Performance of a Machine Learning-Based Methicillin Resistance of Staphylococcus aureus Identification System Using MALDI-TOF MS and Comparison of the Accuracy according to SCCmec Types
Abstract
:1. Introduction
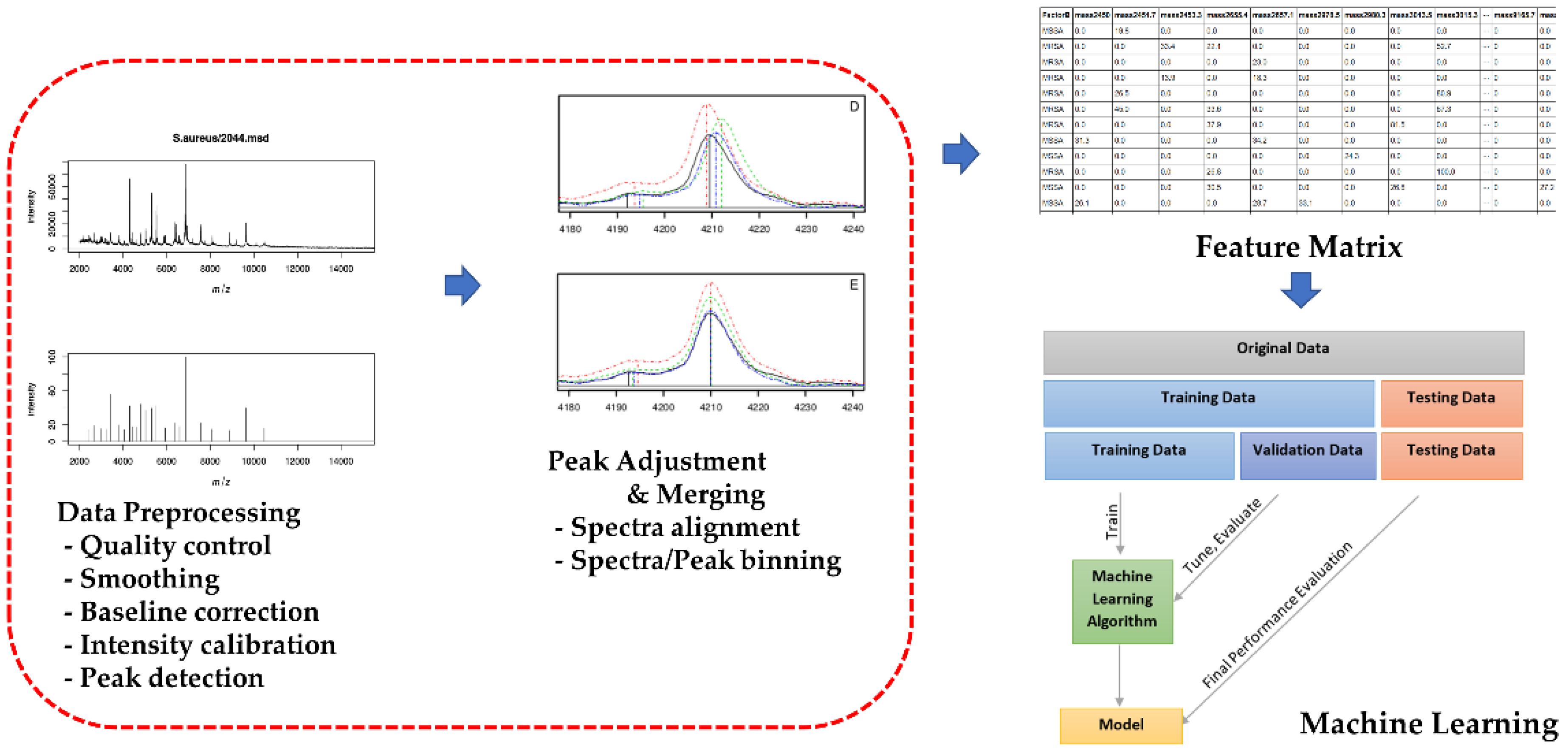
2. Materials and Methods
2.1. Clinical S. aureus Isolates
2.2. MALDI-TOF MS Analysis
2.3. AMRQuest Machine Learning Model for the Identification of MRSA
2.4. PCR for the mecA gene and SCCmec types of S. aureus
2.5. Statistical Analyses
3. Results
3.1. Characteristics of Staphylococcus Aureus Isolates
3.2. Accuracy, Sensitivity, and Specificity of AMRQuest (v.2.1) for MRSA Prediction
3.3. MRSA Prediction of AMRQuest According to SCCmec Types
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fair, R.J.; Tor, Y. Antibiotics and bacterial resistance in the 21st century. Perspect. Med. Chem. 2014, 6, 25–64. [Google Scholar] [CrossRef] [PubMed]
- WHO. Antimicrobial Resistance. Available online: https://www.who.int/news-room/fact-sheets/detail/antimicrobial-resistance (accessed on 5 August 2022).
