EbfC/YbaB: A Widely Distributed Nucleoid-Associated Protein in Prokaryotes
Abstract
:1. Introduction
2. Operon Structure and Expression
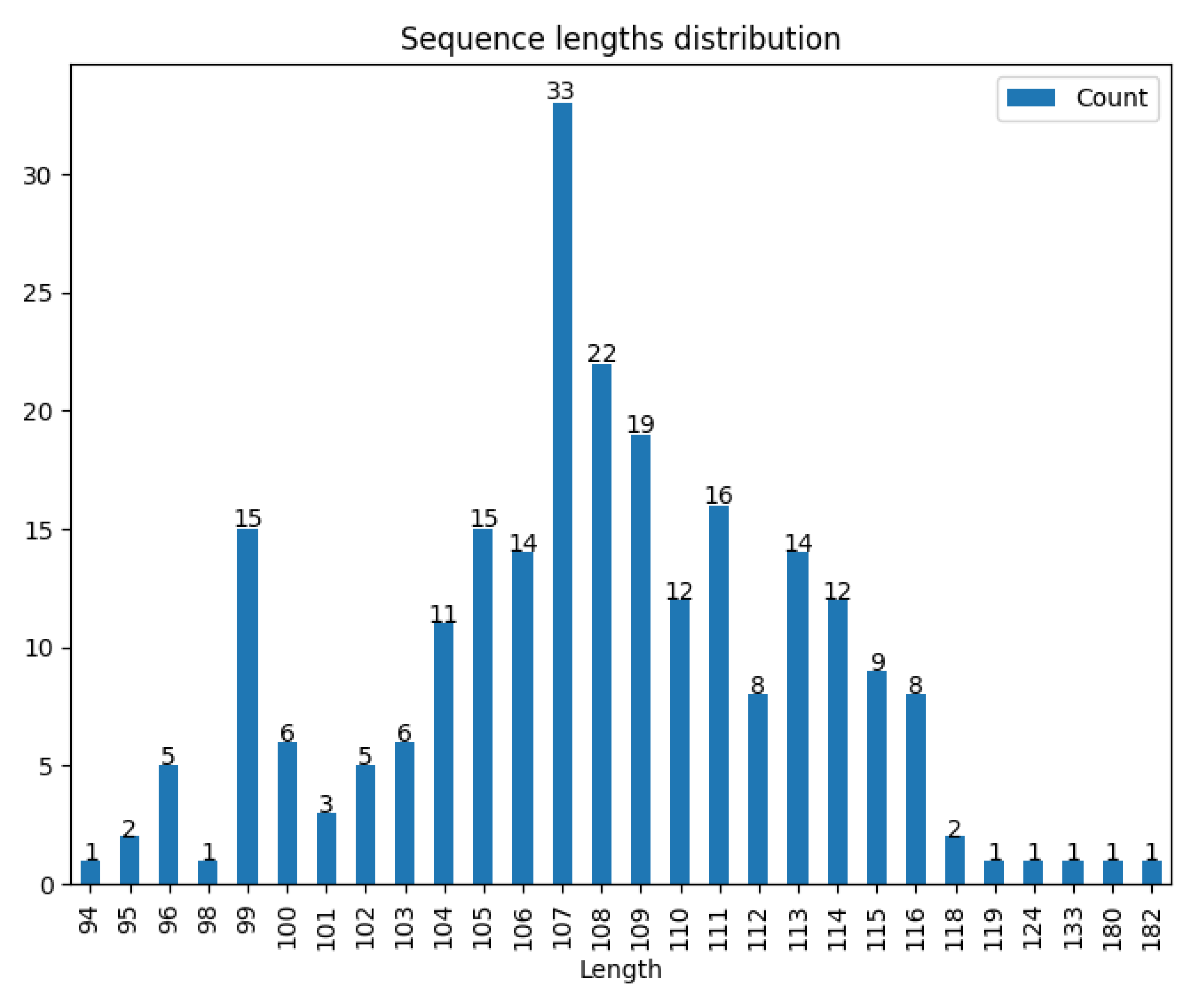
3. EbfC/YbaB Orthologs Are Widely Distributed in Prokaryotes
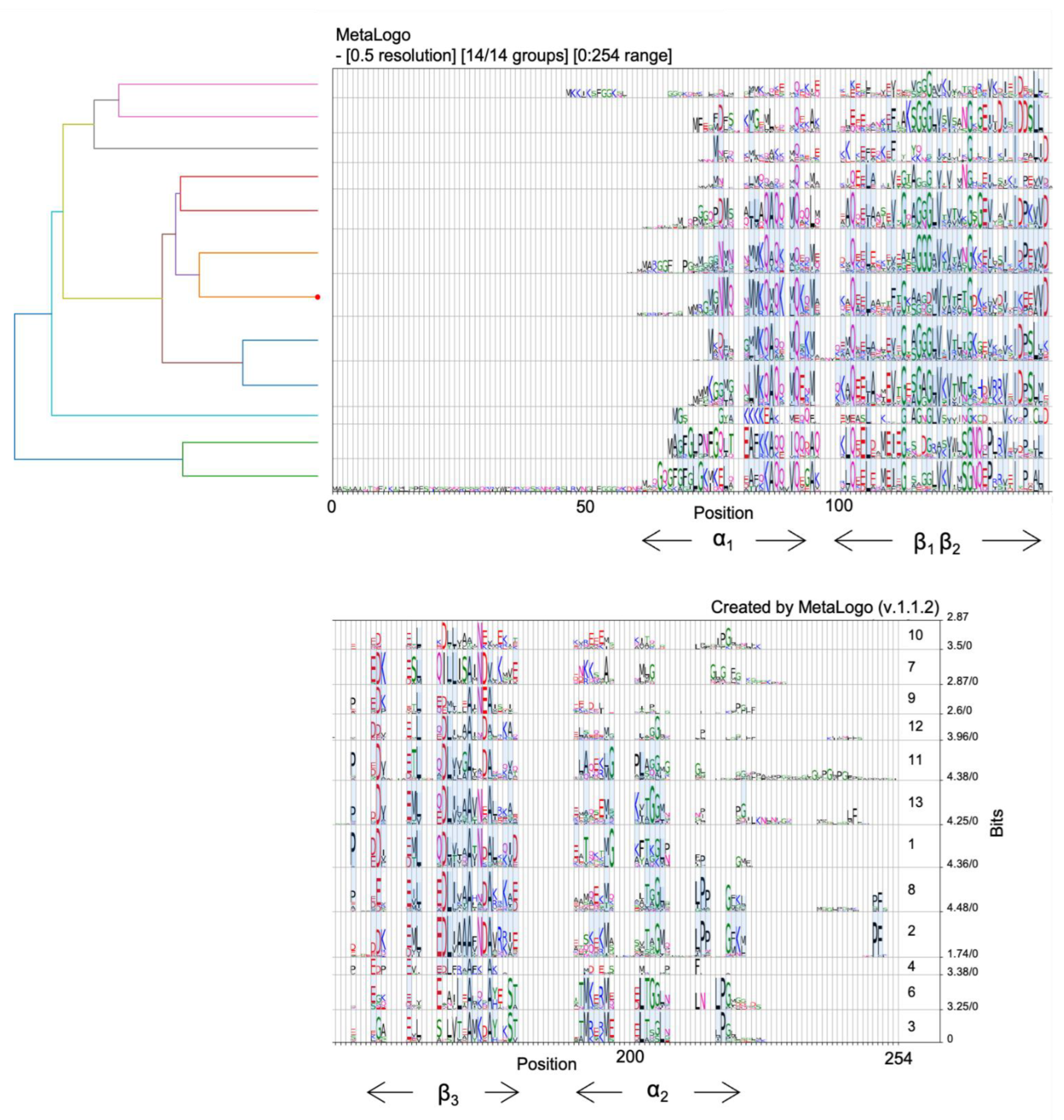
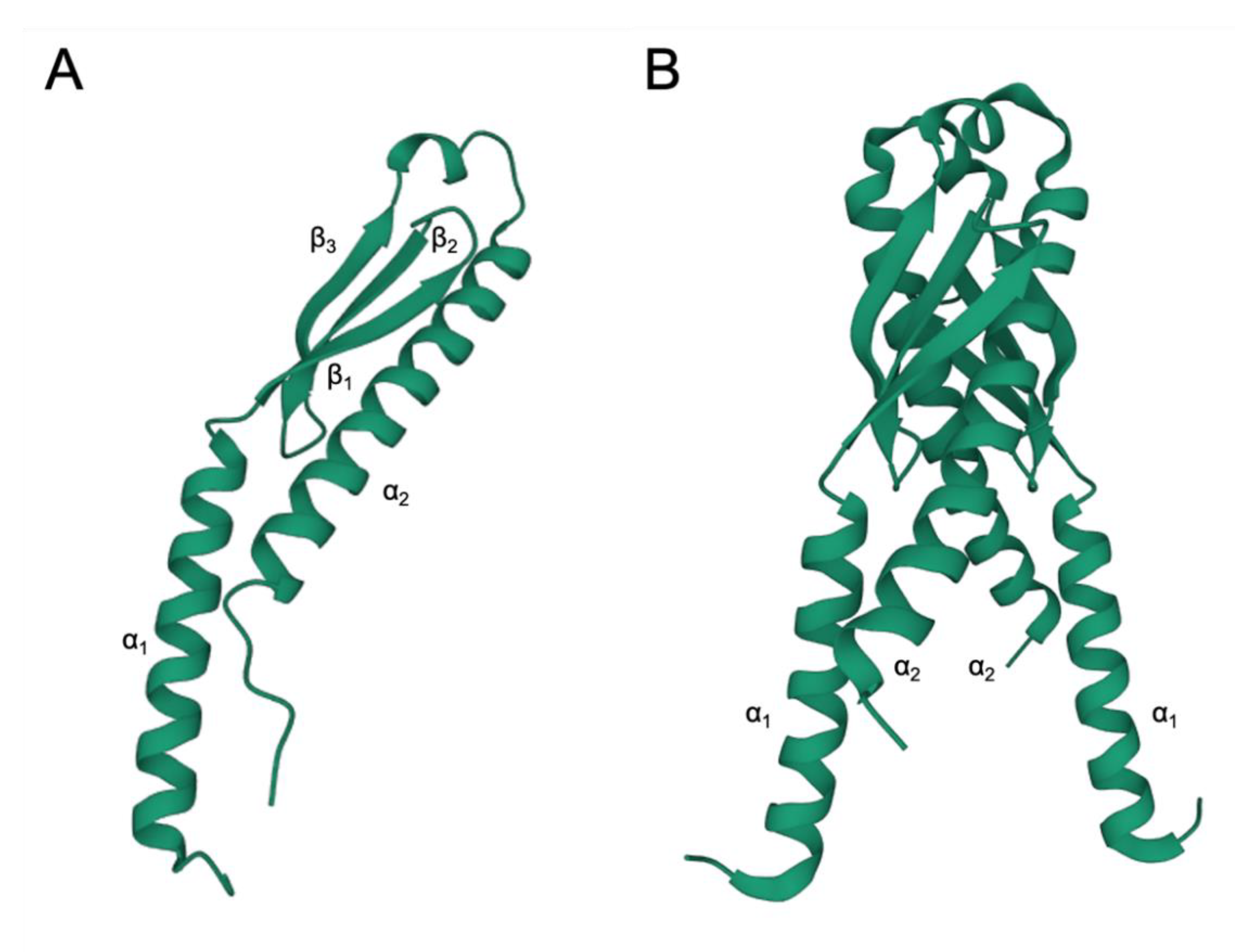
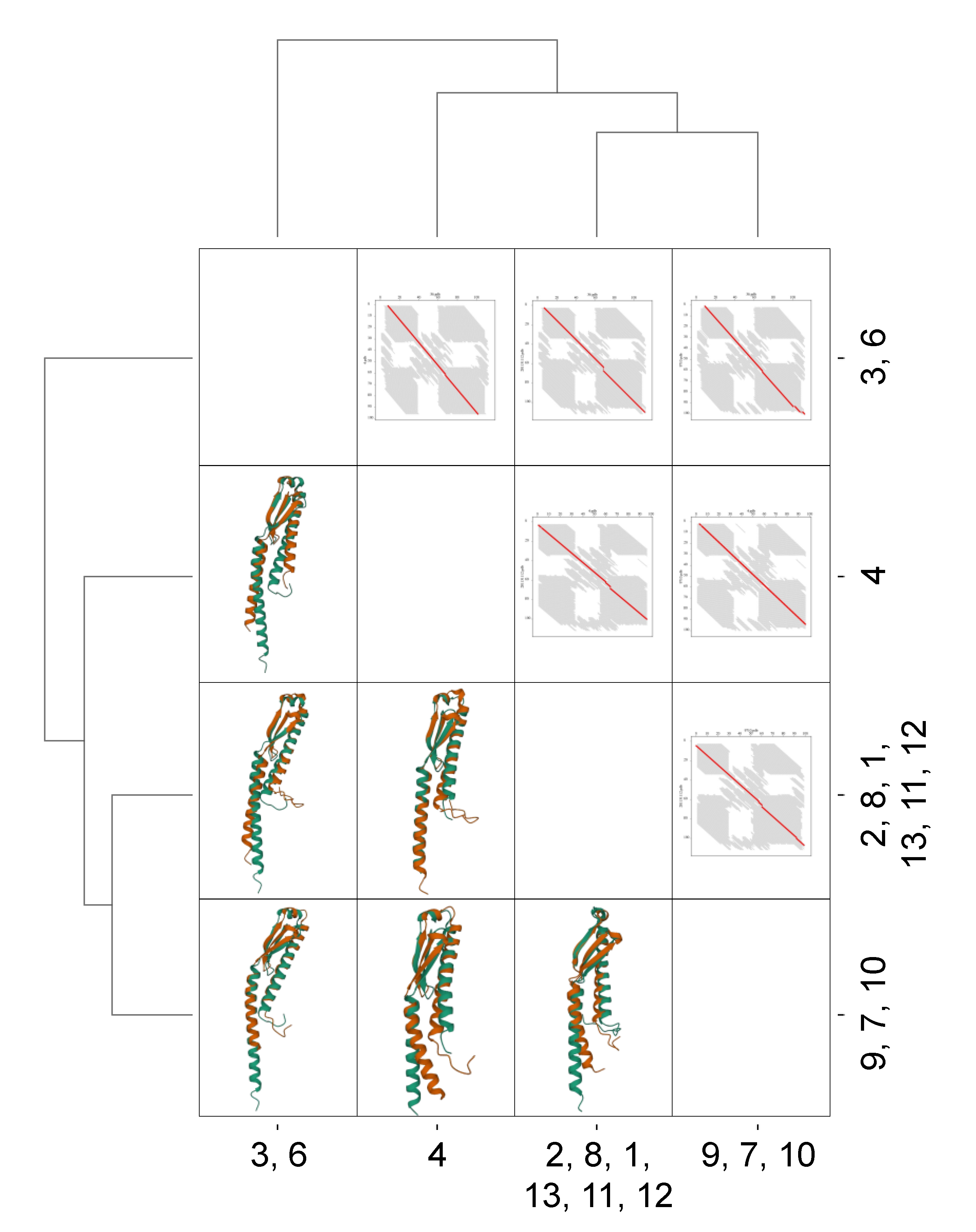
4. EbfC/YbaB Structure
5. Ability to Bind to DNA
6. EbfC/YbaB Has Important Biological Functions
| Biological Roles of EbfC/YbaB | Bacterial Species | Ref. |
|---|---|---|
| Regulatory functions and DNA Repair/Protection | ||
| Component of sigma E regulon (envelop stress response) | Escherichia coli | [41] |
| Regulation of pathogenic factors (EbfC as anti-repressor, stimulating the transcription of the erp operon) | Borrelia burgdorferi | [29] |
| Global regulator of gene expression | Borrelia burgdorferi | [30] |
| UV survival | Deinococcus radiodurans Escherichia coli | [55,61] |
| DNA protection from enzymatic action | Deinococcus radiodurans Caulobacter crescentus | [47,55] |
| DNA protection under iron-limiting conditions | Paenibacillus riograndensis | [62] |
| Cellular processes involving EbfC/YbaB | ||
| Higher expression of several heterologous membrane proteins | Escherichia coli | [43] |
| EbfC/YbaB target of ClpYQ protease | Escherichia coli | [63] |
7. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Dame, R.T.; Rashid, F.-Z.M.; Grainger, D.C. Chromosome Organization in Bacteria: Mechanistic Insights into Genome Structure and Function. Nat. Rev. Genet. 2020, 21, 227–242. [Google Scholar] [CrossRef]
- Dillon, S.C.; Dorman, C.J. Bacterial Nucleoid-Associated Proteins, Nucleoid Structure and Gene Expression. Nat. Rev. Microbiol. 2010, 8, 185–195. [Google Scholar] [CrossRef]
- Amemiya, H.M.; Schroeder, J.; Freddolino, P.L. Nucleoid-Associated Proteins Shape Chromatin Structure and Transcriptional Regulation across the Bacterial Kingdom. Transcription 2021, 12, 182–218. [Google Scholar] [CrossRef]
- Ó Cróinín, T.; Carroll, R.K.; Kelly, A.; Dorman, C.J. Roles for DNA Supercoiling and the Fis Protein in Modulating Expression of Virulence Genes during Intracellular Growth of Salmonella enterica Serovar Typhimurium. Mol. Microbiol. 2006, 62, 869–882. [Google Scholar] [CrossRef]
