacteria and fungi in associated microbiomes [45,46,47,48,49,50,51]. Yet, the extent to which plants exert genetic control over their microbiomes is difficult to disentangle from other natural exogenous stimuli. Natural environments can have biotic and abiotic influences that can mask the effects of host genes that vary between locations and years, demanding multi-year, large-scale field experiments [52,53]. Reductionist approaches using laboratory-controlled conditions can eliminate these confounding factors; however, these studies may also overestimate the influence of certain plant genes and fail to identify genotypic signatures associated with plasticity under context-dependent requirements [54]. Nevertheless, remarkable progress has been made in our understanding of how host genetics drives the composition and function of associated microbiomes [55,56,57].
The contribution of microbial genes to host function and adaptation is similarly dependent on ecological context [58,59]. Yet, the contribution of microbial genetics in holobionts remains difficult to assess simply because the vast majority of microbes lack reference genomes, and this is because most microorganisms are challenging to grow under laboratory conditions. While recent advancements in experimental technologies promise to close this gap using microbial culturomics [60] to isolate and sequence individual genomes, amplicon-based studies (e.g., 16S rRNA, 18S rRNA and ITS) will likely remain the most broadly applied genomic technique to study microbiome diversity and its impact on host function and adaptation [61]. Relevant to the concept of a holobiont, amplicon studies have also begun to shed light on the importance of microbe–microbe interactions within plant communities [61,62]. Yet, as mentioned previously, while amplicon studies are useful for estimating microbial diversity, they fail to provide evidence pertaining to the functional potential and activity of the sampled microbiota. With decreasing costs for massively parallel DNA and RNA sequencing, metagenomic and metatranscriptomic approaches promise to fill this knowledge gap. Today, entire microbial genomes can be reconstructed from metagenome sequencing [63], yielding metagenome-assembled genomes (MAGs) that have increased our functional knowledge of specific microbes within many plant species [64,65,66]. For instance, the study by Xu et al. [66] is an exemplar study demonstrating the utility of genome-resolved metagenomics coupled with downstream reductionist experiments for dissecting plant–microbe interactions in the root-associated microbiome [66]. On the basis of assessing the activity of microbes in mixed communities, isotopic labeling with DNA-based sequencing can be used [67,68,69], but metatranscriptomics remains the most widely adopted technology to assess the functional responses of both plants and microbes to interactions with each other [70,71,72,73] and external environmental stresses [74,75,76].
Moving forward, the combination of host and microbial genomic and transcriptomic information is critically important to improving our understanding of the plant holobiont. In addition to the experimental advancements that further the coverage and resolution of DNA and RNA sequencing in mixed communities [77,78,79,80], innovative computational approaches that improve taxonomic classification [81,82] as well as assembly-based and mapping-based meta-genomic and -transcriptomic profiling are equally important [83,84]. With continual improvements, the integrated study of the genetic features of a plant host alongside that of its associated microbes is becoming a more feasible, though still underdeveloped, approach to understanding plant holobionts [85].
3.1.2. Proteomics
Proteins are considered the central intermediates between a genotype and phenotype, serving as the effectors of function in biology [86]. Currently, it is important to recognize that proteins are no longer considered to be simple translations of genetic code. The extent of chemical diversity proteins can obtain after translation is quite remarkable [87,88], whereby disparate sources of biological variation (e.g., alternative splicing of RNA and post-translational modifications) will affect the fidelity and robustness of a protein structure and function. In general, attempts to compositionally map proteins and their abundances is largely achieved by mass spectrometry-based approaches [89,90]. Over the past several decades, advancements in proteomics have led to new mechanistic insights into how plant hosts recognize their associated microbes and regulate their establishment, persistence, and function [51,91,92]. Beyond the large-scale endeavors that compositionally map and quantify proteome expression changes, the field of proteomics research is recognized as the key data layer to defining the dynamic signal exchange between organisms that allows for recognition between friend and foe [93]. For instance, proteomics has advanced our understanding of early recognition events in the classic example of a plant–microbe interaction—Legume-Rhizobium symbiosis [94]. Measuring cellular and subcellular proteomes not only gives information about what happens to a particular host cellular compartment under symbiotic relationships [51,95,96,97,98], but also includes information necessary to monitor signaling events occurring during the early stages of symbiotic interactions [99,100,101,102,103].
With respect to studying the plant holobiont, and similar to the other omics, the ability to integrate host and microbe omics data together is a challenging yet necessary step forward. Unlike animal and human host-microbe systems [104,105,106], only in the past recent years has the field of plant–microbe research started to implement metaproteomics into experimental designs [107,108,109]. This is largely explained by experimental and technical aspects that challenge the depth and coverage of mapping plant-associated metaproteomes, especially those that are endophytic. As such, early attempts to apply metaproteomics so far largely consist of reductionist approaches to study plant–microbe [110] and microbe–microbe interactions [111]. Moving forward, innovations to improve the experimental [112,113,114,115], technical [116], and computational extraction and analysis of metaproteome data [28,117] from more complex environmental matrices, such as native soil, will be crucial to advancing our understanding of plant holobionts.
3.1.3. Metabolomics
Within plant holobionts, metabolites are the immediate effectors underlying the basic processes of recognition and communication between organisms and they are a currency and commodity shared between symbiotic, commensal, parasitic and pathogenic relationships. Based on genome predictions, we know that plants, and even their associated microbes, have the ability to produce thousands of molecules that together interact with and influence ecosystems [118]. Currently, mass spectrometry-based metabolomics is one of the key technologies used to characterize these diverse complex chemical inventories [119]. In the past decade, the improved accessibility and knowledge obtained from metabolomics has been crucial to understanding how plant metabolites shape their microbial communities and how microbially derived molecules affect plant hosts and ecosystems [120,121,122]. In general, metabolomics is frequently applied to (i) characterize plant root exudation and its impact on microbes in the surrounding environment and (ii) the effect of associated microbes on host metabolism.
Direct analysis of plant–microbe relationships in situ could provide the most relevant data for understanding these biological phenomena, but this can be incredibly difficult to reproduce, replicate and standardize because of high variability in environmental factors, such as soil properties, and it can be easily confounded by the surrounding breakdown of unrelated organisms and other biomaterials [123]. Therefore, the vast majority of metabolomic research employs a reductionist approach. To date, the rhizospheric effect on microbiome composition and function has been studied mostly in sterile or (semi)sterile soil or artificial environmental matrices and habitats, such as hydroponic growth systems [124,125,126]. Because of major technical advancements in mass spectrometry, we now know a great deal about root exudate composition in model species such as Arabidopsis [127,128,129], maize [130], and rice [131,132] and their effects on associated microbiota [133,134,135]. Today, there are still a small number of examples of metabolomics being applied to study root exudates in either field [136,137] or greenhouse soils [138,139]; however, recent experimental advancements promise to address challenges related to non-sterile soil matrices [140].
Crucial to understanding metabolic linkages in plant holobionts, metabolomics can provide a deep appreciation and understanding of how microbially derived metabolites impact plant phenotypes [141,142,143], either by specific pairwise plant–microbe interactions [144,145,146,147] or microbiome-driven changes in plant metabolomes [148]. At present, there is still little, or no effort made to differentiate the origin of metabolites analyzed in plant–microbe co-cultures and this makes linking metabolome information to a particular phenotype challenging. Attempts to distinguish metabolites will benefit our understanding of plant–microbe and microbe–microbe metabolic interactions and thus the use of stable-isotope labeling [149,150] is considered as a promising technical advancement towards our understanding of plant holobionts.
