Impact of a Single-Tube PCR Assay for the Detection of Haemophilus influenzae Serotypes a, c, d, e and f on the Epidemiological Surveillance in Greece
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Source
2.2. Identification of H. Influenzae Serotype in Bacterial Isolates
2.3. DNA Isolation
2.4. Identification of H. influenzae in Clinical Samples
2.5. Development of the Multiplex PCR Assay for the Identification of H. influenzae Serotypes
2.5.1. PCR Primers
2.5.2. Amplification Protocol
2.6. Sensitivity and Specificity Assessment
3. Results
3.1. Assessment of the Multiplex PCR Assay Performance
3.1.1. Sensitivity and Specificity
3.1.2. Identification of H. influenzae in Clinical Samples and Isolates from Patients with Meningitis
3.2. Application to the Epidemiological Surveillance (2003–2020)
3.2.1. Serotype Distribution by Clinical Presentation
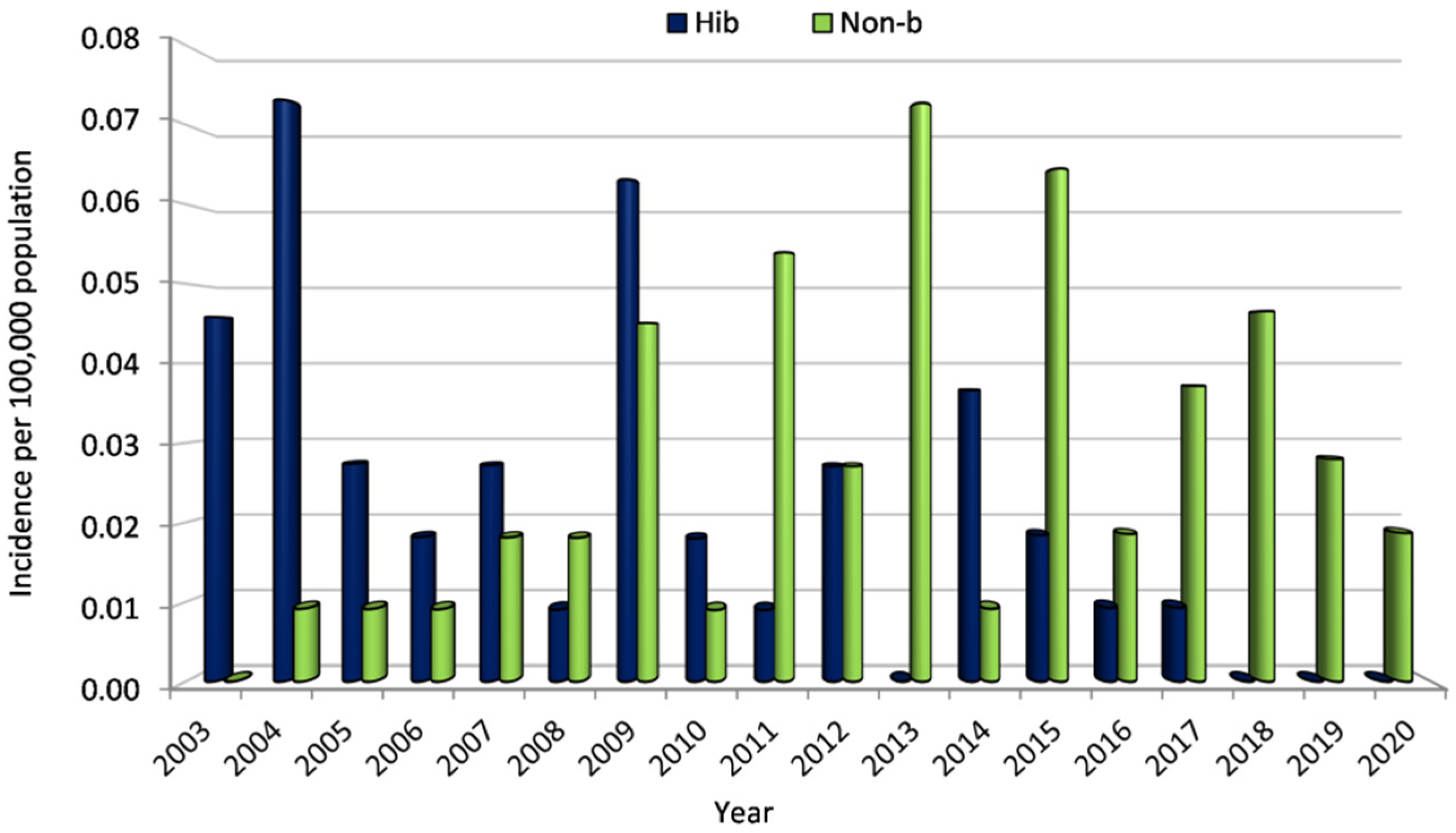
3.2.2. Incidence
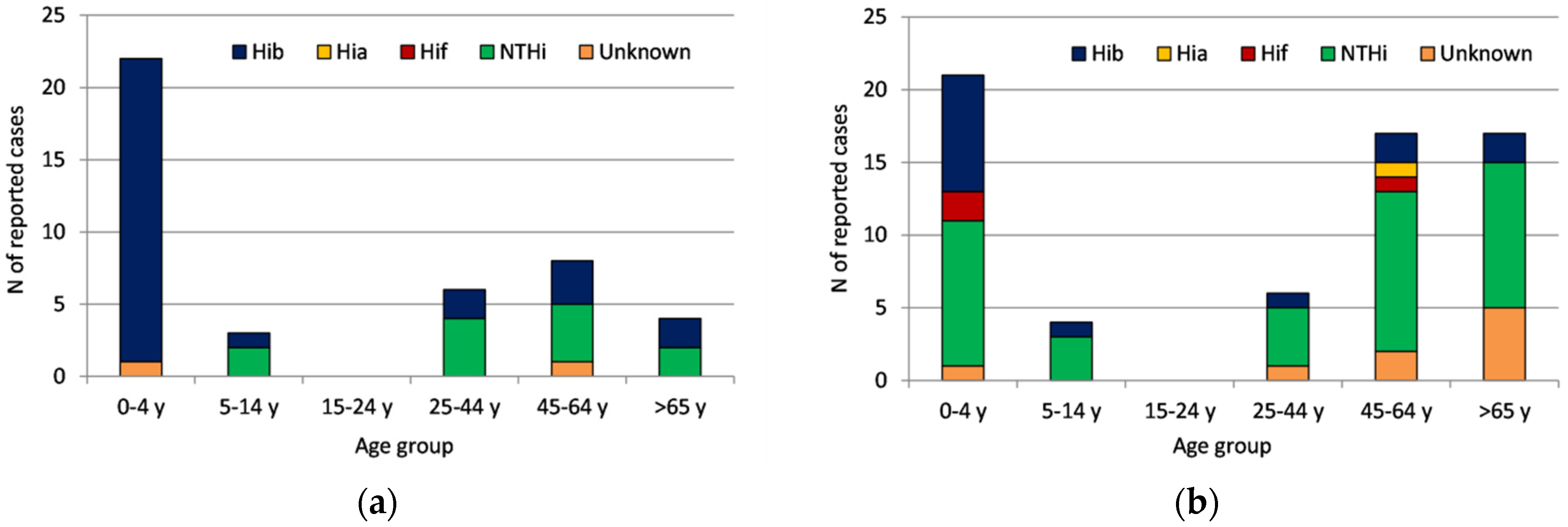
3.2.3. Serotype Distribution by Age Group and Time Period
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Pittman, M. Variation and Type Specificity in the Bacterial Species Hemophilus influenzae. J. Exp. Med. 1931, 53, 471–492. [Google Scholar] [CrossRef] [PubMed]
- Peltola, H.; Salo, E.; Saxén, H. Incidence of Haemophilus influenzae type b meningitis during 18 years of vaccine use: Observational study using routine hospital data. BMJ 2004, 330, 18–19. [Google Scholar] [CrossRef] [PubMed]
- Ulanova, M.; Tsang, R.S.W. Invasive Haemophilus influenzae disease: Changing epidemiology and host-parasite interactions in the 21st century. Infect. Genet. Evol. 2009, 9, 594–605. [Google Scholar] [CrossRef] [PubMed]
- St Geme, J.W., 3rd. The pathogenesis of nontypable Haemophilus influenzae otitis media. Vaccine 2000, 19 (Suppl. S1), S41–S50. [Google Scholar] [CrossRef]
- Jalalvand, F.; Riesbeck, K. Haemophilus influenzae: Recent advances in the understanding of molecular pathogenesis and polymicrobial infections. Curr. Opin. Infect. Dis. 2014, 27, 268–274. [Google Scholar] [CrossRef]
