NFκB1 Polymorphisms Are Associated with Severe Influenza A (H1N1) Virus Infection in a Canadian Population
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Studied Subjects
2.2. PCR Amplification and Sequencing of NFκB1 Gene
2.3. Sequence Analysis
2.4. Statistical Analysis
3. Results
3.1. Population Description
3.2. The Polymorphism of the NFκB1 Gene in the ICU Patients
3.3. Association of SNPs Genotypes with Severe Response to H1N1 Infection
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Writing Committee of the WHO Consultation on Clinical Aspects of Pandemic (H1N1) 2009 Influenza. Clinical Aspects of Pandemic 2009 Influenza A (H1N1) Virus Infection. N. Engl. J. Med. 2010, 362, 1708–1719. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Chan, P.K.; Lui, G.C.; Wong, B.C.; Sin, W.W.; Choi, K.W.; Wong, R.Y.K.; Lee, E.L.Y.; Yeung, A.C.M.; Ngai, K.L.K.; et al. Complications and outcomes of pandemic 2009 influenza A (H1N1) virus infection in hos-pitalized adults: How do they differ from those in seasonal influenza? J. Infect. Dis. 2011, 203, 1739–1747. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Ramirez, R.A.; Ramirez-Venegas, A.; Quintana-Carrillo, R.; Camarena, E.; Falfán-Valencia, R.; Mejía-Aranguré, J.M. TNF, IL6, and IL1B Polymorphisms Are Associated with Severe Influenza A (H1N1) Virus Infection in the Mexican Population. PLoS ONE 2015, 10, e0144832. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, X.; Zhao, B.; Wang, J.; Zhu, Z.; Teng, Z.; Shao, J.; Shen, J.; Gao, Y.; Yuan, Z.; et al. Intensive Cytokine induction in Pandemic H1N1 Influenza Virus Infection Accompanied by Robust Production of IL-10 and IL-6. PLoS ONE 2011, 6, e28680. [Google Scholar] [CrossRef] [PubMed]
- Bermejo-Martin, J.F.; Martin-Loeches, I.; Rello, J.; Anton, A.; Almansa, R.; Xu, L.; Lopez-Campos, G.; Pumarola, T.; Ran, L.; Ramirez, P.; et al. Host adaptive immunity deficiency in severe pandemic influenza. Crit. Care 2010, 14, R167. [Google Scholar] [CrossRef]
- Esposito, S.; Molteni, C.G.; Giliani, S.; Mazza, C.; Scala, A.; Tagliaferri, L.; Pelucchi, C.; Fossali, E.; Plebani, A.; Principi, N. Toll-like receptor 3 gene polymorphisms and severity of pandemic A/H1N1/2009 influenza in otherwise healthy children. Virol. J. 2012, 9, 270. [Google Scholar] [CrossRef]
- Pinto, R.; Herold, S.; Cakarova, L.; Hoegner, K.; Lohmeyer, J.; Planz, O.; Pleschka, S. Inhibition of influenza virus-induced NF-kappaB and Raf/MEK/ERK activation can reduce both virus titers and cytokine expression simultaneously in vitro and in vivo. Antiviral Res. 2011, 92, 45–56. Available online: https://pubmed.ncbi.nlm.nih.gov/21641936/ (accessed on 6 September 2021). [CrossRef]
- Singh, V.; Gupta, D.; Arora, R. NF-κB as a key player in regulation of cellular radiation responses and identification of radiation countermeasures. Discoveries 2015, 3, e35. [Google Scholar] [CrossRef]
- Kircheis, R.; Haasbach, E.; Lueftenegger, D.; Heyken, W.T.; Ocker, M.; Planz, O. NF-κB Pathway as a Potential Target for Treatment of Critical Stage COVID-19 Patients. Front. Immunol. 2020, 11, 598444. [Google Scholar] [CrossRef]
