Engineering Cell Polarization Improves Protein Production in Saccharomyces cerevisiae
Abstract
1. Introduction
2. Materials and Methods
2.1. Strain and Media
2.2. Plasmid and Strain Construction
2.3. CRISPR−Cas9 Mediated Gene Insertion
2.4. Enzymatic Assays
2.5. FACS
3. Results
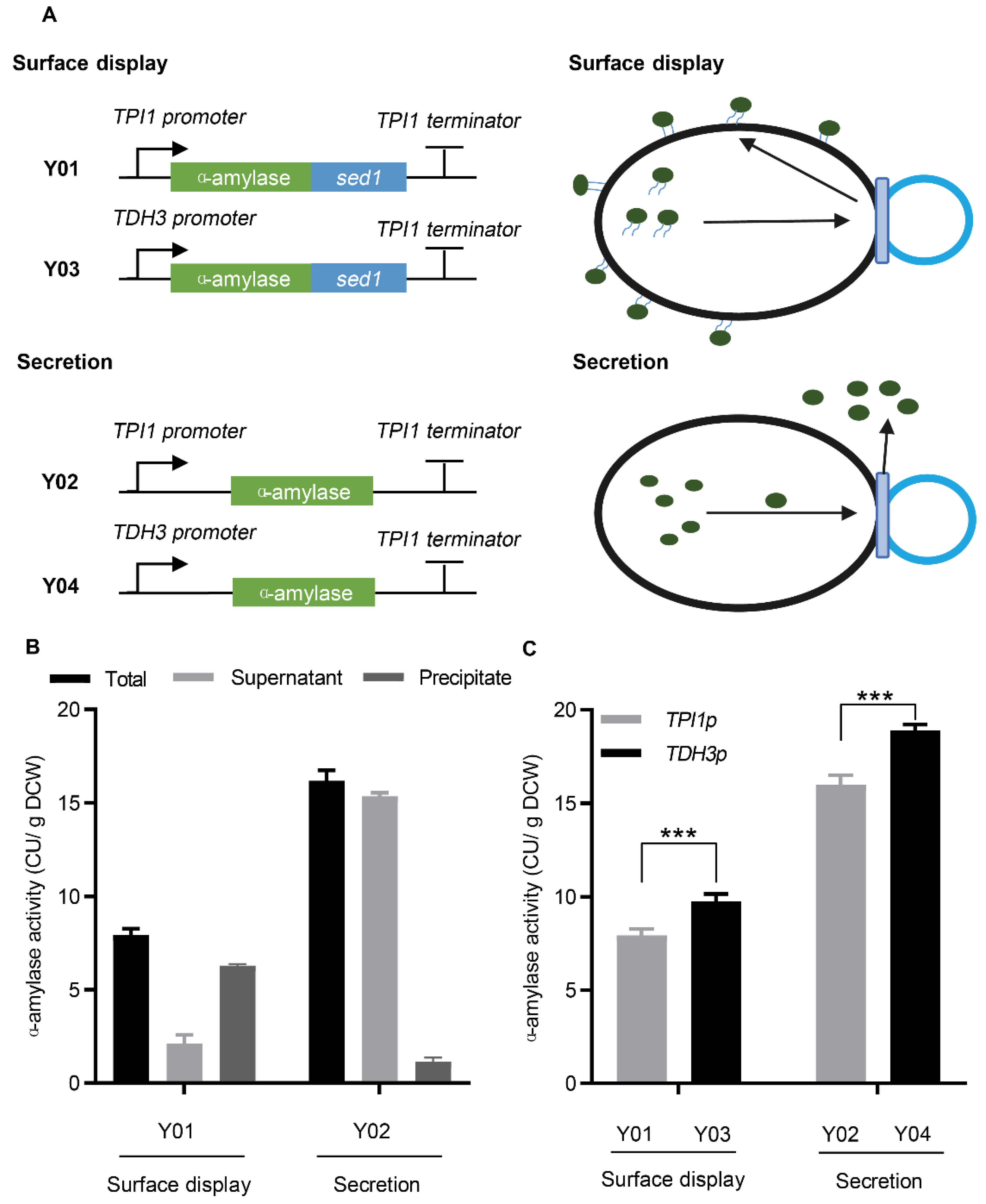
3.1. Secretion and Surface Display of α-Amylase in S. cerevisiae
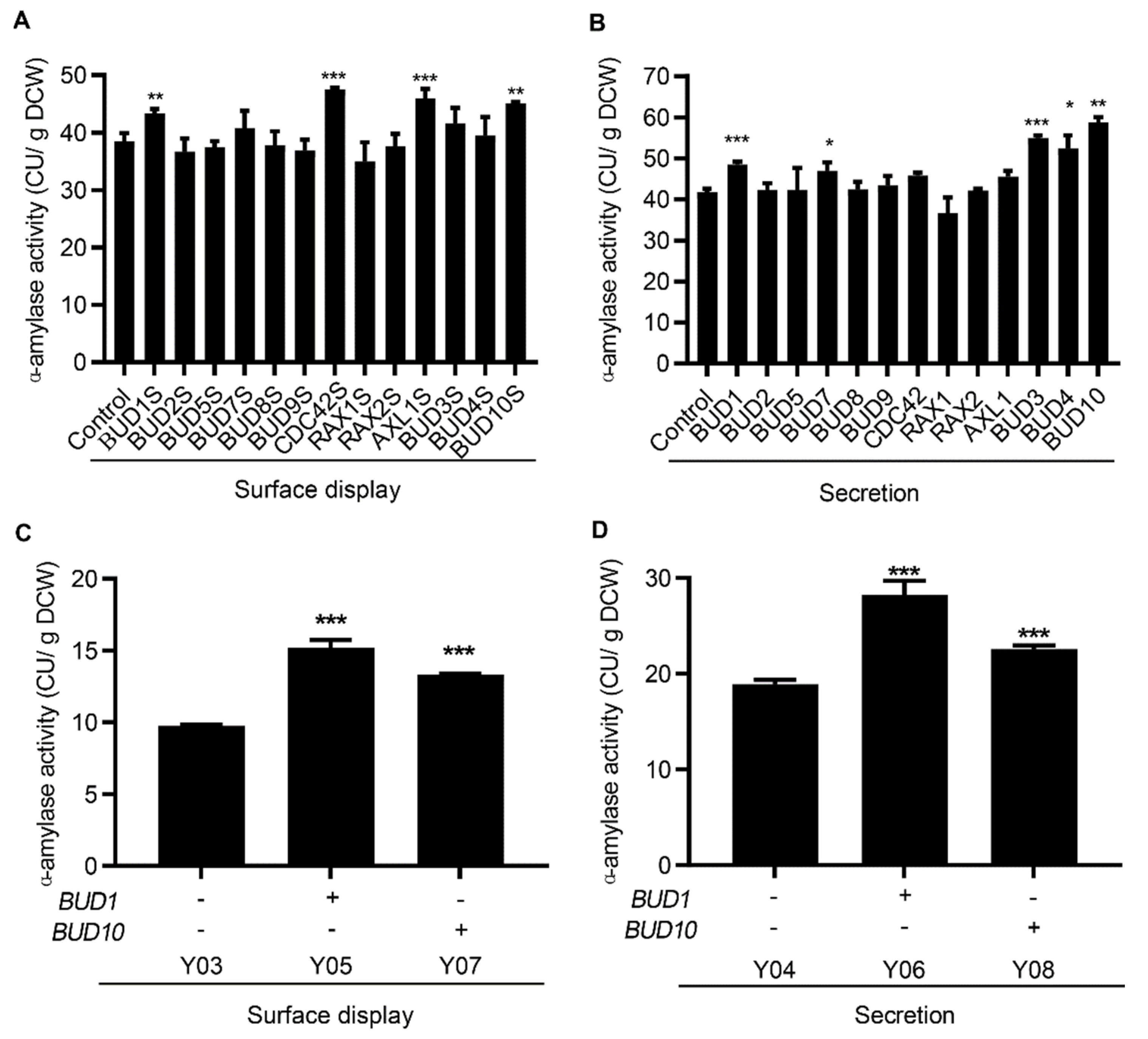
3.2. Cell Polarization Engineering Improved Protein Surface Display and Secretion
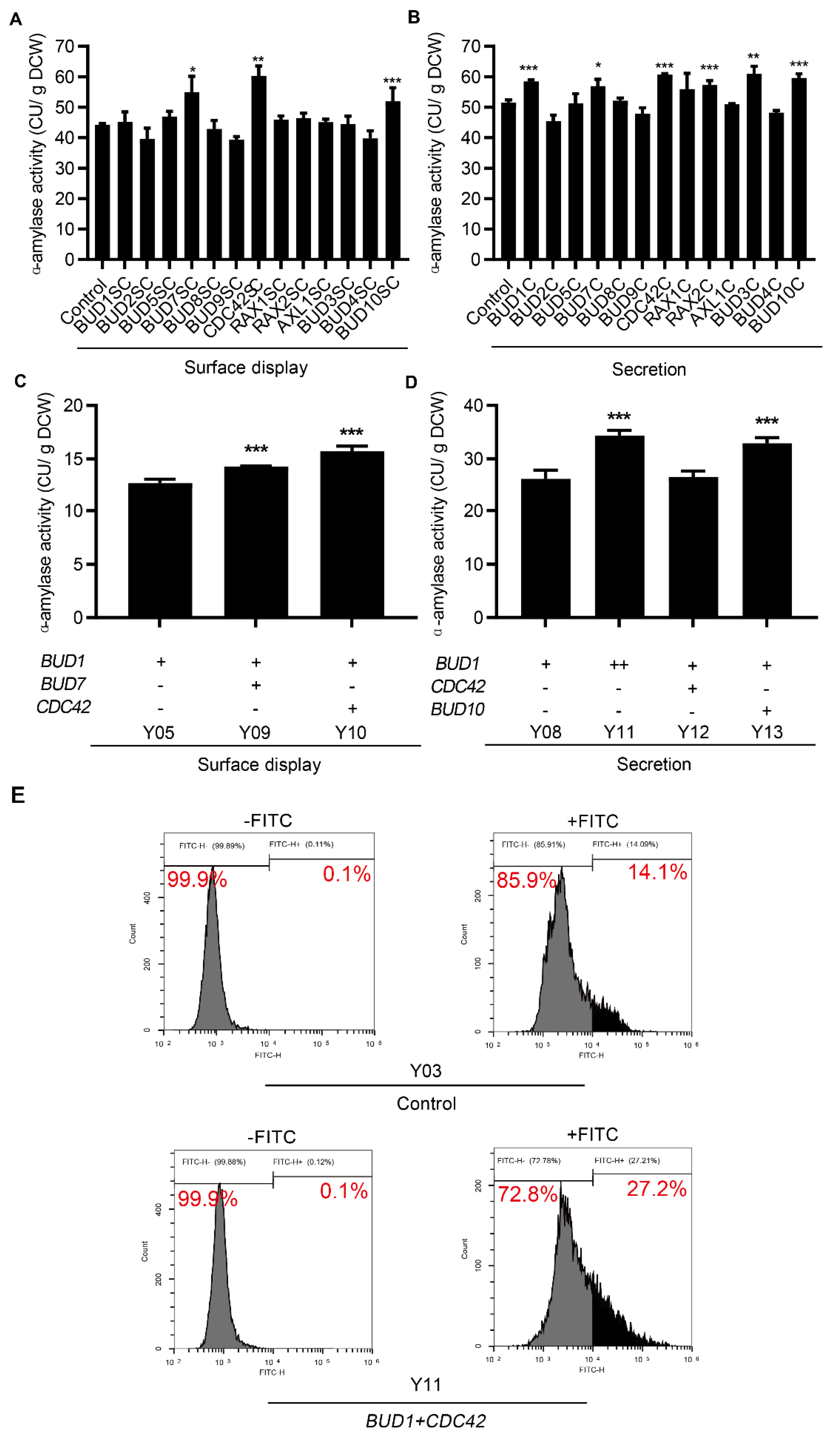
3.3. Effect of Combinatorial Modifications on Protein Surface Display and Secretion
3.4. Applicability Testing of Engineering Cell Polarization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Love, K.R.; Dalvie, N.C.; Love, J.C. The yeast stands alone: The future of protein biologic production. Curr. Opin. Biotechnol. 2018, 53, 50–58. [Google Scholar] [CrossRef]
- Huang, M.; Bao, J.; Nielsen, J.B. Biopharmaceutical protein production by Saccharomyces cerevisiae: Current state and future prospects. Pharm. Bioprocess. 2014, 2, 167–182. [Google Scholar] [CrossRef]
- Sanchez-Garcia, L.; Martín, L.; Mangues, R.; Ferrer-Miralles, N.; Vázquez, E.; Villaverde, A. Recombinant pharmaceuticals from microbial cells: A 2015 update. Microb. Cell Fact. 2016, 15, 33. [Google Scholar] [CrossRef]
- Çelik, E.; Çalık, P. Production of recombinant proteins by yeast cells. Biotechnol. Adv. 2012, 30, 1108–1118. [Google Scholar] [CrossRef]
- Cherf, G.M.; Cochran, J.R. Applications of Yeast Surface Display for Protein Engineering. Methods Mol. Biol. 2015, 1319, 155–175. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kondo, A. Cell-surface display of enzymes by the yeast Saccharomyces cerevisiae for synthetic biology. FEMS Yeast Res. 2015, 15, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, N.A.; Srikrishnan, S. Introduction and expression of genes for metabolic engineering applications in Saccharomyces cerevisiae. FEMS Yeast Res. 2012, 12, 197–214. [Google Scholar] [CrossRef]