3.1.4. Integrative Systems Biology
Combining multiple-omic technologies can be challenging due to the extent of data, lack of consensus between data types, and the different scales at which each technology measures the plant holobiont. When successfully integrated, multi-omic studies offer unprecedented insights into the mechanistic interplay between plants and microbes [137,151,152,153]. Advancements in computational tools and deep-learning applications that account for the increased and varied data types are improving interpretability (reviewed in [154,155,156] while network analyses continue to be a useful approach to analyze the integration of multiple data set types [157,158]. Moving forward, efforts by the research community to extend the utility and accessibility of computations tools and bioinformatic workflows will be a key factor in our scientific advancement of the plant holobiont concept into practical applications. For instance, the open-source data science platform KBase (http://kbase.us, accessed on 15 September 2022) [159], a freely available community resource offering a suite of tools and workflows designed as a “one-stop-shop” to integrate and analyze complex data types, has seen tremendous growth in utilization by the research community. Equally important to the accessibility of tools is the underlying data. Effective data sharing, using FAIR principles [160], is important for this growing research community. While proper data sharing for DNA and RNA sequences is becoming more routine, the ability to “FAIRify” mass spectrometry data from proteomics and metabolomics still represents a significant challenge for the research community. The development of data infrastructures such as GNPS (https://gnps.ucsd.edu/, accessed on 15 September 2022) [161] represented, and continues to be, an exciting and important step in the right direction for capturing and retaining knowledge obtained by mass spectrometry.
4. Conclusions
Understanding the mechanistic principles central to plant holobiont theory provides an opportunity to predict and augment beneficial and detrimental plant–microbe interactions to improve the sustainability and productivity of natural and agricultural systems [162,163]. The interrelatedness between biological and technical advancements has always had important implications on major breakthroughs and scientific advancements. We anticipate that the incredible complexity of plant holobionts will serve as fertile ground for new innovations in omics techniques and related technologies that will pioneer new advances in plant biology. Therefore, we hope that this perspective serves to stimulate new multidisciplinary research conducted in an environment that embraces the complexity of plant holobionts in order to catalyze new advancements to open up new biological questions for the plant–microbe research community.
This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy (DOE). The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The DOE will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan.
Author Contributions
Conceptualization, P.E.A. and D.L.C.; writing—original draft preparation, P.E.A.; writing—review and editing, P.E.A., D.L.C., M.R.A., S.M., H.K.S. and R.L.H.; visualization, P.E.A. All authors have read and agreed to the published version of the manuscript.
Funding
The manuscript is supported by Secure Ecosystem Engineering and Design (SEED) project funded by the Genomic Science Program of the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (BER) as part of the Secure Biosystems Design Science Focus Area (SFA) (https://seed-sfa.ornl.gov/, accessed on 15 September 2022), the Plant-Microbe Interfaces Science Focus Area (https://pmiweb.ornl.gov/, accessed on 15 September 2022), and the Center for Bioenergy Innovation (CBI) (https://cbi.ornl.gov/, accessed on 15 September 2022), a U.S. Department of Energy (DOE) Research Center. Oak Ridge National Laboratory is managed by UT-Battelle, LLC for the U.S. Department of Energy under Contract Number DE-AC05-00OR22725.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Dai, X.; Shen, L. Advances and Trends in Omics Technology Development. Front. Med. 2022, 9, 911861. [Google Scholar] [CrossRef]
- Gamalero, E.; Bona, E.; Glick, B.R. Current Techniques to Study Beneficial Plant-Microbe Interactions. Microorganisms 2022, 10, 1380. [Google Scholar] [CrossRef]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G.; et al. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nat. Rev. Microbiol. 2020, 18, 607–621. [Google Scholar] [CrossRef]
- Baedke, J.; Fabregas-Tejeda, A.; Nieves Delgado, A. The holobiont concept before Margulis. J. Exp. Zool. B Mol. Dev. Evol. 2020, 334, 149–155. [Google Scholar] [CrossRef]
- Lyu, D.; Zajonc, J.; Page, A.; Tanney, C.A.S.; Shah, A.; Monjezi, N.; Msimbira, L.A.; Antar, M.; Nazari, M.; Backer, R.; et al. Plant Holobiont Theory: The Phytomicrobiome Plays a Central Role in Evolution and Success. Microorganisms 2021, 9, 675. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Canizares, C.; Jorrin, B.; Poole, P.S.; Tkacz, A. Understanding the holobiont: The interdependence of plants and their microbiome. Curr. Opin. Microbiol. 2017, 38, 188–196. [Google Scholar] [CrossRef]
- Rosenberg, E. Holistic Fitness: Microbiomes are Part of the Holobiont’s Fitness. In Microbiomes; Springer: Berlin/Heidelberg, Germany, 2021; pp. 101–160. [Google Scholar]
- Vishwakarma, K.; Kumar, N.; Shandilya, C.; Mohapatra, S.; Bhayana, S.; Varma, A. Revisiting Plant-Microbe Interactions and Microbial Consortia Application for Enhancing Sustainable Agriculture: A Review. Front. Microbiol. 2020, 11, 560406. [Google Scholar] [CrossRef]
- Phour, M.; Sehrawat, A.; Sindhu, S.S.; Glick, B.R. Interkingdom signaling in plant-rhizomicrobiome interactions for sustainable agriculture. Microbiol. Res. 2020, 241, 126589. [Google Scholar] [CrossRef]