- Peltola, H. Worldwide Haemophilus influenzae Type b disease at the beginning of the 21st century: Global analysis of the disease burden 25 years after the use of the polysaccharide vaccine and a decade after the advent of conjugates. Clin. Microbiol. Rev. 2000, 13, 302–317. [Google Scholar] [CrossRef]
- Sarangi, J.; Cartwright, K.; Stuart, J.; Brookes, S.; Morris, R.; Slack, M. Invasive Haemophilus influenzae disease in adults. Epidemiol. Infect. 2000, 124, 441–447. [Google Scholar]
- Murphy, T.F. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr. Opin. Infect. Dis. 2003, 16, 129–134. [Google Scholar] [CrossRef]
- Resman, F.; Ristovski, M.; Ahl, J.; Forsgren, A.; Gilsdorf, J.R.; Jasir, A.; Kaijser, B.; Kronvall, G.; Riesbeck, K. Invasive disease caused by Haemophilus influenzae in Sweden 1997-2009; evidence of increasing incidence and clinical burden of non-type b strains. Clin. Microbiol. Infect. 2011, 7, 1638–1645. [Google Scholar] [CrossRef]
- Perdue, D.G.; Bulkow, L.R.; Gellin, B.G.; Davidson, M.; Petersen, K.M.; Singleton, R.J.; Parkinson, A.J. Invasive Haemophilus influenzae disease in Alaskan residents aged 10 years and older before and after infant vaccination programs. JAMA 2000, 283, 3089–3094. [Google Scholar] [CrossRef]
- Heath, P.T.; McVernon, J. The Uk Hib vaccine experience. Arch. Dis Child. 2002, 86, 396–399. [Google Scholar] [CrossRef]
- Bruun, B.; Gahrm-Hansen, B.; Westh, H.; Kilian, M. Clonal relationship of recent invasive Haemophilus influenzae serotype f isolates from Denmark and the United States. J. Med. Microbiol. 2004, 53, 1161–1165. [Google Scholar] [CrossRef]
- Whittaker, R.; Economopoulou, A.; Gomes Dias, J.; Bancroft, E.; Ramliden, M.; Pastore Celentano, L.; European Centre for Disease Prevention and Control Country Experts for Invasive Haemophilus influenzae Disease. Epidemiology of Invasive Haemophilus influenzae Disease, Europe, 2007–2014. Emerg. Infect. Dis. 2017, 23, 396–404. [Google Scholar] [CrossRef]