- Huang, M.; Wang, G.; Qin, J.; Petranovic, D.; Nielsen, J. Engineering the protein secretory pathway of Saccharomyces cerevisiae enables improved protein production. Proc. Natl. Acad. Sci. USA 2018, 115, E11025–E11032. [Google Scholar] [CrossRef]
- Rakestraw, J.A.; Sazinsky, S.L.; Piatesi, A.; Antipov, E.; Wittrup, K.D. Directed evolution of a secretory leader for the improved expression of heterologous proteins and full-length antibodies in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2009, 103, 1192–1201. [Google Scholar] [CrossRef]
- Rodríguez-Limas, W.A.; Tannenbaum, V.; Tyo, K.E. Blocking endocytotic mechanisms to improve heterologous protein titers in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2015, 112, 376–385. [Google Scholar] [CrossRef]
- Bao, J.; Huang, M.; Petranovic, D.; Nielsen, J. Moderate Expression of SEC16 Increases Protein Secretion by Saccharomyces cerevisiae. Appl. Environ. Microbiol. 2017, 83, e03400-16. [Google Scholar] [CrossRef] [PubMed]
- De Ruijter, J.C.; Koskela, E.V.; Frey, A.D. Enhancing antibody folding and secretion by tailoring the Saccharomyces cerevisiae endoplasmic reticulum. Microb. Cell Fact. 2016, 15, 87. [Google Scholar] [CrossRef] [PubMed]
- Schönholzer, F.; Schweingruber, A.M.; Trachsel, H.; Schweingruber, M.E. Intracellular maturation and secretion of acid phosphatase of Saccharomyces cerevisiae. Eur. J. Biochem. 1985, 147, 273–279. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Song, M.; He, Y.; Wang, J.; Wang, S.; Shen, Y.; Hou, J.; Bao, X. Engineering vesicle trafficking improves the extracellular activity and surface display efficiency of cellulases in Saccharomyces cerevisiae. Biotechnol. Biofuels 2017, 10, 53. [Google Scholar] [CrossRef] [PubMed]
- Pruyne, D.; Bretscher, A. Polarization of cell growth in yeast. J. Cell Sci. 2000, 113 Pt 4, 571–585. [Google Scholar] [CrossRef] [PubMed]
- Lord, M.; Yang, M.C.; Mischke, M.; Chant, J. Cell cycle programs of gene expression control morphogenetic protein localization. J. Cell Biol. 2000, 151, 1501–1512. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Glick, B.S. The mechanisms of vesicle budding and fusion. Cell 2004, 116, 153–166. [Google Scholar] [CrossRef]