- Matyssek, R.; Lüttge, U.; zu Castell, W. Evolution of Holobiont-Like Systems: From Individual to Composed Ecological and Global Units. In Progress in Botany; Springer: Berlin/Heidelberg, Germany, 2022. [Google Scholar]
- Harris, J.M.; Balint-Kurti, P.; Bede, J.C.; Day, B.; Gold, S.; Goss, E.M.; Grenville-Briggs, L.J.; Jones, K.M.; Wang, A.; Wang, Y.; et al. What are the Top 10 Unanswered Questions in Molecular Plant-Microbe Interactions? Mol. Plant. Microbe Interact. 2020, 33, 1354–1365. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Michalski, A.; Damoc, E.; Hauschild, J.P.; Lange, O.; Wieghaus, A.; Makarov, A.; Nagaraj, N.; Cox, J.; Mann, M.; Horning, S. Mass spectrometry-based proteomics using Q Exactive, a high-performance benchtop quadrupole Orbitrap mass spectrometer. Mol. Cell Proteom. 2011, 10, M111 011015. [Google Scholar] [CrossRef]
- Xu, L.; Pierroz, G.; Wipf, H.M.; Gao, C.; Taylor, J.W.; Lemaux, P.G.; Coleman-Derr, D. Holo-omics for deciphering plant-microbiome interactions. Microbiome 2021, 9, 69. [Google Scholar] [CrossRef]
- Shade, A. Diversity is the question, not the answer. ISME J. 2017, 11, 1–6. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Ryberg, M.; Abarenkov, K.; Sjokvist, E.; Kristiansson, E. The ITS region as a target for characterization of fungal communities using emerging sequencing technologies. FEMS Microbiol. Lett. 2009, 296, 97–101. [Google Scholar] [CrossRef]
- Bellemain, E.; Carlsen, T.; Brochmann, C.; Coissac, E.; Taberlet, P.; Kauserud, H. ITS as an environmental DNA barcode for fungi: An in silico approach reveals potential PCR biases. BMC Microbiol. 2010, 10, 189. [Google Scholar] [CrossRef]
- Begerow, D.; Nilsson, H.; Unterseher, M.; Maier, W. Current state and perspectives of fungal DNA barcoding and rapid identification procedures. Appl. Microbiol. Biotechnol. 2010, 87, 99–108. [Google Scholar] [CrossRef]
- Lucking, R.; Aime, M.C.; Robbertse, B.; Miller, A.N.; Ariyawansa, H.A.; Aoki, T.; Cardinali, G.; Crous, P.W.; Druzhinina, I.S.; Geiser, D.M.; et al. Unambiguous identification of fungi: Where do we stand and how accurate and precise is fungal DNA barcoding? IMA Fungus 2020, 11, 14. [Google Scholar] [CrossRef]
- Shade, A.; Stopnisek, N. Abundance-occupancy distributions to prioritize plant core microbiome membership. Curr. Opin. Microbiol. 2019, 49, 50–58. [Google Scholar] [CrossRef]
- Shade, A.; Jones, S.E.; Caporaso, J.G.; Handelsman, J.; Knight, R.; Fierer, N.; Gilbert, J.A. Conditionally rare taxa disproportionately contribute to temporal changes in microbial diversity. mBio 2014, 5, e01371-14. [Google Scholar] [CrossRef]
- Beilsmith, K.; Thoen, M.P.M.; Brachi, B.; Gloss, A.D.; Khan, M.H.; Bergelson, J. Genome-wide association studies on the phyllosphere microbiome: Embracing complexity in host-microbe interactions. Plant J. 2019, 97, 164–181. [Google Scholar] [CrossRef]
- Meyer, F.; Fritz, A.; Deng, Z.L.; Koslicki, D.; Lesker, T.R.; Gurevich, A.; Robertson, G.; Alser, M.; Antipov, D.; Beghini, F.; et al. Critical Assessment of Metagenome Interpretation: The second round of challenges. Nat. Methods 2022, 19, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Shakya, M.; Lo, C.C.; Chain, P.S.G. Advances and Challenges in Metatranscriptomic Analysis. Front. Genet. 2019, 10, 904. [Google Scholar] [CrossRef]
- Van Den Bossche, T.; Kunath, B.J.; Schallert, K.; Schape, S.S.; Abraham, P.E.; Armengaud, J.; Arntzen, M.O.; Bassignani, A.; Benndorf, D.; Fuchs, S.; et al. Critical Assessment of MetaProteome Investigation (CAMPI): A multi-laboratory comparison of established workflows. Nat. Commun. 2021, 12, 7305. [Google Scholar] [CrossRef]
- Jacoby, R.P.; Chen, L.; Schwier, M.; Koprivova, A.; Kopriva, S. Recent advances in the role of plant metabolites in shaping the root microbiome. F1000Research 2020, 9, 151. [Google Scholar] [CrossRef] [PubMed]
- Roume, H.; Muller, E.E.; Cordes, T.; Renaut, J.; Hiller, K.; Wilmes, P. A biomolecular isolation framework for eco-systems biology. ISME J. 2013, 7, 110–121. [Google Scholar] [CrossRef]
- Levy, A.; Conway, J.M.; Dangl, J.L.; Woyke, T. Elucidating Bacterial Gene Functions in the Plant Microbiome. Cell Host Microbe 2018, 24, 475–485. [Google Scholar] [CrossRef]
- Lambais, M.R.; Barrera, S.E.; Santos, E.C.; Crowley, D.E.; Jumpponen, A. Phyllosphere Metaproteomes of Trees from the Brazilian Atlantic Forest Show High Levels of Functional Redundancy. Microb. Ecol. 2017, 73, 123–134. [Google Scholar] [CrossRef]
- Abram, F. Systems-based approaches to unravel multi-species microbial community functioning. Comput. Struct. Biotechnol. J. 2015, 13, 24–32. [Google Scholar] [CrossRef]
- Vayssier-Taussat, M.; Albina, E.; Citti, C.; Cosson, J.F.; Jacques, M.A.; Lebrun, M.H.; Le Loir, Y.; Ogliastro, M.; Petit, M.A.; Roumagnac, P.; et al. Shifting the paradigm from pathogens to pathobiome: New concepts in the light of meta-omics. Front. Cell. Infect. Microbiol. 2014, 4, 29. [Google Scholar] [CrossRef]
- Nicholson, J.K.; Lindon, J.C. Systems biology: Metabonomics. Nature 2008, 455, 1054–1056. [Google Scholar] [CrossRef]
- Ghannam, R.B.; Techtmann, S.M. Machine learning applications in microbial ecology, human microbiome studies, and environmental monitoring. Comput. Struct. Biotechnol. J. 2021, 19, 1092–1107. [Google Scholar] [CrossRef]
- Leite, M.F.; Kuramae, E.E. You must choose, but choose wisely: Model-based approaches for microbial community analysis. Soil Biol. Biochem. 2020, 151, 108042. [Google Scholar] [CrossRef]
- Tecon, R.; Mitri, S.; Ciccarese, D.; Or, D.; van der Meer, J.R.; Johnson, D.R. Bridging the Holistic-Reductionist Divide in Microbial Ecology. mSystems 2019, 4, e00265-18. [Google Scholar] [CrossRef]
- Swenson, N.G.; Jones, F.A. Community transcriptomics, genomics and the problem of species co-occurrence. J. Ecol. 2017, 105, 563–568. [Google Scholar] [CrossRef]