- Kelly, A.; Goldberg, M.D.; Carroll, R.K.; Danino, V.; Hinton, J.C.D.; Dorman, C.J. A Global Role for Fis in the Transcriptional Control of Metabolism and Type III Secretion in Salmonella enterica Serovar Typhimurium. Microbiology 2004, 150, 2037–2053. [Google Scholar] [CrossRef]
- Mangan, M.W.; Lucchini, S.; Danino, V.; Cróinín, T.Ó.; Hinton, J.C.D.; Dorman, C.J. The Integration Host Factor (IHF) Integrates Stationary-Phase and Virulence Gene Expression in Salmonella enterica Serovar Typhimurium. Mol. Microbiol. 2006, 59, 1831–1847. [Google Scholar] [CrossRef]
- Dorman, C.J. Function of Nucleoid-Associated Proteins in Chromosome Structuring and Transcriptional Regulation. J. Mol. Microbiol. Biotechnol. 2014, 24, 316–331. [Google Scholar] [CrossRef]
- Dorman, C.J. Co-Operative Roles for DNA Supercoiling and Nucleoid-Associated Proteins in the Regulation of Bacterial Transcription. Biochem. Soc. Trans. 2013, 41, 542–547. [Google Scholar] [CrossRef]
- Navarre, W.W.; Porwollik, S.; Wang, Y.; McClelland, M.; Rosen, H.; Libby, S.J.; Fang, F.C. Selective Silencing of Foreign DNA with Low GC Content by the H-NS Protein in Salmonella. Science 2006, 313, 236–238. [Google Scholar] [CrossRef]
- Lucchini, S.; Rowley, G.; Goldberg, M.D.; Hurd, D.; Harrison, M.; Hinton, J.C.D. H-NS Mediates the Silencing of Laterally Acquired Genes in Bacteria. PLoS Pathog. 2006, 2, e81. [Google Scholar] [CrossRef]
- Dillon, S.C.; Cameron, A.D.S.; Hokamp, K.; Lucchini, S.; Hinton, J.C.D.; Dorman, C.J. Genome-Wide Analysis of the H-NS and Sfh Regulatory Networks in Salmonella Typhimurium Identifies a Plasmid-Encoded Transcription Silencing Mechanism. Mol. Microbiol. 2010, 76, 1250–1265. [Google Scholar] [CrossRef]
- Will, W.R.; Navarre, W.W.; Fang, F.C. Integrated Circuits: How Transcriptional Silencing and Counter-Silencing Facilitate Bacterial Evolution. Curr. Opin. Microbiol. 2015, 23, 8–13. [Google Scholar] [CrossRef]
- Dorman, M.J.; Dorman, C.J. Regulatory Hierarchies Controlling Virulence Gene Expression in Shigella flexneri and Vibrio cholerae. Front. Microbiol. 2018, 9, 2686. [Google Scholar] [CrossRef]
- Dorman, C.J.; Schumacher, M.A.; Bush, M.J.; Brennan, R.G.; Buttner, M.J. When Is a Transcription Factor a NAP? Curr. Opin. Microbiol. 2020, 55, 26–33. [Google Scholar] [CrossRef]