- Pruyne, D.; Bretscher, A. Polarization of cell growth in yeast. I. Establishment and maintenance of polarity states. J. Cell Sci. 2000, 113 Pt 3, 365–375. [Google Scholar] [CrossRef]
- Donovan, K.W.; Bretscher, A. Myosin-V is activated by binding secretory cargo and released in coordination with Rab/exocyst function. Dev. Cell 2012, 23, 769–781. [Google Scholar] [CrossRef]
- Schott, D.; Ho, J.; Pruyne, D.; Bretscher, A. The COOH-terminal domain of Myo2p, a yeast myosin V, has a direct role in secretory vesicle targeting. J. Cell Biol. 1999, 147, 791–808. [Google Scholar] [CrossRef]
- Finger, F.P.; Novick, P. Spatial regulation of exocytosis: Lessons from yeast. J. Cell Biol. 1998, 142, 609–612. [Google Scholar] [CrossRef]
- Adamo, J.E.; Moskow, J.J.; Gladfelter, A.S.; Viterbo, D.; Lew, D.J.; Brennwald, P.J. Yeast Cdc42 functions at a late step in exocytosis, specifically during polarized growth of the emerging bud. J. Cell Biol. 2001, 155, 581–592. [Google Scholar] [CrossRef] [PubMed]
- Zheng, T.; Zhang, M.; Wu, L.; Guo, S.; Liu, X.; Zhao, J.; Xue, W.; Li, J.; Liu, C.; Li, X.; et al. Upcycling CO2 into energy-rich long-chain compounds via electrochemical and metabolic engineering. Nat. Catal. 2022, 5, 388–396. [Google Scholar] [CrossRef]
- Wittrup, K.; Benig, V. Optimization of amino acid supplements for heterologous protein secretion in Saccharomyces cerevisiae. Biotechnol. Tech. 1994, 8, 161–166. [Google Scholar] [CrossRef]
- Liu, Z.; Tyo, K.E.; Martínez, J.L.; Petranovic, D.; Nielsen, J. Different expression systems for production of recombinant proteins in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2012, 109, 1259–1268. [Google Scholar] [CrossRef]
- Tang, H.; Hou, J.; Shen, Y.; Xu, L.; Yang, H.; Fang, X.; Bao, X. High β-glucosidase secretion in Saccharomyces cerevisiae improves the efficiency of cellulase hydrolysis and ethanol production in simultaneous saccharification and fermentation. J. Microbiol. Biotechnol. 2013, 23, 1577–1585. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Bao, X.; Shen, Y.; Song, M.; Wang, S.; Wang, C.; Hou, J. Engineering protein folding and translocation improves heterologous protein secretion in Saccharomyces cerevisiae. Biotechnol. Bioeng. 2015, 112, 1872–1882. [Google Scholar] [CrossRef]