- Wang, Y.; Song, T.; Li, K.; Jin, Y.; Yue, J.; Ren, H.; Liang, L. Different Subtypes of Influenza Viruses Target Different Human Proteins and Pathways Leading to Different Pathogenic Phenotypes. BioMed Res. Int. 2019, 2019, 4794910. [Google Scholar] [CrossRef]
- Gao, S.; Song, L.; Li, J.; Zhang, Z.; Peng, H.; Jiang, W.; Wang, Q.; Kang, T.; Chen, S.; Huang, W. Influenza A virus-encoded NS1 virulence factor protein inhibits innate immune response by targeting IKK. Cell. Microbiol. 2012, 14, 1849–1866. [Google Scholar] [CrossRef] [PubMed]
- Nogales, A.; DeDiego, M.L. Host Single Nucleotide Polymorphisms Modulating Influenza A Virus Disease in Humans. Pathogens 2019, 8, 168. Available online: http://www.ncbi.nlm.nih.gov/pubmed/31574965 (accessed on 10 June 2020). [CrossRef]
- Ehrhardt, C.; Rückle, A.; Hrincius, E.R.; Haasbach, E.; Anhlan, D.; Ahmann, K.; Banning, C.; Reiling, S.J.; Kühn, J.; Strobl, S.; et al. The NF-κB inhibitor SC75741 efficiently blocks influenza virus propagation and confers a high barrier for development of viral resistance. Cell. Microbiol. 2013, 15, 1198–1211. [Google Scholar] [CrossRef] [PubMed]
- Schmolke, M.; Viemann, D.; Roth, J.; Ludwig, S. Essential Impact of NF-κB Signaling on the H5N1 Influenza A Virus-Induced Transcriptome. J. Immunol. 2009, 183, 5180–5189. Available online: https://www-jimmunol-org.uml.idm.oclc.org/content/183/8/5180 (accessed on 6 September 2021). [CrossRef] [PubMed]
- Lee, N.; Cao, B.; Ke, C.; Lu, H.; Hu, Y.; Tam, C.H.T.; Ma, R.C.W.; Guan, D.; Zhu, Z.; Li, H.; et al. IFITM3, TLR3, and CD55 Gene SNPs and Cumulative Genetic Risks for Severe Outcomes in Chinese Patients with H7N9/H1N1 pdm09 Influenza. J. Infect. Dis. 2017, 216, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Strauss, M.; Palma-Vega, M.; Casares-Marfil, D.; Bosch-Nicolau, P.; Presti, M.S.L.; Molina, I.; González, C.I.; Network, C.G.C.; Martín, J.; Acosta-Herrera, M.; et al. Genetic polymorphisms of IL17A associated with Chagas disease: Results from a meta-analysis in Latin American populations. Sci. Rep. 2020, 10, 5015. [Google Scholar] [CrossRef] [PubMed]
- Horby, P.; Nguyen, N.; Dunstan, S.; Baillie, J.K. The Role of Host Genetics in Susceptibility to Influenza: A Systematic Review. PLoS ONE 2012, 7, e33180. [Google Scholar] [CrossRef] [PubMed]
- Keshavarz, M.; Namdari, H.; Farahmand, M.; Mehrbod, P.; Mokhtari-Azad, T.; Rezaei, F. Association of polymorphisms in in-flammatory cytokines encoding genes with severe cases of influenza A/H1N1 and B in an Iranian population. Virol. J. 2019, 16, 79. [Google Scholar] [CrossRef]
- Rogo, L.D.; Rezaei, F.; Marashi, S.M.; Yekaninejad, M.S.; Naseri, M.; Ghavami, N.; Mokhtari-Azad, T. Seasonal influenza A/H3N2 virus infection and IL-1Β, IL-10, IL-17, and IL-28 poly-morphisms in Iranian population. J. Med. Virol. 2016, 88, 2078–2084. Available online: http://www.ncbi.nlm.nih.gov/pubmed/27155288 (accessed on 10 June 2020). [CrossRef]
- Go, Y.M.; Kang, S.M.; Roede, J.R.; Orr, M.; Jones, D.P. Increased inflammatory signaling and lethality of influenza h1n1 by nuclear thioredoxin-1. PLoS ONE 2011, 6, e18918. [Google Scholar]