- Zengler, K.; Hofmockel, K.; Baliga, N.S.; Behie, S.W.; Bernstein, H.C.; Brown, J.B.; Dinneny, J.R.; Floge, S.A.; Forry, S.P.; Hess, M.; et al. EcoFABs: Advancing microbiome science through standardized fabricated ecosystems. Nat. Methods 2019, 16, 567–571. [Google Scholar] [CrossRef]
- Oyserman, B.O.; Cordovez, V.; Flores, S.S.; Leite, M.F.A.; Nijveen, H.; Medema, M.H.; Raaijmakers, J.M. Extracting the GEMs: Genotype, Environment, and Microbiome Interactions Shaping Host Phenotypes. Front. Microbiol. 2020, 11, 574053. [Google Scholar] [CrossRef]
- Stopnisek, N.; Shade, A. Persistent microbiome members in the common bean rhizosphere: An integrated analysis of space, time, and plant genotype. ISME J. 2021, 15, 2708–2722. [Google Scholar] [CrossRef]
- Raes, J.; Letunic, I.; Yamada, T.; Jensen, L.J.; Bork, P. Toward molecular trait-based ecology through integration of biogeochemical, geographical and metagenomic data. Mol. Syst. Biol. 2011, 7, 473. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Salas-Gonzalez, I.; Conway, J.M.; Finkel, O.M.; Gilbert, S.; Russ, D.; Teixeira, P.; Dangl, J.L. The Plant Microbiome: From Ecology to Reductionism and Beyond. Annu. Rev. Microbiol. 2020, 74, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Larrainzar, E.; Riely, B.K.; Kim, S.C.; Carrasquilla-Garcia, N.; Yu, H.J.; Hwang, H.J.; Oh, M.; Kim, G.B.; Surendrarao, A.K.; Chasman, D.; et al. Deep Sequencing of the Medicago truncatula Root Transcriptome Reveals a Massive and Early Interaction between Nodulation Factor and Ethylene Signals. Plant Physiol. 2015, 169, 233–265. [Google Scholar] [CrossRef]
- Petre, B.; Lorrain, C.; Stukenbrock, E.H.; Duplessis, S. Host-specialized transcriptome of plant-associated organisms. Curr. Opin. Plant Biol. 2020, 56, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Tabrett, A.; Horton, M.W. The influence of host genetics on the microbiome. F1000Research 2020, 9, 84. [Google Scholar] [CrossRef]
- Clouse, K.M.; Wagner, M.R. Plant Genetics as a Tool for Manipulating Crop Microbiomes: Opportunities and Challenges. Front. Bioeng. Biotechnol. 2021, 9, 567548. [Google Scholar] [CrossRef]
- Lebeis, S.L.; Paredes, S.H.; Lundberg, D.S.; Breakfield, N.; Gehring, J.; McDonald, M.; Malfatti, S.; Glavina del Rio, T.; Jones, C.D.; Tringe, S.G.; et al. Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 2015, 349, 860–864. [Google Scholar] [CrossRef]
- Bodenhausen, N.; Bortfeld-Miller, M.; Ackermann, M.; Vorholt, J.A. A synthetic community approach reveals plant genotypes affecting the phyllosphere microbiota. PLoS Genet. 2014, 10, e1004283. [Google Scholar] [CrossRef]
- Qiao, Z.; Yates, T.B.; Shrestha, H.K.; Engle, N.L.; Flanagan, A.; Morrell-Falvey, J.L.; Sun, Y.; Tschaplinski, T.J.; Abraham, P.E.; Labbe, J.; et al. Towards engineering ectomycorrhization into switchgrass bioenergy crops via a lectin receptor-like kinase. Plant Biotechnol. J. 2021, 19, 2454–2468. [Google Scholar] [CrossRef]
- Edwards, J.A.; Santos-Medellin, C.M.; Liechty, Z.S.; Nguyen, B.; Lurie, E.; Eason, S.; Phillips, G.; Sundaresan, V. Compositional shifts in root-associated bacterial and archaeal microbiota track the plant life cycle in field-grown rice. PLoS Biol. 2018, 16, e2003862. [Google Scholar] [CrossRef]
- Wagner, M.R.; Lundberg, D.S.; Del Rio, T.G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef]
- Anderson, J.T.; Wagner, M.R.; Rushworth, C.A.; Prasad, K.V.; Mitchell-Olds, T. The evolution of quantitative traits in complex environments. Heredity 2014, 112, 4–12. [Google Scholar] [CrossRef]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjalmsson, B.J.; Nordborg, M.; Gordon, J.I.; et al. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 2014, 5, 5320. [Google Scholar] [CrossRef]
- Chen, T.; Nomura, K.; Wang, X.; Sohrabi, R.; Xu, J.; Yao, L.; Paasch, B.C.; Ma, L.; Kremer, J.; Cheng, Y.; et al. A plant genetic network for preventing dysbiosis in the phyllosphere. Nature 2020, 580, 653–657. [Google Scholar] [CrossRef]
- Xiong, C.; Zhu, Y.G.; Wang, J.T.; Singh, B.; Han, L.L.; Shen, J.P.; Li, P.P.; Wang, G.B.; Wu, C.F.; Ge, A.H.; et al. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef]
- Henry, L.P.; Bruijning, M.; Forsberg, S.K.G.; Ayroles, J.F. The microbiome extends host evolutionary potential. Nat. Commun. 2021, 12, 5141. [Google Scholar] [CrossRef] [PubMed]
- Laforest-Lapointe, I.; Paquette, A.; Messier, C.; Kembel, S.W. Leaf bacterial diversity mediates plant diversity and ecosystem function relationships. Nature 2017, 546, 145–147. [Google Scholar] [CrossRef]
- Diakite, A.; Dubourg, G.; Dione, N.; Afouda, P.; Bellali, S.; Ngom, I.I.; Valles, C.; Tall, M.L.; Lagier, J.C.; Raoult, D. Optimization and standardization of the culturomics technique for human microbiome exploration. Sci. Rep. 2020, 10, 9674. [Google Scholar] [CrossRef]
- Regalado, J.; Lundberg, D.S.; Deusch, O.; Kersten, S.; Karasov, T.; Poersch, K.; Shirsekar, G.; Weigel, D. Combining whole-genome shotgun sequencing and rRNA gene amplicon analyses to improve detection of microbe-microbe interaction networks in plant leaves. ISME J. 2020, 14, 2116–2130. [Google Scholar] [CrossRef]
- Niu, B.; Paulson, J.N.; Zheng, X.; Kolter, R. Simplified and representative bacterial community of maize roots. Proc. Natl. Acad. Sci. USA 2017, 114, E2450–E2459. [Google Scholar] [CrossRef] [PubMed]
- Quince, C.; Walker, A.W.; Simpson, J.T.; Loman, N.J.; Segata, N. Shotgun metagenomics, from sampling to analysis. Nat. Biotechnol. 2017, 35, 833–844. [Google Scholar] [CrossRef]
- Wicaksono, W.A.; Cernava, T.; Berg, C.; Berg, G. Bog ecosystems as a playground for plant-microbe coevolution: Bryophytes and vascular plants harbour functionally adapted bacteria. Microbiome 2021, 9, 170. [Google Scholar] [CrossRef]
- Zheng, Y.; Xu, Z.; Liu, H.; Liu, Y.; Zhou, Y.; Meng, C.; Ma, S.; Xie, Z.; Li, Y.; Zhang, C.S. Patterns in the Microbial Community of Salt-Tolerant Plants and the Functional Genes Associated with Salt Stress Alleviation. Microbiol. Spectr. 2021, 9, e0076721. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Chiniquy, D.; Pierroz, G.; Deng, S.; Gao, C.; Diamond, S.; Simmons, T.; Wipf, H.M.; Caddell, D.; et al. Genome-resolved metagenomics reveals role of iron metabolism in drought-induced rhizosphere microbiome dynamics. Nat. Commun. 2021, 12, 3209. [Google Scholar] [CrossRef]