- LaClaire, L.L.; Tondella, M.L.C.; Beall, D.S.; Noble, C.A.; Raghunathan, P.L.; Rosenstein, N.E.; Popovic, T.; Active Bacterial Core Surveillance Team Members. Identification of Haemophilus influenzae by Standard Slide agglutination Serotyping and PCR-Based Capsule Typing. J. Clin. Microbiol. 2003, 41, 393–396. [Google Scholar] [CrossRef]
- Maaroufi, Y.; De Bruyne, J.M.; Heymans, C.; Crokaert, F. Real-time PCR for determining capsular serotypes of Haemophilus influenzae. J. Clin. Microbiol. 2007, 45, 2305–2308. [Google Scholar] [CrossRef]
- Wroblewski, D.; Halse, T.A.; Hayes, J.; Kohlerschmidt, D.; Musser, K.A. Utilization of a real-time PCR approach for Haemophilus influenzae serotype determination as analternative to the slide agglutination test. Mol. Cell Probes. 2013, 27, 86–89. [Google Scholar] [CrossRef]
- Takano, C.; Seki, M.; Kim, D.W.; Kilgore, P.E.; Fuwa, K.; Takahashi, K.; Inazaki, T.; Hayakawa, S. Molecular Serotype-Specific Identification of Non-type b Haemophilus influenzae by Loop-Mediated Isothermal Amplification. Front. Microbiol. 2017, 8, 1877. [Google Scholar] [CrossRef]
- Potts, C.C.; Topaz, N.; Rodriguez-Rivera, L.D.; Hu, F.; Chang, H.Y.; Whaley, M.J.; Schmink, S.; Retchless, A.C.; Chen, A.; Ramos, E.; et al. Genomic characterization of Haemophilus influenzae: A focus on the capsule locus. BMC Genom. 2019, 20, 733. [Google Scholar] [CrossRef]
- Tzanakaki, G.; Tsopanomichalou, M.; Kesanopoulos, K.; Matzourani, R.; Sioumala, M.; Tabaki, A.; Kremastinou, J. Simultaneous single-tube PCR assay for the detection of Neisseria meningitis, Haemophilus influenzae type b and Streptococcus pneumoniae. Clin. Microbiol. Infect. 2005, 11, 386–390. [Google Scholar] [CrossRef]
- Xirogianni, A.; Tzanakaki, G.; Karagianni, E.; Markoulatos, P.; Kourea-Kremastinou, J. Development of a single- tube polymerase chain reaction assay for the simultaneous detection of Haemophilus influenzae, Pseudomonas aeruginosa, Staphylococcus aureus and Streptococcus spp. directly in clinical samples. Diagn. Microbiol. Infect. Dis. 2009, 63, 121–126. [Google Scholar] [CrossRef]
- Falla, T.J.; Crook, D.W.M.; Brophy, L.N.; Maskell, D.; Croll, J.S.; Moxon, E.R. PCR for Capsular Typing of Haemophilus influenzae. J. Clin. Microbiol. 1994, 32, 2382–2386. [Google Scholar] [CrossRef]