- Deng, J.; Wu, Y.; Zheng, Z.; Chen, N.; Luo, X.; Tang, H.; Keasling, J.D. A synthetic promoter system for well-controlled protein expression with different carbon sources in Saccharomyces cerevisiae. Microb. Cell Fact. 2021, 20, 202. [Google Scholar] [CrossRef] [PubMed]
- Yu, T.; Zhou, Y.J.; Wenning, L.; Liu, Q.; Krivoruchko, A.; Siewers, V.; Nielsen, J.; David, F. Metabolic engineering of Saccharomyces cerevisiae for production of very long chain fatty acid-derived chemicals. Nat. Commun. 2017, 8, 15587. [Google Scholar] [CrossRef]
- Mans, R.; van Rossum, H.M.; Wijsman, M.; Backx, A.; Kuijpers, N.G.; van den Broek, M.; Daran-Lapujade, P.; Pronk, J.T.; van Maris, A.J.; Daran, J.M. CRISPR/Cas9: A molecular Swiss army knife for simultaneous introduction of multiple genetic modifications in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15, fov004. [Google Scholar] [CrossRef]
- Kitayama, C.; Sugimoto, A.; Yamamoto, M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J. Cell Biol. 1997, 137, 1309–1319. [Google Scholar] [CrossRef]
- Springer, S.; Chen, E.; Duden, R.; Marzioch, M.; Rowley, A.; Hamamoto, S.; Merchant, S.; Schekman, R. The p24 proteins are not essential for vesicular transport in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2000, 97, 4034–4039. [Google Scholar] [CrossRef]
- Castillon, G.A.; Aguilera-Romero, A.; Manzano-Lopez, J.; Epstein, S.; Kajiwara, K.; Funato, K.; Watanabe, R.; Riezman, H.; Muñiz, M. The yeast p24 complex regulates GPI-anchored protein transport and quality control by monitoring anchor remodeling. Mol. Biol. Cell 2011, 22, 2924–2936. [Google Scholar] [CrossRef]
- Belden, W.J.; Barlowe, C. Erv25p, a Component of COPII-coated Vesicles, Forms a Complex with Emp24p That Is Required for Efficient Endoplasmic Reticulum to Golgi Transport*. J. Biol. Chem. 1996, 271, 26939–26946. [Google Scholar] [CrossRef]