- Starr, E.P.; Shi, S.; Blazewicz, S.J.; Probst, A.J.; Herman, D.J.; Firestone, M.K.; Banfield, J.F. Stable isotope informed genome-resolved metagenomics reveals that Saccharibacteria utilize microbially-processed plant-derived carbon. Microbiome 2018, 6, 122. [Google Scholar] [CrossRef]
- Zhou, J.; Chai, X.; Zhang, L.; George, T.S.; Wang, F.; Feng, G. Different Arbuscular Mycorrhizal Fungi Cocolonizing on a Single Plant Root System Recruit Distinct Microbiomes. mSystems 2020, 5, e00929-20. [Google Scholar] [CrossRef]
- Starr, E.P.; Shi, S.; Blazewicz, S.J.; Koch, B.J.; Probst, A.J.; Hungate, B.A.; Pett-Ridge, J.; Firestone, M.K.; Banfield, J.F. Stable-Isotope-Informed, Genome-Resolved Metagenomics Uncovers Potential Cross-Kingdom Interactions in Rhizosphere Soil. mSphere 2021, 6, e0008521. [Google Scholar] [CrossRef]
- Mark, G.L.; Dow, J.M.; Kiely, P.D.; Higgins, H.; Haynes, J.; Baysse, C.; Abbas, A.; Foley, T.; Franks, A.; Morrissey, J.; et al. Transcriptome profiling of bacterial responses to root exudates identifies genes involved in microbe-plant interactions. Proc. Natl. Acad. Sci. USA 2005, 102, 17454–17459. [Google Scholar] [CrossRef]
- Spaepen, S.; Bossuyt, S.; Engelen, K.; Marchal, K.; Vanderleyden, J. Phenotypical and molecular responses of Arabidopsis thaliana roots as a result of inoculation with the auxin-producing bacterium Azospirillum brasilense. New Phytol. 2014, 201, 850–861. [Google Scholar] [CrossRef]
- Santos-Medellin, C.; Edwards, J.; Nguyen, B.; Sundaresan, V. Acquisition of a complex root microbiome reshapes the transcriptomes of rice plants. New Phytol. 2022, 235, 2008–2021. [Google Scholar] [CrossRef]
- Nerva, L.; Garcia, J.F.; Favaretto, F.; Giudice, G.; Moffa, L.; Sandrini, M.; Cantu, D.; Zanzotto, A.; Gardiman, M.; Velasco, R.; et al. The hidden world within plants: Metatranscriptomics unveils the complexity of wood microbiomes. J. Exp. Bot. 2022, 73, 2682–2697. [Google Scholar] [CrossRef]
- Yamazaki, S.; Mardani-Korrani, H.; Kaida, R.; Ochiai, K.; Kobayashi, M.; Nagano, A.J.; Fujii, Y.; Sugiyama, A.; Aoki, Y. Field multi-omics analysis reveals a close association between bacterial communities and mineral properties in the soybean rhizosphere. Sci. Rep. 2021, 11, 8878. [Google Scholar] [CrossRef]
- Zhang, Y.; Xu, J.; Wang, E.; Wang, N. Mechanisms Underlying the Rhizosphere-To-Rhizoplane Enrichment of Cellvibrio Unveiled by Genome-Centric Metagenomics and Metatranscriptomics. Microorganisms 2020, 8, 583. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, E.; Pitre, F.E.; Page, A.P.; Marleau, J.; Guidi Nissim, W.; St-Arnaud, M.; Labrecque, M.; Joly, S.; Yergeau, E.; Brereton, N.J.B. Trees, fungi and bacteria: Tripartite metatranscriptomics of a root microbiome responding to soil contamination. Microbiome 2018, 6, 53. [Google Scholar] [CrossRef]
- Ikeda, S.; Kaneko, T.; Okubo, T.; Rallos, L.E.; Eda, S.; Mitsui, H.; Sato, S.; Nakamura, Y.; Tabata, S.; Minamisawa, K. Development of a bacterial cell enrichment method and its application to the community analysis in soybean stems. Microb. Ecol. 2009, 58, 703–714. [Google Scholar] [CrossRef]
- Jiao, J.Y.; Wang, H.X.; Zeng, Y.; Shen, Y.M. Enrichment for microbes living in association with plant tissues. J. Appl. Microbiol. 2006, 100, 830–837. [Google Scholar] [CrossRef]
- Song, L.; Xie, K. Engineering CRISPR/Cas9 to mitigate abundant host contamination for 16S rRNA gene-based amplicon sequencing. Microbiome 2020, 8, 80. [Google Scholar] [CrossRef]
- Nobori, T.; Tsuda, K. In planta Transcriptome Analysis of Pseudomonas syringae. Bio Protoc. 2018, 8, e2987. [Google Scholar] [CrossRef]
- Garcia, B.J.; Simha, R.; Garvin, M.; Furches, A.; Jones, P.; Gazolla, J.; Hyatt, P.D.; Schadt, C.W.; Pelletier, D.; Jacobson, D. A k-mer based approach for classifying viruses without taxonomy identifies viral associations in human autism and plant microbiomes. Comput. Struct. Biotechnol. J. 2021, 19, 5911–5919. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, A.; Leclercq, M.; Sanabria, M.; Perin, O.; Droit, A. Machine Learning and Deep Learning Applications in Metagenomic Taxonomy and Functional Annotation. Front. Microbiol. 2022, 13, 811495. [Google Scholar] [CrossRef]
- Ghurye, J.S.; Cepeda-Espinoza, V.; Pop, M. Metagenomic Assembly: Overview, Challenges and Applications. Yale J. Biol. Med. 2016, 89, 353–362. [Google Scholar]
- Nissen, J.N.; Johansen, J.; Allesoe, R.L.; Sonderby, C.K.; Armenteros, J.J.A.; Gronbech, C.H.; Jensen, L.J.; Nielsen, H.B.; Petersen, T.N.; Winther, O.; et al. Improved metagenome binning and assembly using deep variational autoencoders. Nat. Biotechnol. 2021, 39, 555–560. [Google Scholar] [CrossRef]
- Alberdi, A.; Andersen, S.B.; Limborg, M.T.; Dunn, R.R.; Gilbert, M.T.P. Disentangling host-microbiota complexity through hologenomics. Nat. Rev. Genet. 2022, 23, 281–297. [Google Scholar] [CrossRef]
- Feussner, I.; Polle, A. What the transcriptome does not tell—Proteomics and metabolomics are closer to the plants’ patho-phenotype. Curr. Opin. Plant Biol. 2015, 26, 26–31. [Google Scholar] [CrossRef]
- Smith, L.M.; Kelleher, N.L. Proteoforms as the next proteomics currency. Science 2018, 359, 1106–1107. [Google Scholar] [CrossRef]
- Smith, L.M.; Agar, J.N.; Chamot-Rooke, J.; Danis, P.O.; Ge, Y.; Loo, J.A.; Pasa-Tolic, L.; Tsybin, Y.O.; Kelleher, N.L.; Consortium for Top-Down, P. The Human Proteoform Project: Defining the human proteome. Sci. Adv. 2021, 7, eabk0734. [Google Scholar] [CrossRef]
- Mergner, J.; Frejno, M.; List, M.; Papacek, M.; Chen, X.; Chaudhary, A.; Samaras, P.; Richter, S.; Shikata, H.; Messerer, M.; et al. Mass-spectrometry-based draft of the Arabidopsis proteome. Nature 2020, 579, 409–414. [Google Scholar] [CrossRef]