- Lam, T.; Elias, J.; Frosch, M.; Vogel, U.; Claus, H. New Diagnostic PCR for Haemophilus influenzae serotype e based on the cap locus of strain ATCC8142. Int. J. Med. Microbiol. 2011, 301, 176–179. [Google Scholar] [CrossRef]
- Korbie, D.J.; Mattick, J.S. Touchdown PCR for increased specificity and sensitivity in PCR amplification. Nat. Protoc. 2008, 3, 1452–1456. [Google Scholar] [CrossRef]
- Tsolia, M.N.; Theodoridou, M.N.; Mostrou, G.J.; Paraskaki, I.I.; Pangali, A.M.; Yelesme, A.S.; Kalambalikis, P.A.; Gaviotaki, A.E.; Zoumboulakis, D.J.; Sinaniotis, C.A. Epidemiology of invasive Haemophilus influenzae type b infections among children in Greece before the introduction of immunization. Scand. J. Infect. Dis. 1998, 30, 165–168. [Google Scholar]
- Papavasileiou, K.; Papavasileiou, E.; Tzanakaki, G.; Voyatzi, A.; Kremastinou, J.; Chatzipanagiotou, S. Acute bacterial meningitis cases diagnosed by culture and PCR in a children’s hospital throughout a 9-Year period (2000–2008) in Athens, Greece. Mol. Diagn. Ther. 2011, 15, 109–113. [Google Scholar] [CrossRef]
- Theodoridou, M.N.; Vasilopoulou, V.A.; Atsali, E.E.; Pangalis, A.M.; Mostrou, G.J.; Syriopoulou, V.P.; Hadjichristodoulou, C.S. Meningitis registry of hospitalized cases in children: Epidemiological patterns of acute bacterial meningitis throughout a 32-year period. BMC Infect. Dis. 2007, 7, 101. [Google Scholar] [CrossRef]
- Ladomenou, F.; Tzanakaki, G.; Kolyva, S.; Katsarakis, I.; Maraki, S.; Galanakis, E. Conjugate vaccines dramatically reshaped the epidemiology of bacterial meningitis in a well-defined child population. Acta Paediatr. 2020, 109, 368–374. [Google Scholar] [CrossRef]
- Kofteridis, D.; Samonis, G.; Mantadakis, E.; Maraki, S.; Chrysofakis, G.; Alegakis, D.; Papadakis, J.; Gikas, A.; Bouros, D. Lower respiratory tract infections caused by Haemophilus influenzae: Clinical features and predictors of outcome. Med. Sci. Monit. 2009, 15, CR135-9. Available online: http://www.medscimonit.com/fulltxt.php?ICID=869612 (accessed on 4 July 2022).