- Prestinaci, F.; Pezzotti, P.; Pantosti, A. Antimicrobial resistance: A global multifaceted phenomenon. Pathog. Glob. Health 2015, 109, 309–318. [Google Scholar] [CrossRef] [PubMed]
- Lewis, K. Platforms for antibiotic discovery. Nat. Rev. Drug Discov. 2013, 12, 371–387. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- De Oliveira, D.M.P.; Forde, B.M.; Kidd, T.J.; Harris, P.N.A.; Schembri, M.A.; Beatson, S.A.; Paterson, D.L.; Walker, M.J. Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 2020, 33, e00181-19. [Google Scholar] [CrossRef]
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef]
- Loffler, C.A.; Macdougall, C. Update on prevalence and treatment of methicillin-resistant Staphylococcus aureus infections. Expert Rev. Anti-Infect. Ther. 2007, 5, 961–981. [Google Scholar] [CrossRef]
- Noll, C.; Nasruddin-Yekta, A.; Sternisek, P.; Weig, M.; Groß, U.; Schilling, A.F.; Beil, F.T.; Bader, O. Rapid direct detection of pathogens for diagnosis of joint infections by MALDI-TOF MS after liquid enrichment in the BacT/Alert blood culture system. PLoS ONE 2020, 15, e0243790. [Google Scholar] [CrossRef]
- Van Belkum, A.; Welker, M.; Pincus, D.; Charrier, J.P.; Girard, V. Matrix-Assisted Laser Desorption ionization time-of-flight mass spectrometry in clinical microbiology: What are the current issues? Ann. Lab. Med. 2017, 37, 475–483. [Google Scholar] [CrossRef]
- Osthoff, M.; Gürtler, N.; Bassetti, S.; Balestra, G.; Marsch, S.; Pargger, H.; Weisser, M.; Egli, A. Impact of MALDI-TOF-MS-based identification directly from positive blood cultures on patient management: A controlled clinical trial. Clin. Microbiol. Infect. 2017, 23, 78–85. [Google Scholar] [CrossRef]
- Weis, C.; Cuénod, A.; Rieck, B.; Dubuis, O.; Graf, S.; Lang, C.; Oberle, M.; Brackmann, M.; Søgaard, K.K.; Osthoff, M.; et al. Direct antimicrobial resistance prediction from clinical MALDI-TOF mass spectra using machine learning. Nat. Med. 2022, 28, 164–174. [Google Scholar] [CrossRef]
- Kasas, S.; Malovichko, A.; Villalba, M.I.; Vela, M.E.; Yantorno, O.; Willaert, R.G. Nanomotion Detection-Based Rapid Antibiotic Susceptibility Testing. Antibiotics 2021, 10, 287. [Google Scholar] [CrossRef] [PubMed]
- Edwards-Jones, V.; Claydon, M.A.; Evason, D.J.; Walker, J.; Fox, A.J.; Gordon, D.B. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 2000, 49, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Josten, M.; Dischinger, J.; Szekat, C.; Reif, M.; Al-Sabti, N.; Sahl, H.G.; Parcina, M.; Bekeredjian-Ding, I.; Bierbaum, G. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 2014, 304, 1018–1023. [Google Scholar] [CrossRef] [PubMed]
- Du, Z.; Yang, R.; Guo, Z.; Song, Y.; Wang, J. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2002, 74, 5487–5491. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.S.; de Lencastre, H.; Garau, J.; Kluytmans, J.; Malhotra-Kumar, S.; Peschel, A.; Harbarth, S. Methicillin-resistant Staphylococcus aureus. Nat. Rev. Dis. Prim. 2018, 4, 18033. [Google Scholar] [CrossRef]