- Thul, P.J.; Lindskog, C. The human protein atlas: A spatial map of the human proteome. Protein Sci. 2018, 27, 233–244. [Google Scholar] [CrossRef]
- Marx, H.; Minogue, C.E.; Jayaraman, D.; Richards, A.L.; Kwiecien, N.W.; Siahpirani, A.F.; Rajasekar, S.; Maeda, J.; Garcia, K.; Del Valle-Echevarria, A.R.; et al. A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotechnol. 2016, 34, 1198–1205. [Google Scholar] [CrossRef]
- Sebastiana, M.; Martins, J.; Figueiredo, A.; Monteiro, F.; Sardans, J.; Penuelas, J.; Silva, A.; Roepstorff, P.; Pais, M.S.; Coelho, A.V. Oak protein profile alterations upon root colonization by an ectomycorrhizal fungus. Mycorrhiza 2017, 27, 109–128. [Google Scholar] [CrossRef]
- Jayaraman, D.; Forshey, K.L.; Grimsrud, P.A.; Ane, J.M. Leveraging proteomics to understand plant-microbe interactions. Front. Plant Sci. 2012, 3, 44. [Google Scholar] [CrossRef]
- Khatabi, B.; Gharechahi, J.; Ghaffari, M.R.; Liu, D.; Haynes, P.A.; McKay, M.J.; Mirzaei, M.; Salekdeh, G.H. Plant-Microbe Symbiosis: What Has Proteomics Taught Us? Proteomics 2019, 19, e1800105. [Google Scholar] [CrossRef]
- Elmore, J.M.; Liu, J.; Smith, B.; Phinney, B.; Coaker, G. Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol. Cell. Proteom. 2012, 11, M111.014555. [Google Scholar] [CrossRef]
- Jones, A.M.; Thomas, V.; Bennett, M.H.; Mansfield, J.; Grant, M. Modifications to the Arabidopsis defense proteome occur prior to significant transcriptional change in response to inoculation with Pseudomonas syringae. Plant Physiol. 2006, 142, 1603–1620. [Google Scholar] [CrossRef]
- Lee, S.J.; Saravanan, R.S.; Damasceno, C.M.; Yamane, H.; Kim, B.D.; Rose, J.K. Digging deeper into the plant cell wall proteome. Plant Physiol. Biochem. 2004, 42, 979–988. [Google Scholar] [CrossRef]
- Shrivastava, N.; Jiang, L.; Li, P.; Sharma, A.K.; Luo, X.; Wu, S.; Pandey, R.; Gao, Q.; Lou, B. Proteomic approach to understand the molecular physiology of symbiotic interaction between Piriformospora indica and Brassica napus. Sci. Rep. 2018, 8, 5773. [Google Scholar] [CrossRef]
- Rose, C.M.; Venkateshwaran, M.; Volkening, J.D.; Grimsrud, P.A.; Maeda, J.; Bailey, D.J.; Park, K.; Howes-Podoll, M.; den Os, D.; Yeun, L.H.; et al. Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteom. 2012, 11, 724–744. [Google Scholar] [CrossRef]
- Riley, N.M.; Coon, J.J. Phosphoproteomics in the Age of Rapid and Deep Proteome Profiling. Anal. Chem. 2016, 88, 74–94. [Google Scholar] [CrossRef]
- Benschop, J.J.; Mohammed, S.; O’Flaherty, M.; Heck, A.J.; Slijper, M.; Menke, F.L. Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteom. 2007, 6, 1198–1214. [Google Scholar] [CrossRef]
- Rayapuram, N.; Bigeard, J.; Alhoraibi, H.; Bonhomme, L.; Hesse, A.M.; Vinh, J.; Hirt, H.; Pflieger, D. Quantitative Phosphoproteomic Analysis Reveals Shared and Specific Targets of Arabidopsis Mitogen-Activated Protein Kinases (MAPKs) MPK3, MPK4, and MPK6. Mol. Cell. Proteom. 2018, 17, 61–80. [Google Scholar] [CrossRef]
- Nakagami, H.; Sugiyama, N.; Mochida, K.; Daudi, A.; Yoshida, Y.; Toyoda, T.; Tomita, M.; Ishihama, Y.; Shirasu, K. Large-scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 2010, 153, 1161–1174. [Google Scholar] [CrossRef]
- Long, S.; Yang, Y.; Shen, C.; Wang, Y.; Deng, A.; Qin, Q.; Qiao, L. Metaproteomics characterizes human gut microbiome function in colorectal cancer. NPJ Biofilms Microbiomes 2020, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Xiong, W.; Abraham, P.E.; Li, Z.; Pan, C.; Hettich, R.L. Microbial metaproteomics for characterizing the range of metabolic functions and activities of human gut microbiota. Proteomics 2015, 15, 3424–3438. [Google Scholar] [CrossRef] [PubMed]
- Wilmes, P.; Heintz-Buschart, A.; Bond, P.L. A decade of metaproteomics: Where we stand and what the future holds. Proteomics 2015, 15, 3409–3417. [Google Scholar] [CrossRef] [PubMed]
- Bao, Z.; Okubo, T.; Kubota, K.; Kasahara, Y.; Tsurumaru, H.; Anda, M.; Ikeda, S.; Minamisawa, K. Metaproteomic identification of diazotrophic methanotrophs and their localization in root tissues of field-grown rice plants. Appl. Environ. Microbiol. 2014, 80, 5043–5052. [Google Scholar] [CrossRef]
- Knief, C.; Delmotte, N.; Chaffron, S.; Stark, M.; Innerebner, G.; Wassmann, R.; von Mering, C.; Vorholt, J.A. Metaproteogenomic analysis of microbial communities in the phyllosphere and rhizosphere of rice. ISME J. 2012, 6, 1378–1390. [Google Scholar] [CrossRef]
- Salvato, F.; Vintila, S.; Finkel, O.M.; Dangl, J.; Kleiner, M. Evaluation of protein extraction methods for metaproteomic analyses of root-associated microbes. Mol. Plant Microbe Interact. 2022. [Google Scholar] [CrossRef]
- Appidi, M.R.; Bible, A.N.; Carper, D.L.; Jawdy, S.S.; Giannone, R.J.; Hettich, R.L.; Morrell-Falvey, J.; Abraham, P.E. Development of an Experimental Approach to Achieve Spatially Resolved Plant Root-Associated Metaproteomics Using an Agar-Plate System. Mol. Plant Microbe Interact. 2022, 35, 639–649. [Google Scholar] [CrossRef]
- Shrestha, H.K.; Appidi, M.R.; Villalobos Solis, M.I.; Wang, J.; Carper, D.L.; Burdick, L.; Pelletier, D.A.; Doktycz, M.J.; Hettich, R.L.; Abraham, P.E. Metaproteomics reveals insights into microbial structure, interactions, and dynamic regulation in defined communities as they respond to environmental disturbance. BMC Microbiol. 2021, 21, 308. [Google Scholar] [CrossRef]
- Chourey, K.; Jansson, J.; VerBerkmoes, N.; Shah, M.; Chavarria, K.L.; Tom, L.M.; Brodie, E.L.; Hettich, R.L. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J. Proteome Res. 2010, 9, 6615–6622. [Google Scholar] [CrossRef]
- Qian, C.; Hettich, R.L. Optimized Extraction Method to Remove Humic Acid Interferences from Soil Samples Prior to Microbial Proteome Measurements. J. Proteome Res. 2017, 16, 2537–2546. [Google Scholar] [CrossRef] [PubMed]