- Georgakopoulou, T.; Menegas, D.; Katsioulis, A.; Theodoridou, M.; Kremastinou, J.; Hadjichristodoulou, C. A cross-sectional vaccination coverage study in preschool children attending nurseries-kindergartens: Implications on economic crisis effect. Hum. Vaccin. Immunother. 2017, 13, 190–197. [Google Scholar] [CrossRef]
- Larson, H.; de Figueiredo, A.; Karafillakis, E.; Rawal, M. State of Vaccine Confidence in the EU 2018; Publications Office of the European Union: Luxembourg, 2018. Available online: https://ec.europa.eu/health/system/files/2018-11/2018_vaccine_confidence_en_0.pdf (accessed on 13 November 2018).
- Monge, S.; Mollema, L.; de Melker, H.; Sanders, E.; van der Ende, A.; Knol, M. Clinical Characterization of Invasive Disease Caused by Haemophilus influenzae Serotype b in a High Vaccination Coverage Setting. J. Pediatric. Infect. Dis. Soc. 2019, 8, 261–264. [Google Scholar] [CrossRef]
- Giufrè, M.; Lindh, E.; Cardines, R.; Pezzotti, P.; Cerquetti, M. Invasive Haemophilus influenzae type b (Hib) disease in children in Italy, after 20 years of routine use of conjugate Hib vaccines. Vaccine 2020, 38, 6533–6538. [Google Scholar] [CrossRef]
- Takla, A.; Schönfeld, V.; Claus, H.; Krone, M.; An der Heiden, M.; Koch, J.; Vogel, U.; Wichmann, O.; Lâm, T.T. Invasive Haemophilus influenzae Infections in Germany After the Introduction of Routine Childhood Immunization, 2001–2016. Open Forum Infect. Dis. 2020, 7, ofaa444. [Google Scholar] [CrossRef]
- Koelman, D.L.H.; van Kassel, M.N.; Bijlsma, M.W.; Brouwer, M.C.; van de Beek, D.; van der Ende, A. Changing Epidemiology of Bacterial Meningitis Since Introduction of Conjugate Vaccines: Three Decades of National Meningitis Surveillance in The Netherlands. Clin. Infect. Dis. 2021, 73, e1099–e1107. [Google Scholar] [CrossRef]
- Giufrè, M.; Fabiani, M.; Cardines, R.; Riccardo, F.; Caporali, M.G.; D’Ancona, F.; Pezzotti, P.; Cerquetti, M. Increasing trend in invasive non-typeable Haemophilus influenzae disease and molecular characterization of the isolates, Italy, 2012–2016. Vaccine 2018, 36, 6615–6622. [Google Scholar] [CrossRef]
- Deghmane, A.E.; Hong, E.; Chehboub, S.; Terrade, A.; Falguières, M.; Sort, M.; Harrison, O.; Jolley, K.A.; Taha, M.K. High diversity of invasive Haemophilus influenzae isolates in France and the emergence of resistance to third generation cephalosporins by alteration of ftsI gene. J. Infect. 2019, 79, 7–14. [Google Scholar] [CrossRef]
- McElligott, M.; Meyler, K.; Bennett, D.; Mulhall, R.; Drew, R.J.; Cunney, R. Epidemiology of Haemophilus influenzae in the Republic of Ireland, 2010–2018. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 2335–2344. [Google Scholar] [CrossRef]
- Ladhani, S.; Slack, M.P.; Heath, P.T.; von Gottberg, A.; Chandra, M.; Ramsay, M.E.; European Union Invasive Bacterial Infection Surveillance participants. Invasive Haemophilus influenzae Disease, Europe, 1996–2006. Emerg. Infect. Dis. 2010, 16, 455–463. [Google Scholar] [CrossRef]