- Arcaroli, J.; Silva, E.; Maloney, J.P.; He, Q.; Svetkauskaite, D.; Murphy, J.R.; Abraham, E. Variant IRAK-1 haplotype is associated with increased nuclear factor-κB activation and worse outcomes in sepsis. Am. J. Respir. Crit. Care Med. 2006, 173, 1335–1341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yates, A.D.; Achuthan, P.; Akanni, W.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Benett, R.; et al. Ensembl 2020. Nucleic Acids Res. 2020, 48, D682–D688. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef] [PubMed]
- Garrison, E.; Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv 2012, arXiv:12073907. [Google Scholar]
- Milne, I.; Stephen, G.; Bayer, M.; Cock, P.; Pritchard, L.; Cardle, L.; Shaw, P.D.; Marshall, D. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013, 14, 193–202. [Google Scholar] [CrossRef] [PubMed]
- Sherry, S.T.; Ward, M.; Sirotkin, K. dbSNP—Database for single nucleotide polymorphisms and other classes of minor genetic variation. Genome Res. 1999, 9, 677–679. [Google Scholar] [CrossRef]
- Mogensen, T.H.; Paludan, S.R. Molecular Pathways in Virus-Induced Cytokine Production. Microbiol. Mol. Biol. Rev. 2001, 65, 131–150. [Google Scholar] [CrossRef]
- Silva, M.D.J.; De Santana, M.B.R.; Tosta, B.R.; Espinheira, R.P.; Alcantara-Neves, N.M.; Barreto, M.L.; Figueiredo, C.A.; Costa, R.D.S. Variants in the IL17 pathway genes are associated with atopic asthma and atopy makers in a South American population. Allergy Asthma Clin. Immunol. 2019, 15, 28. [Google Scholar] [CrossRef]
- Curjuric, I.; Imboden, M.; Nadif, R.; Kumar, A.; Schindler, C.; Haun, M.; Kronenberg, F.; Künzli, N.; Phuleria, H.; Postma, D.S.; et al. Different Genes Interact with Particulate Matter and Tobacco Smoke Exposure in Affecting Lung Function Decline in the General Population. PLoS ONE 2012, 7, e40175. [Google Scholar]
- Rojas, J.; Fernandez, I.; Pastor, J.C.; MacLaren, R.E.; Ramkissoon, Y.; Harsum, S.; Charteris, D.G.; Van Meurs, J.C.; Amarakoon, S.; Ruiz-Moreno, J.M.; et al. A Genetic Case-Control Study Confirms the Implication of SMAD7 and TNF Locus in the Development of Proliferative Vitreoretinopathy. Investig. Ophthalmol. Vis. Sci. 2013, 54, 1665–1678. [Google Scholar] [CrossRef]
- Paquette, S.G.; Banner, D.; Chi, L.T.B.; Leόn, A.J.; Xu, L.; Ran, L.; Huang, S.S.; Farooqui, A.; Kelvin, D.J.; Kelvin, A.A. Pandemic H1N1 influenza A directly induces a robust and acute inflammatory gene signature in primary human bronchial epithelial cells downstream of membrane fusion. Virology 2013, 448, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, M.L.; Kracht, M.; Saul, V.V. The intricate interplay between RNA viruses and NF-κB. Biochim. Biophys. Acta Mol. Cell Res. 2014, 1843, 2754–2764. [Google Scholar] [CrossRef] [PubMed]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.S.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef] [Green Version]
- Silva, R.Q.; Coto-Segura, P.; González-Lara, L.; Alonso, B.; Gómez, J.; Cuesta-Llavona, E.; Coto, E. FRI0461 genetic analysis of the nf-kb pathway can be useful to distinguish patients at risk of psoriatic arthritis within the spectrum of psoriatic disease. Ann. Rheum. Dis. 2019, 78 (Suppl. S2), 924. Available online: https://ard-bmj-com.uml.idm.oclc.org/content/78/Suppl_2/924.1 (accessed on 7 November 2021).