- Flåtten, I.; Skarstad, K. The Fis Protein Has a Stimulating Role in Initiation of Replication in Escherichia coli In Vivo. PLoS ONE 2013, 8, e83562. [Google Scholar] [CrossRef]
- Dorman, C.J.; Colgan, A.; Dorman, M.J. Bacterial Pathogen Gene Regulation: A DNA-Structure-Centred View of a Protein-Dominated Domain. Clin. Sci. 2016, 130, 1165–1177. [Google Scholar] [CrossRef]
- Leite, B.; Werle, C.H.; do Carmo, C.P.; Nóbrega, D.B.; Milanez, G.P.; Culler, H.F.; Sircili, M.P.; Alvarez-Martinez, C.E.; Brocchi, M. Integration Host Factor Is Important for Biofilm Formation by Salmonella enterica Enteritidis. Pathog. Dis. 2017, 75, ftx074. [Google Scholar] [CrossRef]
- Fitzgerald, S.; Dillon, S.C.; Chao, T.-C.; Wiencko, H.L.; Hokamp, K.; Cameron, A.D.S.; Dorman, C.J. Re-Engineering Cellular Physiology by Rewiring High-Level Global Regulatory Genes. Sci. Rep. 2015, 5, 17653. [Google Scholar] [CrossRef]
- Ali, B.M.; Amit, R.; Braslavsky, I.; Oppenheim, A.B.; Gileadi, O.; Stavans, J. Compaction of Single DNA Molecules Induced by Binding of Integration Host Factor (IHF). Proc. Natl. Acad. Sci. USA 2001, 98, 10658–10663. [Google Scholar] [CrossRef]
- Duprey, A.; Reverchon, S.; Nasser, W. Bacterial Virulence and Fis: Adapting Regulatory Networks to the Host Environment. Trends Microbiol. 2014, 22, 92–99. [Google Scholar] [CrossRef]
- Cameron, A.D.S.; Stoebel, D.M.; Dorman, C.J. DNA Supercoiling Is Differentially Regulated by Environmental Factors and FIS in Escherichia coli and Salmonella enterica. Mol. Microbiol. 2011, 80, 85–101. [Google Scholar] [CrossRef]
- Mangan, M.W.; Lucchini, S.; Ó Cróinín, T.; Fitzgerald, S.; Hinton, J.C.D.; Dorman, C.J. Nucleoid-Associated Protein HU Controls Three Regulons That Coordinate Virulence, Response to Stress and General Physiology in Salmonella enterica Serovar Typhimurium. Microbiology 2011, 157, 1075–1087. [Google Scholar] [CrossRef]
- Lucchini, S.; McDermott, P.; Thompson, A.; Hinton, J.C.D. The H-NS-like Protein StpA Represses the RpoS (Sigma 38) Regulon during Exponential Growth of Salmonella Typhimurium. Mol. Microbiol. 2009, 74, 1169–1186. [Google Scholar] [CrossRef]