- Ladhani, S.N.; Collins, S.; Vickers, A.; Litt, D.J.; Crawford, C.; Ramsay, M.E.; Slack, M.P. Invasive Haemophilus influenzae serotype e and f disease, England and Wales. Emerg. Infect. Dis 2012, 18, 725–732. [Google Scholar] [CrossRef]
- Kalies, H.; Siedler, A.; Gröndahl, B.; Grote, V.; Milde-Busch, A.; von Kries, R. Invasive Haemophilus influenzae infections in Germany: Impact of non-type b serotypes in the post-vaccine era. BMC Infect. Dis. 2009, 9, 45. [Google Scholar] [CrossRef]
- Ulu-Kilic, A.; Altay, F.A.; Gürbüz, Y.; Otgun, S.N.; Sencan, I. Haemophilus influenzae serotype e meningitis in an adult. J. Infect. Dev. Ctries. 2010, 4, 253–255. [Google Scholar] [CrossRef][Green Version]

| Serotype | Primers | Sequences (5′-3′) | Amplicon Size (bp) | Publication |
|---|---|---|---|---|
| a | haf | ATCTTACAACTTAGCGAATAC | 1180 | modified [15] (6 nucleotides from the 5′ edge have been deleted) |
| ha2 | GAATATGACCTGATCTTCTG | [21] | ||
| c | hc2 | CAGAGGCAAGCTATTAGTGA | 200 | [21] |
| hc3 | TGGCAGCGTAAATATCCTAA | |||
| d | hd1 | TGATGACCGATACAACCTGT | 150 | |
| hd2 | TCCACTCTTCAAACCATTCT | |||
| e | TTL 63 | GAGCAATTCCATCGTAGTAAC | 592 | [22] |
| TTL 64 | TTCTTCATATCCTCGCAATTG | |||
| f | hf1 | GCTACTATCAAGTCCAAATC | 456 | [21] |
| hf2 | CGCAATTATGGAAGAAAGCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xirogianni, A.; Georgakopoulou, T.; Patsourakos, V.; Magaziotou, I.; Papandreou, A.; Simantirakis, S.; Tzanakaki, G. Impact of a Single-Tube PCR Assay for the Detection of Haemophilus influenzae Serotypes a, c, d, e and f on the Epidemiological Surveillance in Greece. Microorganisms 2022, 10, 1367. https://doi.org/10.3390/microorganisms10071367
Xirogianni A, Georgakopoulou T, Patsourakos V, Magaziotou I, Papandreou A, Simantirakis S, Tzanakaki G. Impact of a Single-Tube PCR Assay for the Detection of Haemophilus influenzae Serotypes a, c, d, e and f on the Epidemiological Surveillance in Greece. Microorganisms. 2022; 10(7):1367. https://doi.org/10.3390/microorganisms10071367
Chicago/Turabian StyleXirogianni, Athanasia, Theano Georgakopoulou, Vassileios Patsourakos, Ioanna Magaziotou, Anastasia Papandreou, Stelmos Simantirakis, and Georgina Tzanakaki. 2022. "Impact of a Single-Tube PCR Assay for the Detection of Haemophilus influenzae Serotypes a, c, d, e and f on the Epidemiological Surveillance in Greece" Microorganisms 10, no. 7: 1367. https://doi.org/10.3390/microorganisms10071367
APA StyleXirogianni, A., Georgakopoulou, T., Patsourakos, V., Magaziotou, I., Papandreou, A., Simantirakis, S., & Tzanakaki, G. (2022). Impact of a Single-Tube PCR Assay for the Detection of Haemophilus influenzae Serotypes a, c, d, e and f on the Epidemiological Surveillance in Greece. Microorganisms, 10(7), 1367. https://doi.org/10.3390/microorganisms10071367