- Miragaia, M. Factors contributing to the evolution of mecA-mediated beta-lactam resistance in staphylococci: Update and new insights from whole genome sequencing (WGS). Front. Microbiol. 2018, 9, 2723. [Google Scholar] [CrossRef]
- Urushibara, N.; Aung, M.S.; Kawaguchiya, M.; Kobayashi, N. Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite islands carrying speG in ST5 MRSA in Japan. J. Antimicrob. Chemother. 2020, 75, 46–50. [Google Scholar] [CrossRef]
- Uehara, Y. Current status of staphylococcal cassette chromosome mec (SCCmec). Antibiotics 2022, 11, 86. [Google Scholar] [CrossRef]
- Lakhundi, S.; Zhang, K. Methicillin-resistant Staphylococcus aureus: Molecular characterization, evolution, and epidemiology. Clin. Microbiol. Rev. 2018, 31, e00020-18. [Google Scholar] [CrossRef]
- Sogawa, K.; Watanabe, M.; Ishige, T.; Segawa, S.; Miyabe, A.; Murata, S.; Saito, T.; Sanda, A.; Furuhata, K.; Nomura, F. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus using MALDI-TOF mass spectrometry. Biocontrol. Sci. 2017, 22, 163–169. [Google Scholar] [CrossRef] [Green Version]
- Josten, M.; Reif, M.; Szekat, C.; Al-Sabti, N.; Roemer, T.; Sparbier, K.; Kostrzewa, M.; Rohde, H.; Sahl, H.G.; Bierbaum, G. Analysis of the matrix-assisted laser desorption ionization-time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J. Clin. Microbiol. 2013, 51, 1809–1817. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.Y.; Lee, T.Y.; Tseng, Y.J.; Liu, T.P.; Huang, K.Y.; Chang, Y.T.; Chen, C.H.; Lu, J.J. A new scheme for strain typing of methicillin-resistant Staphylococcus aureus on the basis of matrix-assisted laser desorption ionization time-of-flight mass spectrometry by using machine learning approach. PLoS ONE 2018, 13, e0194289. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.R.; Washington, J.A. Microscopic and bacteriologic analysis of expectorated sputum. Mayo Clin. Proc. 1975, 50, 339–344. [Google Scholar] [PubMed]
- Wayne, P.A. Performance Standards for Antimicrobial Susceptibility Testing, 27th ed.; Clinical and Laboratory Standards Institute (CLSI): Pittsburgh, PA, USA, 2016; Volume M100s. [Google Scholar]
- Ito, T.; Kuwahara-Arai, K.; Katayama, Y.; Uehara, Y.; Han, X.; Kondo, Y.; Hiramatsu, K. Staphylococcal cassette chromosome mec (SCCmec) analysis of MRSA. Methods Mol. Biol. 2014, 1085, 131–148. [Google Scholar] [CrossRef] [PubMed]
- Shreffler, J.; Huecker, M.R. Diagnostic Testing Accuracy: Sensitivity, Specificity, Predictive Values and Likelihood Ratios. In StatPearls; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2022. [Google Scholar]