- Cho, B.-K.; Knight, E.M.; Barrett, C.L.; Palsson, B.Ø. Genome-Wide Analysis of Fis Binding in Escherichia coli Indicates a Causative Role for A-/AT-Tracts. Genome Res. 2008, 18, 900–910. [Google Scholar] [CrossRef]
- Boubrik, F.; Bonnefoy, E.; Rouvière-Yaniv, J. HU and IHF: Similarities and Differences. In Escherichia coli, the Lack of HU Is Not Compensated for by IHF. Res. Microbiol. 1991, 142, 239–247. [Google Scholar] [CrossRef]
- Castaing, B.; Zelwer, C.; Laval, J.; Boiteux, S. HU Protein of Escherichia coli Binds Specifically to DNA That Contains Single-Strand Breaks or Gaps. J. Biol. Chem. 1995, 270, 10291–10296. [Google Scholar] [CrossRef]
- Perez-Rueda, E.; Ibarra, J.A. Distribution of Putative Xenogeneic Silencers in Prokaryote Genomes. Comput. Biol. Chem. 2015, 58, 167–172. [Google Scholar] [CrossRef]
- Riley, S.P.; Bykowski, T.; Cooley, A.E.; Burns, L.H.; Babb, K.; Brissette, C.A.; Bowman, A.; Rotondi, M.; Miller, M.C.; DeMoll, E.; et al. Borrelia burgdorferi EbfC Defines a Newly-Identified, Widespread Family of Bacterial DNA-Binding Proteins. Nucleic Acids Res. 2009, 37, 1973–1983. [Google Scholar] [CrossRef]
- Babb, K.; Bykowski, T.; Riley, S.P.; Miller, M.C.; Demoll, E.; Stevenson, B. Borrelia burgdorferi EbfC, a Novel, Chromosomally Encoded Protein, Binds Specific DNA Sequences Adjacent to erp Loci on the Spirochete’s Resident Cp32 Prophages. J. Bacteriol. 2006, 188, 4331–4339. [Google Scholar] [CrossRef]
- Jutras, B.L.; Bowman, A.; Brissette, C.A.; Adams, C.A.; Verma, A.; Chenail, A.M.; Stevenson, B. EbfC (YbaB) Is a New Type of Bacterial Nucleoid-Associated Protein and a Global Regulator of Gene Expression in the Lyme Disease Spirochete. J. Bacteriol. 2012, 194, 3395–3406. [Google Scholar] [CrossRef] [Green Version]
- Jutras, B.L.; Verma, A.; Adams, C.A.; Brissette, C.A.; Burns, L.H.; Whetstine, C.R.; Bowman, A.; Chenail, A.M.; Zückert, W.R.; Stevenson, B. BpaB and EbfC DNA-Binding Proteins Regulate Production of the Lyme Disease Spirochete’s Infection-Associated Erp Surface Proteins. J. Bacteriol. 2012, 194, 778–786. [Google Scholar] [CrossRef]
- Steere, A.C.; Strle, F.; Wormser, G.P.; Hu, L.T.; Branda, J.A.; Hovius, J.W.R.; Li, X.; Mead, P.S. Lyme Borreliosis. Nat. Rev. Dis. Primers 2016, 2, 16090. [Google Scholar] [CrossRef]