- Li, P.; Guo, M.; Wang, C.; Liu, X.; Zou, Q. An overview of SNP interactions in genome-wide association studies. Brief. Funct. Genom. 2014, 14, 143–155. [Google Scholar] [CrossRef] [PubMed]
- Leeman, J.R.; Gilmore, T.D. Alternative Splicing in the NF-κB Signaling Pathway. Available online: www.nf-kb.org (accessed on 30 July 2021).
- Hitomi, Y.; Nakatani, K.; Kojima, K.; Nishida, N.; Kawai, Y.; Kawashima, M.; Aiba, Y.; Nagasaki, M.; Nakamura, M.; Tokunaga, K. NFKB1 and MANBA Confer Disease Susceptibility to Primary Biliary Cholangitis via Independent Putative Primary Functional Variants. Cell. Mol. Gastroenterol. Hepatol. 2019, 7, 515–532. [Google Scholar] [CrossRef]
- Gounder, A.P.; Boon, A.C.M. Influenza Pathogenesis: The Effect of Host Factors on Severity of Disease. J. Immunol. 2019, 202, 341–350. [Google Scholar] [CrossRef]
- Tian, C.; Hromatka, B.S.; Kiefer, A.K.; Eriksson, N.; Noble, S.M.; Tung, J.Y.; Hinds, D.A. Genome-wide association and HLA region fine-mapping studies identify susceptibility loci for multiple common infections. Nat. Commun. 2017, 8, 599. [Google Scholar] [CrossRef]
- Curk, T.; Rot, G.; Zupan, B. SNPsyn: Detection and exploration of SNP–SNP interactions. Nucleic Acids Res. 2011, 39, W444. [Google Scholar] [CrossRef]
- Yang, X.R.; Jacobs, K.; Kerstann, K.F.; Bergen, A.W.; Goldstein, A.M.; Goldin, L.R. Linkage analysis of the GAW14 simulated dataset with microsatellite and single-nucleotide polymorphism markers in large pedigrees. BMC Genet. 2005, 6, S14. [Google Scholar] [CrossRef] [Green Version]
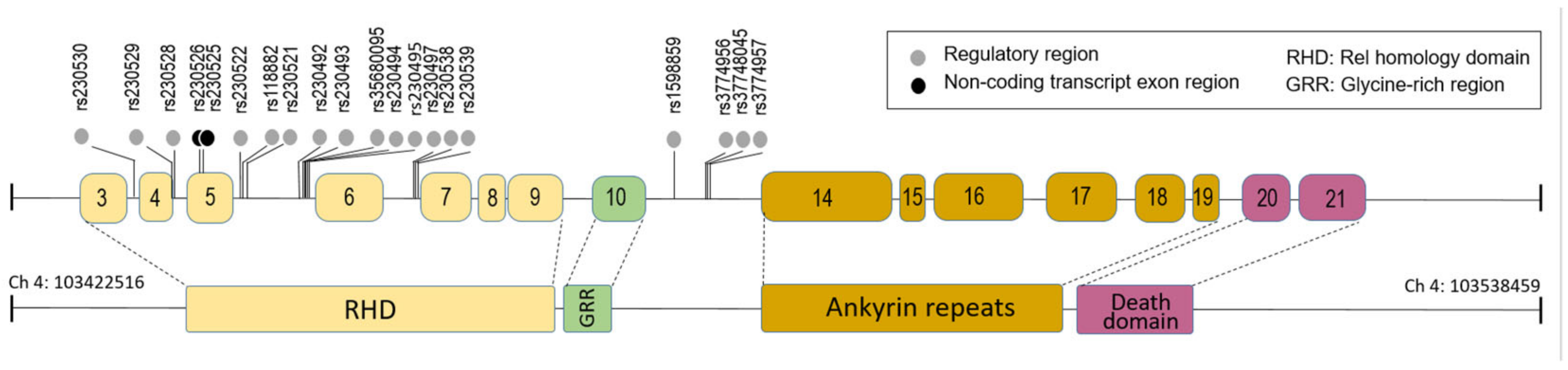
| Variables | H1N1pdm09 (n = 36) |
|---|---|
| Ethnicity (%) | Caucasian (100%) |
| Gender | |
| Male | 19 (52.78%) |
| Female | 17 (47.22%) |
| Age (years) | |
| <25 | 2 (5.67%) |
| 25–49 | 12 (33.33%) |
| 50–64 | 17 (47.22%) |
| >65 | 5 (13.89%) |
| Average | 50.76 + 14.13 |
| Pregnancy | 2 (5.67%) |
| Comorbidity | |
| Obesity | 5 (13.89%) |
| Diabetes Mellitus | 9 (25.00%) |
| Hypertension | 17 (47.22%) |
| Coronary heart disease | 1 (2.78%) |
| Congestive heart failure | 3 (8.33%) |
| Chronic obstructive | 5 (13.89%) |
| pulmonary disease | |
| Asthma | 7 (19.44%) |
| Immunosuppression | 1 (2.78%) |
| Invasive Ventilation (%) | |