- Wolters, M.; Rohde, H.; Maier, T.; Belmar-Campos, C.; Franke, G.; Scherpe, S.; Aepfelbacher, M.; Christner, M. MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages. Int. J. Med. Microbiol. 2011, 301, 64–68. [Google Scholar] [CrossRef]
- Lasch, P.; Fleige, C.; Stämmler, M.; Layer, F.; Nübel, U.; Witte, W.; Werner, G. Insufficient discriminatory power of MALDI-TOF mass spectrometry for typing of Enterococcus faecium and Staphylococcus aureus isolates. J. Microbiol. Methods 2014, 100, 58–69. [Google Scholar] [CrossRef]
- Walker, J.; Fox, A.J.; Edwards-Jones, V.; Gordon, D.B. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: Media effects and inter-laboratory reproducibility. J. Microbiol. Methods 2002, 48, 117–126. [Google Scholar] [CrossRef]
- Croxatto, A.; Prod’Hom, G.; Greub, G. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol. Rev. 2012, 36, 380–407. [Google Scholar] [CrossRef]
- Zitnik, M.; Nguyen, F.; Wang, B.; Leskovec, J.; Goldenberg, A.; Hoffman, M.M. Machine learning for integrating data in biology and medicine: Principles, practice, and opportunities. Inf. Fusion 2019, 50, 71–91. [Google Scholar] [CrossRef]
- Yu, J.; Tien, N.; Liu, Y.C.; Cho, D.Y.; Chen, J.W.; Tsai, Y.T.; Huang, Y.C.; Chao, H.J.; Chen, C.J. Rapid identification of methicillin-resistant Staphylococcus aureus using MALDI-TOF MS and machine learning from over 20,000 clinical isolates. Microbiol. Spectr. 2022, 10, e0048322. [Google Scholar] [CrossRef]
- Tang, K.; Tang, D.; Wang, Q.; Li, C. MALDI-TOF MS platform combined with machine learning to establish a model for rapid identification of methicillin-resistant Staphylococcus aureus. J. Microbiol. Methods 2021, 180, 106109. [Google Scholar] [CrossRef] [PubMed]
- Zetola, N.; Francis, J.S.; Nuermberger, E.L.; Bishai, W.R. Community-acquired meticillin-resistant Staphylococcus aureus: An emerging threat. Lancet Infect. Dis. 2005, 5, 275–286. [Google Scholar] [CrossRef]
- Robinson, D.A.; Enright, M.C. Evolutionary models of the emergence of methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2003, 47, 3926–3934. [Google Scholar] [CrossRef] [PubMed]
- Vandenesch, F.; Naimi, T.; Enright, M.C.; Lina, G.; Nimmo, G.R.; Heffernan, H.; Liassine, N.; Bes, M.; Greenland, T.; Reverdy, M.E.; et al. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: Worldwide emergence. Emerg. Infect. Dis. 2003, 9, 978–984. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jeong, J.H.; Cha, H.Y.; Jin, J.S.; Lee, J.C.; Lee, Y.C.; Seol, S.Y.; Cho, D.T. Detection of diverse SCCmec variants in methicillin-resistant Staphylococcus aureus and comparison of SCCmec typing methods. Clin. Microbiol. Infect. 2007, 13, 1128–1130. [Google Scholar] [CrossRef] [PubMed]