- Lim, K.; Tempczyk, A.; Parsons, J.F.; Bonander, N.; Toedt, J.; Kelman, Z.; Howard, A.; Eisenstein, E.; Herzberg, O. Crystal Structure of YbaB from Haemophilus influenzae (HI0442), a Protein of Unknown Function Coexpressed with the Recombinational DNA Repair Protein RecR. Proteins Struct. Funct. Bioinform. 2003, 50, 375–379. [Google Scholar] [CrossRef]
- Cooley, A.E.; Riley, S.P.; Kral, K.; Miller, M.C.; DeMoll, E.; Fried, M.G.; Stevenson, B. DNA-Binding by Haemophilus influenzae and Escherichia coli YbaB, Members of a Widely-Distributed Bacterial Protein Family. BMC Microbiol. 2009, 9, 137. [Google Scholar] [CrossRef]
- Peláez, A.I.; Ribas-Aparicio, R.M.; Gómez, A.; Rodicio, M.R. Structural and Functional Characterization of the recR Gene of Streptomyces. Mol. Genet. Genom. 2001, 265, 663–672. [Google Scholar] [CrossRef]
- Yeung, T.; Mullin, D.A.; Chen, K.S.; Craig, E.A.; Bardwell, J.C.; Walker, J.R. Sequence and Expression of the Escherichia coli recR Locus. J. Bacteriol. 1990, 172, 6042–6047. [Google Scholar] [CrossRef]
- Mahdi, A.A.; Lloyd, R.G. The recR Locus of Escherichia coli K-12: Molecular Cloning, DNA Sequencing and Identification of the Gene Product. Nucleic Acids Res. 1989, 17, 6781–6794. [Google Scholar] [CrossRef]
- Stevenson, B.; Krusenstjerna, A.C.; Castro-Padovani, T.N.; Savage, C.R.; Jutras, B.L.; Saylor, T.C. The Consistent Tick-Vertebrate Infectious Cycle of the Lyme Disease Spirochete Enables Borrelia burgdorferi to Control Protein Expression by Monitoring Its Physiological Status. J. Bacteriol. 2022, 204, e0060621. [Google Scholar] [CrossRef]
- Michel, B.; Leach, D. Homologous Recombination—Enzymes and Pathways. EcoSal Plus 2012, 5, 21. [Google Scholar] [CrossRef]
- Oakley, A.J. A Structural View of Bacterial DNA Replication. Protein Sci. 2019, 28, 990–1004. [Google Scholar] [CrossRef] [Green Version]
- Rezuchova, B.; Miticka, H.; Homerova, D.; Roberts, M.; Kormanec, J. New Members of the Escherichia coli σE Regulon Identified by a Two-Plasmid System. FEMS Microbiol. Lett. 2003, 225, 1–7. [Google Scholar] [CrossRef]
- Saha, S.; Lach, S.R.; Konovalova, A. Homeostasis of the Gram-Negative Cell Envelope. Curr. Opin. Microbiol. 2021, 61, 99–106. [Google Scholar] [CrossRef] [PubMed]