- Mandalakis, M.; Panikov, N.S.; Polymenakou, P.N.; Sizova, M.V.; Stamatakis, A. A simple cleanup method for the removal of humic substances from soil protein extracts using aluminum coagulation. Environ. Sci. Pollut. Res. Int. 2018, 25, 23845–23856. [Google Scholar] [CrossRef] [PubMed]
- Tartaglia, M.; Bastida, F.; Sciarrillo, R.; Guarino, C. Soil Metaproteomics for the Study of the Relationships Between Microorganisms and Plants: A Review of Extraction Protocols and Ecological Insights. Int. J. Mol. Sci. 2020, 21, 8455. [Google Scholar] [CrossRef]
- Kleiner, M.; Thorson, E.; Sharp, C.E.; Dong, X.; Liu, D.; Li, C.; Strous, M. Assessing species biomass contributions in microbial communities via metaproteomics. Nat. Commun. 2017, 8, 1558. [Google Scholar] [CrossRef] [PubMed]
- Jouffret, V.; Miotello, G.; Culotta, K.; Ayrault, S.; Pible, O.; Armengaud, J. Increasing the power of interpretation for soil metaproteomics data. Microbiome 2021, 9, 195. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Mannochio-Russo, H.; Costa-Lotufo, L.V.; Jarmusch, A.K.; Dorrestein, P.C. Mass spectrometry-based metabolomics in microbiome investigations. Nat. Rev. Microbiol. 2022, 20, 143–160. [Google Scholar] [CrossRef]
- Aksenov, A.A.; da Silva, R.; Knight, R.; Lopes, N.P.; Dorrestein, P.C. Global chemical analysis of biology by mass spectrometry. Nat. Rev. Chem. 2017, 1, 0054. [Google Scholar] [CrossRef]
- Sasse, J.; Martinoia, E.; Northen, T. Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef]
- Hiruma, K. Roles of Plant-Derived Secondary Metabolites during Interactions with Pathogenic and Beneficial Microbes under Conditions of Environmental Stress. Microorganisms 2019, 7, 362. [Google Scholar] [CrossRef]
- Gupta, S.; Schillaci, M.; Roessner, U. Metabolomics as an emerging tool to study plant-microbe interactions. Emerg. Top. Life Sci. 2022, 6, 175–183. [Google Scholar] [CrossRef]
- Oburger, E.; Jones, D.L. Sampling root exudates–mission impossible? Rhizosphere 2018, 6, 116–133. [Google Scholar] [CrossRef]
- Kawasaki, A.; Okada, S.; Zhang, C.; Delhaize, E.; Mathesius, U.; Richardson, A.E.; Watt, M.; Gilliham, M.; Ryan, P.R. A sterile hydroponic system for characterising root exudates from specific root types and whole-root systems of large crop plants. Plant Methods 2018, 14, 114. [Google Scholar] [CrossRef]
- Lopez-Guerrero, M.G.; Wang, P.; Phares, F.; Schachtman, D.P.; Alvarez, S.; van Dijk, K. A glass bead semi-hydroponic system for intact maize root exudate analysis and phenotyping. Plant Methods 2022, 18, 25. [Google Scholar] [CrossRef]
- Sasse, J.; Kant, J.; Cole, B.J.; Klein, A.P.; Arsova, B.; Schlaepfer, P.; Gao, J.; Lewald, K.; Zhalnina, K.; Kosina, S.; et al. Multilab EcoFAB study shows highly reproducible physiology and depletion of soil metabolites by a model grass. New Phytol. 2019, 222, 1149–1160. [Google Scholar] [CrossRef]
- Ziegler, J.; Schmidt, S.; Chutia, R.; Muller, J.; Bottcher, C.; Strehmel, N.; Scheel, D.; Abel, S. Non-targeted profiling of semi-polar metabolites in Arabidopsis root exudates uncovers a role for coumarin secretion and lignification during the local response to phosphate limitation. J. Exp. Bot. 2016, 67, 1421–1432. [Google Scholar] [CrossRef]
- Chaparro, J.M.; Badri, D.V.; Bakker, M.G.; Sugiyama, A.; Manter, D.K.; Vivanco, J.M. Root exudation of phytochemicals in Arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS ONE 2013, 8, e55731. [Google Scholar] [CrossRef]
- Strehmel, N.; Bottcher, C.; Schmidt, S.; Scheel, D. Profiling of secondary metabolites in root exudates of Arabidopsis thaliana. Phytochemistry 2014, 108, 35–46. [Google Scholar] [CrossRef]
- Carvalhais, L.C.; Dennis, P.G.; Fedoseyenko, D.; Hajirezaei, M.R.; Borriss, R.; von Wirén, N. Root exudation of sugars, amino acids, and organic acids by maize as affected by nitrogen, phosphorus, potassium, and iron deficiency. J. Plant Nutr. Soil Sci. 2011, 174, 3–11. [Google Scholar] [CrossRef]
- Suzuki, K.; Okazaki, K.; Tawaraya, K.; Osaki, M.; Shinano, T. Gas chromatography–mass spectrometry associated global analysis of rice root exudates under aseptical conditions. Soil Sci. Plant Nutr. 2009, 55, 505–513. [Google Scholar] [CrossRef]
- Tawaraya, K.; Horie, R.; Wagatsuma, T.; Saito, K.; Oikawa, A. Metabolite profiling of shoot extract, root extract, and root exudate of rice under nitrogen and phosphorus deficiency. Soil Sci. Plant Nutr. 2018, 64, 312–322. [Google Scholar] [CrossRef]
- Stringlis, I.A.; Yu, K.; Feussner, K.; de Jonge, R.; Van Bentum, S.; Van Verk, M.C.; Berendsen, R.L.; Bakker, P.; Feussner, I.; Pieterse, C.M.J. MYB72-dependent coumarin exudation shapes root microbiome assembly to promote plant health. Proc. Natl. Acad. Sci. USA 2018, 115, E5213–E5222. [Google Scholar] [CrossRef]
- Zhang, N.; Yang, D.; Wang, D.; Miao, Y.; Shao, J.; Zhou, X.; Xu, Z.; Li, Q.; Feng, H.; Li, S.; et al. Whole transcriptomic analysis of the plant-beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 during enhanced biofilm formation regulated by maize root exudates. BMC Genom. 2015, 16, 685. [Google Scholar] [CrossRef]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef]
- Phillips, R.P.; Erlitz, Y.; Bier, R.; Bernhardt, E.S. New approach for capturing soluble root exudates in forest soils. Funct. Ecol. 2008, 22, 990–999. [Google Scholar] [CrossRef]
- Wilson, R.M.; Tfaily, M.M.; Kolton, M.; Johnston, E.R.; Petro, C.; Zalman, C.A.; Hanson, P.J.; Heyman, H.M.; Kyle, J.E.; Hoyt, D.W.; et al. Soil metabolome response to whole-ecosystem warming at the Spruce and Peatland Responses under Changing Environments experiment. Proc. Natl. Acad. Sci. USA 2021, 118, e2004192118. [Google Scholar] [CrossRef]
- Li, Y.; Xu, L.; Letuma, P.; Lin, W. Metabolite profiling of rhizosphere soil of different allelopathic potential rice accessions. BMC Plant Biol. 2020, 20, 265. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Yang, Y.; Ling, W.; Kong, H.; Zhu, X. Gradient distribution of root exudates and polycyclic aromatic hydrocarbons in rhizosphere soil. Soil Sci. Soc. Am. J. 2011, 75, 1694–1703. [Google Scholar] [CrossRef]