- Gurunathan, S.; David, D.; Gerst, J.E. Dynamin and clathrin are required for the biogenesis of a distinct class of secretory vesicles in yeast. EMBO J. 2002, 21, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Harsay, E.; Bretscher, A. Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 1995, 131, 297–310. [Google Scholar] [CrossRef]
- Inokuma, K.; Bamba, T.; Ishii, J.; Ito, Y.; Hasunuma, T.; Kondo, A. Enhanced cell-surface display and secretory production of cellulolytic enzymes with Saccharomyces cerevisiae Sed1 signal peptide. Biotechnol. Bioeng. 2016, 113, 2358–2366. [Google Scholar] [CrossRef]
- Nikawa, J.-i.; Akiyoshi, M.; Hirata, S.; Fukuda, T. Saccharomyces Cerevisiae IRE2/HAC1 Is Involved in IRE1-Mediated KAR2 Expression. Nucleic Acids Res. 1996, 24, 4222–4226. [Google Scholar] [CrossRef]
- Robinson, A.S.; Hines, V.; Wittrup, K.D. Protein disulfide isomerase overexpression increases secretion of foreign proteins in Saccharomyces cerevisiae. Bio/Technology 1994, 12, 381–384. [Google Scholar] [CrossRef]
- Tang, H.; Wang, S.; Wang, J.; Song, M.; Xu, M.; Zhang, M.; Shen, Y.; Hou, J.; Bao, X. N-hypermannose glycosylation disruption enhances recombinant protein production by regulating secretory pathway and cell wall integrity in Saccharomyces cerevisiae. Sci. Rep. 2016, 6, 25654. [Google Scholar] [CrossRef] [PubMed]
- Ash, J.; Dominguez, M.; Bergeron, J.J.; Thomas, D.Y.; Bourbonnais, Y. The yeast proprotein convertase encoded by YAP3 is a glycophosphatidylinositol-anchored protein that localizes to the plasma membrane. J. Biol. Chem. 1995, 270, 20847–20854. [Google Scholar] [CrossRef] [PubMed]
- Hermey, G. The Vps10p-domain receptor family. Cell Mol. Life Sci. 2009, 66, 2677–2689. [Google Scholar] [CrossRef]
- Lemus, L.; Goder, V. Pep4-dependent microautophagy is required for post-ER degradation of GPI-anchored proteins. Autophagy 2022, 18, 223–225. [Google Scholar] [CrossRef]