- Skretas, G.; Georgiou, G. Simple Genetic Selection Protocol for Isolation of Overexpressed Genes That Enhance Accumulation of Membrane-Integrated Human G Protein-Coupled Receptors in Escherichia coli. Appl. Environ. Microbiol. 2010, 76, 5852–5859. [Google Scholar] [CrossRef] [PubMed]
- Blum, M.; Chang, H.-Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Chen, Y.; He, Z.; Men, Y.; Dong, G.; Hu, S.; Ying, X. MetaLogo: A Heterogeneity-Aware Sequence Logo Generator and Aligner. Brief. Bioinform. 2022, 23, bbab591. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for Clustering the next-Generation Sequencing Data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Pal, P.; Modi, M.; Ravichandran, S.; Yennamalli, R.M.; Priyadarshini, R. DNA-Binding Properties of YbaB, a Putative Nucleoid-Associated Protein from Caulobacter crescentus. Front. Microbiol. 2021, 12, 733344. [Google Scholar] [CrossRef]
- Gopalan, A.; Deka, G.; Prabhavathi, M.; Savithri, H.S.; Murthy, M.R.N.; Raja, A. Structural and Biophysical Characterization of Rv3716c, a Hypothetical Protein from Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2018, 495, 982–987. [Google Scholar] [CrossRef]
- Tian, S.; Chen, H.; Sun, T.; Wang, H.; Zhang, X.; Liu, Y.; Xia, J.; Guo, C.; Lin, D. Expression, Purification and Characterization of Esx-1 Secretion-Associated Protein EspL from Mycobacterium tuberculosis. Protein Expr. Purif. 2016, 128, 42–51. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly Accurate Protein Structure Prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Sehnal, D.; Bittrich, S.; Deshpande, M.; Svobodová, R.; Berka, K.; Bazgier, V.; Velankar, S.; Burley, S.K.; Koča, J.; Rose, A.S. Mol* Viewer: Modern Web App for 3D Visualization and Analysis of Large Biomolecular Structures. Nucleic Acids Res. 2021, 49, W431–W437. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Jaroszewski, L.; Iyer, M.; Sedova, M.; Godzik, A. FATCAT 2.0: Towards a Better Understanding of the Structural Diversity of Proteins. Nucleic Acids Res. 2020, 48, W60–W64. [Google Scholar] [CrossRef] [PubMed]
- Carnielli, C.M.; Artier, J.; Franco de Oliveira, J.C.; Novo-Mansur, M.T.M. Interaction Network and Mass Spectrometry Data of Xanthomonas citri Subsp. citri Surface Proteins from Differential Proteomic Analysis of Infectious and Non-Infectious Cells. Data Brief 2016, 8, 1400–1411. [Google Scholar] [CrossRef] [PubMed]