- Park, C.; Lee, D.G.; Choi, S.M.; Choi, J.H.; Park, S.H.; Yoo, J.H.; Shin, W.S. Comment on: A survey of community-associated methicillin-resistant Staphylococcus aureus in Korea. J. Antimicrob. Chemother. 2008, 62, 211–212. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, D.C.; de Lencastre, H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Berglund, C.; Mölling, P.; Sjöberg, L.; Söderquist, B. Predominance of staphylococcal cassette chromosome mec (SCCmec) type IV among methicillin-resistant Staphylococcus aureus (MRSA) in a Swedish county and presence of unknown SCCmec types with Panton-Valentine leukocidin genes. Clin. Microbiol. Infect. 2005, 11, 447–456. [Google Scholar] [CrossRef]
- Kim, J.M.; Kim, I.; Chung, S.H.; Chung, Y.; Han, M.; Kim, J.S. Rapid discrimination of methicillin-resistant Staphylococcus aureus by MALDI-TOF MS. Pathogens 2019, 8, 214. [Google Scholar] [CrossRef] [Green Version]
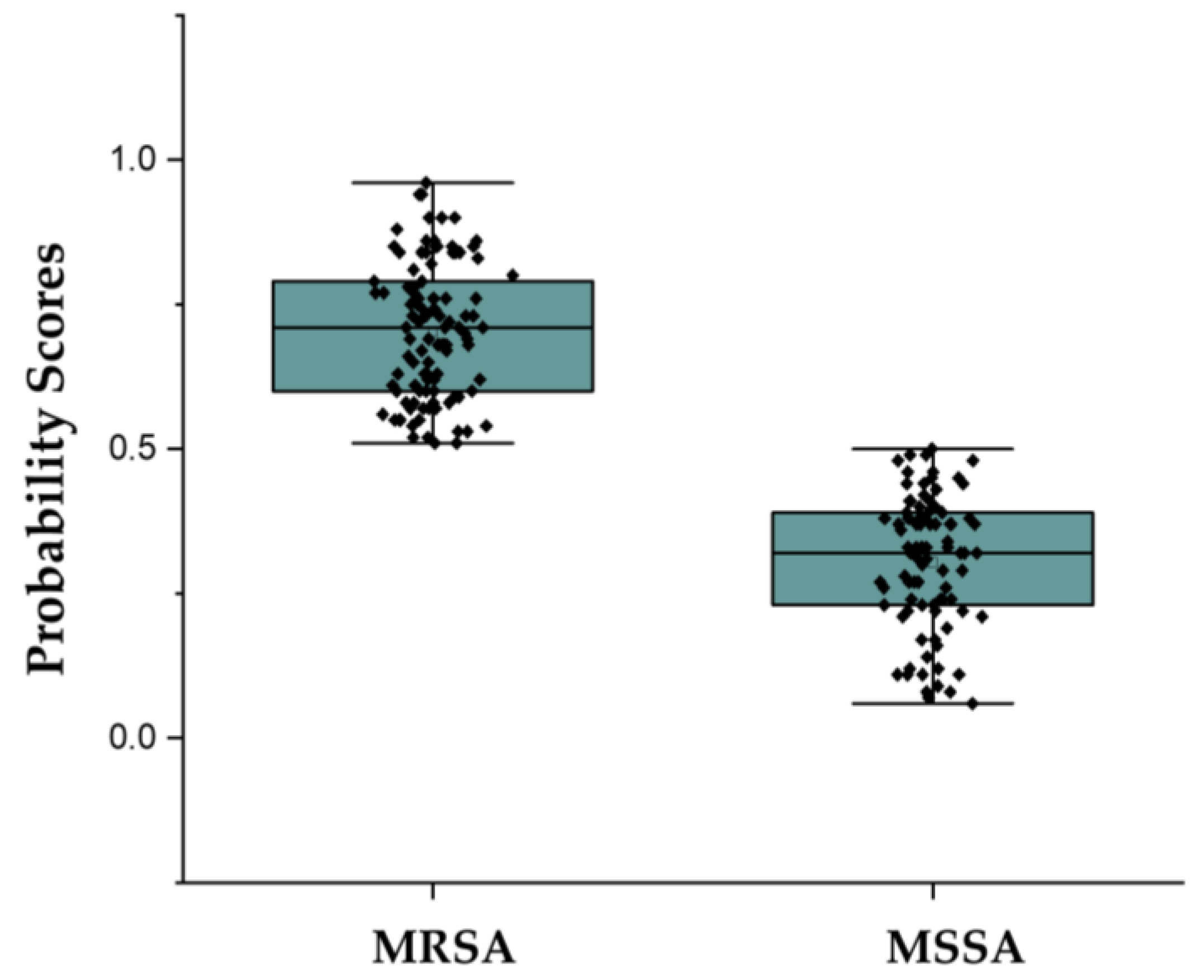

| AMRQuest (v. 2.1) | Total | Accuracy | Sensitivity (95% CI) | Specificity (95% CI) | LR+ 1 | LR− 2 | AUC (95% CI) | Kappa Value | ||
|---|---|---|---|---|---|---|---|---|---|---|
| MRSA | MSSA | |||||||||
| MRSA | 90 | 16 | 106 | 87.6% | 91.8% (84.5–96.4) | 83.3% (74.4–90.2) | 5.510 | 0.098 | 0.876 (0.821–0.919) | 0.750 |
| MSSA | 8 | 80 | 88 | |||||||
| Total | 98 | 96 | 194 | |||||||
| SCCmec Types | AMRQuest (v. 2.1) | Total | Accuracy | |
|---|---|---|---|---|
| MRSA | MSSA | |||
| II | 62 | 3 | 65 | 95.4% |
| IV | 3 | 11 | 14 | 21.4% |
| IVA | 25 | 1 | 26 | 96.1% |
| V | 0 | 1 | 1 | 0.0% |
| MSSA | 8 | 80 | 88 | 90.9% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeon, K.; Kim, J.-M.; Rho, K.; Jung, S.H.; Park, H.S.; Kim, J.-S. Performance of a Machine Learning-Based Methicillin Resistance of Staphylococcus aureus Identification System Using MALDI-TOF MS and Comparison of the Accuracy according to SCCmec Types. Microorganisms 2022, 10, 1903. https://doi.org/10.3390/microorganisms10101903
Jeon K, Kim J-M, Rho K, Jung SH, Park HS, Kim J-S. Performance of a Machine Learning-Based Methicillin Resistance of Staphylococcus aureus Identification System Using MALDI-TOF MS and Comparison of the Accuracy according to SCCmec Types. Microorganisms. 2022; 10(10):1903. https://doi.org/10.3390/microorganisms10101903
Chicago/Turabian StyleJeon, Kibum, Jung-Min Kim, Kyoohyoung Rho, Seung Hee Jung, Hyung Soon Park, and Jae-Seok Kim. 2022. "Performance of a Machine Learning-Based Methicillin Resistance of Staphylococcus aureus Identification System Using MALDI-TOF MS and Comparison of the Accuracy according to SCCmec Types" Microorganisms 10, no. 10: 1903. https://doi.org/10.3390/microorganisms10101903
APA StyleJeon, K., Kim, J.-M., Rho, K., Jung, S. H., Park, H. S., & Kim, J.-S. (2022). Performance of a Machine Learning-Based Methicillin Resistance of Staphylococcus aureus Identification System Using MALDI-TOF MS and Comparison of the Accuracy according to SCCmec Types. Microorganisms, 10(10), 1903. https://doi.org/10.3390/microorganisms10101903