| Apache II Score | 22.12 ± 6.79 |
| Death Rate | 0.41 ± 0.19 |
| SNP ID | Chromosome Location | Minor Allele | MAF a | p Value | Phenotype d | |
|---|---|---|---|---|---|---|
| ICU b | GBR c | |||||
| rs1599961 | 103443569 (intronic) | G | 0.18 | 0.46 | 1.53 × 10−7 | Cardiovascular, hypertension, |
| rs1585215 | 103444474 (intronic) | C | 0.28 | 0.38 | 1.53 × 10−2 | Hodgkin lymphoma |
| rs1585213 | 103444698 (intronic) | T | 0.38 | 0.46 | 9.78 × 10−3 | measles vaccine effectiveness, various diseases |
| rs1598856 | 103446115 (intronic) | A | 0.50 | 0.57 | 4.82 × 10−2 | Cholangitis, ovarian cancer, gastric cancer |
| rs230535 | 103448582 (intronic) | A | 0.51 | 0.62 | 6.11 × 10−3 | Bipolar disease |
| rs170731 | 103448903 (intronic) | T | 0.39 | 0.62 | 1.30 × 10−6 | lymphedema, inflammatory reactions |
| rs230534 | 103449041 (intronic) | T | 0.32 | 0.62 | 3.36 × 10−8 | Maculopathy, biliary and crohn’s disease |
| rs230533 | 103450083 (intronic) | A | 0.44 | 0.62 | 8.10 × 10−5 | crohn’s disease |
| rs230532 | 103450167 (intronic) | T | 0.54 | 0.62 | 3.83 × 10−2 | Hepatitis C, PBC, breast cancer inflammation |
| rs230531 | 103450377 (intronic) | G | 0.35 | 0.62 | 2.24 × 10−9 | PBC, ovarian cancer, gastric cancer |
| rs230530 | 103453980 (regulatory) | G | 0.18 | 0.43 | 2.41 × 10−3 | Hepatitis C, gastric cancer, allergic rhinitis |
| rs230529 | 103457418 (regulatory) | C | 0.19 | 0.55 | 2.09 × 10−7 | drug addictions, schizophrenia, psychosis |
| rs230528 | 103457585 (regulatory) | G | 0.19 | 0.55 | 1.60 × 10−8 | lung disease, coronary artery disease |
| rs230526 | 103458825 (exonic) | G | 0.25 | 0.55 | 7.88 × 10−5 | metabolic variation, cytokine variation |
| rs230525 | 103458877 (exonic) | G | 0.38 | 0.62 | 3.08 × 10−5 | liver cancer |
| rs4648004 | 103461107 (intronic) | G | 0.11 | 0.32 | 6.08 × 10−3 | cerebrovascular, cardiovascular |
| rs230523 | 103462038 (intronic) | C | 0.11 | 0.62 | 0.00 × 100 | various diseases |
| rs118882 | 103463007 (regulatory) | T | 0.39 | 0.62 | 1.23 × 10−6 | allergic rhinitis |
| rs230521 | 103463328 (regulatory) | C | 0.26 | 0.55 | 6.59 × 10−5 | breast cancer, hepatitis c, colorectal cancer, |
| rs230520 | 103465612 (intronic) | G | 0.44 | 0.62 | 6.27 × 10−4 | PBC |