- Turab Naqvi, A.A.; Rahman, S.; Rubi; Zeya, F.; Kumar, K.; Choudhary, H.; Jamal, M.S.; Kim, J.; Hassan, M.I. Genome Analysis of Chlamydia trachomatis for Functional Characterization of Hypothetical Proteins to Discover Novel Drug Targets. Int. J. Biol. Macromol. 2017, 96, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Wang, F.; Hua, X.; Ma, T.; Chen, J.; Xu, X.; Wang, L.; Tian, B.; Hua, Y. Genetic and Biochemical Characteristics of the Histone-like Protein DR0199 in Deinococcus radiodurans. Microbiology 2012, 158, 936–943. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Li, Y.; Zhang, H.; Liu, W.; Dong, Z.; Liu, L.; Liu, S.; Sheng, X.; Min, J.; Huang, R.; et al. The Chloroplast-Localized Protein LTA1 Regulates Tiller Angle and Yield of Rice. Crop J. 2022, 10, 952–961. [Google Scholar] [CrossRef]
- Casjens, S.R.; Di, L.; Akther, S.; Mongodin, E.F.; Luft, B.J.; Schutzer, S.E.; Fraser, C.M.; Qiu, W.-G. Primordial Origin and Diversification of Plasmids in Lyme Disease Agent Bacteria. BMC Genom. 2018, 19, 218. [Google Scholar] [CrossRef]
- Jutras, B.L.; Chenail, A.M.; Carroll, D.W.; Miller, M.C.; Zhu, H.; Bowman, A.; Stevenson, B. Bpur, the Lyme Disease Spirochete’s PUR Domain Protein: Identification as a Transcriptional Modulator and Characterization of Nucleic Acid Interactions. J. Biol. Chem. 2013, 288, 26220–26234. [Google Scholar] [CrossRef]
- Medrano, M.S.; Ding, Y.; Wang, X.-G.; Lu, P.; Coburn, J.; Hu, L.T. Regulators of Expression of the Oligopeptide Permease A Proteins of Borrelia burgdorferi. J. Bacteriol. 2007, 189, 2653–2659. [Google Scholar] [CrossRef]
- Menikpurage, I.P.; Woo, K.; Mera, P.E. Transcriptional Activity of the Bacterial Replication Initiator DnaA. Front. Microbiol. 2021, 12, 662317. [Google Scholar] [CrossRef]
- Sargentini, N.J.; Gularte, N.P.; Hudman, D.A. Screen for Genes Involved in Radiation Survival of Escherichia coli and Construction of a Reference Database. Mutat. Res./Fundam. Mol. Mech. Mutagenes. 2016, 793–794, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sperb, E.R.; Tadra-Sfeir, M.Z.; Sperotto, R.A.; Fernandes, G.D.C.; Pedrosa, F.D.O.; de Souza, E.M.; Passaglia, L.M.P. Iron Deficiency Resistance Mechanisms Enlightened by Gene Expression Analysis in Paenibacillus riograndensis SBR5. Res. Microbiol. 