- Kuroda, K.; Ueda, M. Molecular design of the microbial cell surface toward the recovery of metal ions. Curr. Opin. Biotechnol. 2011, 22, 427–433. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Georgiou, G. Cell-Surface display of heterologous proteins: From high-throughput screening to environmental applications. Biotechnol. Bioeng. 2002, 79, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.Y.; Choi, J.H.; Xu, Z. Microbial cell-surface display. Trends Biotechnol. 2003, 21, 45–52. [Google Scholar] [CrossRef]
- Grabherr, R.; Ernst, W. The baculovirus expression system as a tool for generating diversity by viral surface display. Comb. Chem. High Throughput Screen. 2001, 4, 185–192. [Google Scholar] [CrossRef]
- Wang, G.; Huang, M.; Nielsen, J. Exploring the potential of Saccharomyces cerevisiae for biopharmaceutical protein production. Curr. Opin. Biotechnol. 2017, 48, 77–84. [Google Scholar] [CrossRef]
- Yang, X.; Tang, H.; Song, M.; Shen, Y.; Hou, J.; Bao, X. Development of novel surface display platforms for anchoring heterologous proteins in Saccharomyces cerevisiae. Microb. Cell Fact 2019, 18, 85. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Shen, J.; Deng, J.; Li, H.; Zhao, J.; Tang, H.; Bao, X. Engineering Cell Polarization Improves Protein Production in Saccharomyces cerevisiae. Microorganisms 2022, 10, 2005. https://doi.org/10.3390/microorganisms10102005
Yang S, Shen J, Deng J, Li H, Zhao J, Tang H, Bao X. Engineering Cell Polarization Improves Protein Production in Saccharomyces cerevisiae. Microorganisms. 2022; 10(10):2005. https://doi.org/10.3390/microorganisms10102005
Chicago/Turabian StyleYang, Shuo, Junfeng Shen, Jiliang Deng, Hongxing Li, Jianzhi Zhao, Hongting Tang, and Xiaoming Bao. 2022. "Engineering Cell Polarization Improves Protein Production in Saccharomyces cerevisiae" Microorganisms 10, no. 10: 2005. https://doi.org/10.3390/microorganisms10102005
APA StyleYang, S., Shen, J., Deng, J., Li, H., Zhao, J., Tang, H., & Bao, X. (2022). Engineering Cell Polarization Improves Protein Production in Saccharomyces cerevisiae. Microorganisms, 10(10), 2005. https://doi.org/10.3390/microorganisms10102005