- Petriacq, P.; Williams, A.; Cotton, A.; McFarlane, A.E.; Rolfe, S.A.; Ton, J. Metabolite profiling of non-sterile rhizosphere soil. Plant J. 2017, 92, 147–162. [Google Scholar] [CrossRef]
- Harman, G.; Khadka, R.; Doni, F.; Uphoff, N. Benefits to Plant Health and Productivity from Enhancing Plant Microbial Symbionts. Front. Plant Sci. 2020, 11, 610065. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Vivanco, J.M.; Shen, Q. The unseen rhizosphere root-soil-microbe interactions for crop production. Curr. Opin. Microbiol. 2017, 37, 8–14. [Google Scholar] [CrossRef]
- Huang, W.; Sun, D.; Chen, L.; An, Y. Integrative analysis of the microbiome and metabolome in understanding the causes of sugarcane bitterness. Sci. Rep. 2021, 11, 6024. [Google Scholar] [CrossRef]
- Gust, A.A.; Biswas, R.; Lenz, H.D.; Rauhut, T.; Ranf, S.; Kemmerling, B.; Gotz, F.; Glawischnig, E.; Lee, J.; Felix, G.; et al. Bacteria-derived peptidoglycans constitute pathogen-associated molecular patterns triggering innate immunity in Arabidopsis. J. Biol. Chem. 2007, 282, 32338–32348. [Google Scholar] [CrossRef]
- Camanes, G.; Scalschi, L.; Vicedo, B.; Gonzalez-Bosch, C.; Garcia-Agustin, P. An untargeted global metabolomic analysis reveals the biochemical changes underlying basal resistance and priming in Solanum lycopersicum, and identifies 1-methyltryptophan as a metabolite involved in plant responses to Botrytis cinerea and Pseudomonas syringae. Plant J. 2015, 84, 125–139. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.; Tan, D.X.; Reiter, R.J.; Shi, H. Comparative metabolomic analysis highlights the involvement of sugars and glycerol in melatonin-mediated innate immunity against bacterial pathogen in Arabidopsis. Sci. Rep. 2015, 5, 15815. [Google Scholar] [CrossRef]
- Korenblum, E.; Dong, Y.; Szymanski, J.; Panda, S.; Jozwiak, A.; Massalha, H.; Meir, S.; Rogachev, I.; Aharoni, A. Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. USA 2020, 117, 3874–3883. [Google Scholar] [CrossRef]
- Huberty, M.; Martis, B.; van Kampen, J.; Choi, Y.H.; Vrieling, K.; Klinkhamer, P.G.L.; Bezemer, T.M. Soil Inoculation Alters Leaf Metabolic Profiles in Genetically Identical Plants. J. Chem. Ecol. 2020, 46, 745–755. [Google Scholar] [CrossRef]
- Pang, Q.; Zhang, T.; Wang, Y.; Kong, W.; Guan, Q.; Yan, X.; Chen, S. Metabolomics of Early Stage Plant Cell-Microbe Interaction Using Stable Isotope Labeling. Front. Plant Sci. 2018, 9, 760. [Google Scholar] [CrossRef]
- Chokkathukalam, A.; Kim, D.H.; Barrett, M.P.; Breitling, R.; Creek, D.J. Stable isotope-labeling studies in metabolomics: New insights into structure and dynamics of metabolic networks. Bioanalysis 2014, 6, 511–524. [Google Scholar] [CrossRef] [PubMed]
- Chialva, M.; Salvioli di Fossalunga, A.; Daghino, S.; Ghignone, S.; Bagnaresi, P.; Chiapello, M.; Novero, M.; Spadaro, D.; Perotto, S.; Bonfante, P. Native soils with their microbiotas elicit a state of alert in tomato plants. New Phytol. 2018, 220, 1296–1308. [Google Scholar] [CrossRef]
- Jiang, D.; Lin, R.; Tan, M.; Yan, J.; Yan, S. The mycorrhizal-induced growth promotion and insect resistance reduction in Populus alba x P. berolinensis seedlings: A multi-omics study. Tree Physiol. 2022, 42, 1059–1069. [Google Scholar] [CrossRef]
- Carrell, A.A.; Velickovic, D.; Lawrence, T.J.; Bowen, B.P.; Louie, K.B.; Carper, D.L.; Chu, R.K.; Mitchell, H.D.; Orr, G.; Markillie, L.M.; et al. Novel metabolic interactions and environmental conditions mediate the boreal peatmoss-cyanobacteria mutualism. ISME J. 2022, 16, 1074–1085. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sidharth, S.; Zeng, S.; Jiang, Y.; Chan, Y.O.; Lyu, Z.; McCubbin, T.; Mertz, R.; Sharp, R.E.; Joshi, T. Bioinformatics for plant and agricultural discoveries in the age of multiomics: A review and case study of maize nodal root growth under water deficit. Physiol. Plant 2022, 174, e13672. [Google Scholar] [CrossRef] [PubMed]
- Weighill, D.; Tschaplinski, T.J.; Tuskan, G.A.; Jacobson, D. Data Integration in Poplar: ’Omics Layers and Integration Strategies. Front. Genet. 2019, 10, 874. [Google Scholar] [CrossRef] [PubMed]
- Weighill, D.; Jones, P.; Bleker, C.; Ranjan, P.; Shah, M.; Zhao, N.; Martin, M.; DiFazio, S.; Macaya-Sanz, D.; Schmutz, J.; et al. Multi-Phenotype Association Decomposition: Unraveling Complex Gene-Phenotype Relationships. Front. Genet. 2019, 10, 417. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, A.; Mathe, E.; Merling, M.; Ma, Q.; Liu, B. Network analyses in microbiome based on high-throughput multi-omics data. Brief. Bioinform. 2021, 22, 1639–1655. [Google Scholar] [CrossRef]
- Jones, P.; Weighill, D.; Shah, M.; Climer, S.; Schmutz, J.; Sreedasyam, A.; Tuskan, G.; Jacobson, D. Network Modeling of Complex Data Sets. Methods Mol. Biol. 2020, 2096, 197–215. [Google Scholar] [CrossRef]
- Arkin, A.P.; Cottingham, R.W.; Henry, C.S.; Harris, N.L.; Stevens, R.L.; Maslov, S.; Dehal, P.; Ware, D.; Perez, F.; Canon, S.; et al. KBase: The United States Department of Energy Systems Biology Knowledgebase. Nat. Biotechnol. 2018, 36, 566–569. [Google Scholar] [CrossRef]
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018. [Google Scholar] [CrossRef]
- Wang, M.; Carver, J.J.; Phelan, V.V.; Sanchez, L.M.; Garg, N.; Peng, Y.; Nguyen, D.D.; Watrous, J.; Kapono, C.A.; Luzzatto-Knaan, T.; et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016, 34, 828–837. [Google Scholar] [CrossRef]
- Jack, C.N.; Petipas, R.H.; Cheeke, T.E.; Rowland, J.L.; Friesen, M.L. Microbial Inoculants: Silver Bullet or Microbial Jurassic Park? Trends Microbiol. 2021, 29, 299–308. [Google Scholar] [CrossRef]
- Moore, J.A.M.; Abraham, P.E.; Michener, J.K.; Muchero, W.; Cregger, M.A. Ecosystem consequences of introducing plant growth promoting rhizobacteria to managed systems and potential legacy effects. New Phytol. 2022, 234, 1914–1918. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).