2016, 167, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Tsai, C.-H.; Ho, Y.-H.; Sung, T.-C.; Wu, W.-F.; Chen, C.-S. Escherichia coli Proteome Microarrays Identified the Substrates of ClpYQ Protease. Mol. Cell. Proteom. 2017, 16, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Skovierova, H.; Rowley, G.; Rezuchova, B.; Homerova, D.; Lewis, C.; Roberts, M.; Kormanec, J. Identification of the SigmaE Regulon of Salmonella enterica Serovar Typhimurium. Microbiology 2006, 152, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Overall, C.C.; Johnson, R.C.; Jones, M.B.; McDermott, J.E.; Heffron, F.; Adkins, J.N.; Cambronne, E.D. ChIP-Seq Analysis of the σE Regulon of Salmonella enterica Serovar Typhimurium Reveals New Genes Implicated in Heat Shock and Oxidative Stress Response. PLoS ONE 2015, 10, e0138466. [Google Scholar] [CrossRef]
- Hews, C.L.; Cho, T.; Rowley, G.; Raivio, T.L. Maintaining Integrity Under Stress: Envelope Stress Response Regulation of Pathogenesis in Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 313. [Google Scholar] [CrossRef]
- Molan, K.; Žgur Bertok, D. Small Prokaryotic DNA-Binding Proteins Protect Genome Integrity throughout the Life Cycle. IJMS 2022, 23, 4008. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cordeiro, T.F.V.B.; Gontijo, M.T.P.; Jorge, G.P.; Brocchi, M. EbfC/YbaB: A Widely Distributed Nucleoid-Associated Protein in Prokaryotes. Microorganisms 2022, 10, 1945. https://doi.org/10.3390/microorganisms10101945
Cordeiro TFVB, Gontijo MTP, Jorge GP, Brocchi M. EbfC/YbaB: A Widely Distributed Nucleoid-Associated Protein in Prokaryotes. Microorganisms. 2022; 10(10):1945. https://doi.org/10.3390/microorganisms10101945
Chicago/Turabian StyleCordeiro, Tamires Fernanda Vilas Boas, Marco Túlio Pardini Gontijo, Genesy Perez Jorge, and Marcelo Brocchi. 2022. "EbfC/YbaB: A Widely Distributed Nucleoid-Associated Protein in Prokaryotes" Microorganisms 10, no. 10: 1945. https://doi.org/10.3390/microorganisms10101945
APA StyleCordeiro, T. F. V. B., Gontijo, M. T. P., Jorge, G. P., & Brocchi, M. (2022). EbfC/YbaB: A Widely Distributed Nucleoid-Associated Protein in Prokaryotes. Microorganisms, 10(10), 1945. https://doi.org/10.3390/microorganisms10101945