| rs230519 | 103466749(intronic) | T | 0.44 | 0.62 | 6.10 × 10−4 | PBC, Tuberculosis |
| rs93059 | 103468518 (intronic) | A | 0.11 | 0.42 | 1.51 × 10−5 | various diseases |
| rs230493 | 103486216 (regulatory) | A | 0.42 | 0.62 | 2.89 × 10−4 | thyroid cancer, PBC, antipsychotic response |
| rs35680095 | 103486698 (regulatory) | A | 0.03 | 0.12 | 3.66 × 10−2 | atopic asthma |
| rs230494 | 103486969 (regulatory) | A | 0.29 | 0.55 | 7.03 × 10−4 | various diseases |
| rs230496 | 103488491 (intronic) | G | 0.31 | 0.55 | 3.16 × 10−3 | liver cancer |
| rs230498 | 103489603 (intronic) | A | 0.44 | 0.63 | 2.89 × 10−3 | ovarian cancer, gastric cancer |
| rs230500 | 103491724 (intronic) | A | 0.40 | 0.63 | 8.34 × 10−5 | cytokine regulation |
| rs230539 | 103495532 (regulatory) | G | 0.36 | 0.6 | 3.24 × 10−8 | ovarian cancer, gastric cancer |
| rs1598858 | 103506095 (intronic) | G | 0.18 | 0.4 | 1.82 × 10−4 | kidney inflammation, cytokine regulation, |
| rs3774956 | 103508526 (regulatory) | T | 0.31 | 0.48 | 3.77 × 10−3 | lymphedema, cytokine regulation, |
| rs4648045 | 103508703 (regulatory) | C | 0.29 | 0.41 | 3.80 × 10−3 | various diseases |
| rs3774959 | 103511114 (intronic) | A | 0.19 | 0.41 | 4.48 × 10−5 | Asthma, glioma, cytokine regulation, |
| rs4698858 | 103513073 (intronic) | - | 0.36 | 0.48 | 3.62 × 10−2 | cytokine regulation |
| rs11722146 | 103524629 (intronic) | - | 0.19 | 0.35 | 3.24 × 10−3 | colon and rectal cancer, colorectal cancer, |
| rs12509403 | 103525350 (intronic) | T | 0.18 | 0.35 | 7.08 × 10−4 | kidney inflammation, allergic rhinitis |
| rs12509517 | 103525508 (intronic) | C | 0.19 | 0.35 | 3.18 × 10−3 | colorectal cancer, gastric cancer |
| rs3774968 | 103531112 (intronic) | A | 0.49 | 0.6 | 2.84 × 10−3 | plasma cell leukemia, vte, ovarian cancer, |
| SNP ID | Alleles (Ref/Alt) | ICU | GBR | Adjusted p Value | SNP ID | Alleles (Ref/Alt) | ICU | GBR | Adjusted p Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RAF | AAF | RAF | AAF | RAF | AAF | RAF | AAF | ||||||
| rs1599961 | G/A | 0.03 | 0.2 | 0.5 | 0.2 | 1.53 × 10−7 | rs230497 | T/A | 0.06 | 0.2 | 0.5 | 0.4 | 0.00 × 100 |
| rs1585215 | T/C | 0.22 | 0.2 | 0.5 | 0.1 | 1.53 × 10−2 | rs230498 | A/G | 0.28 | 0.3 | 0.5 | 0.4 | 2.89 × 10−3 |
| rs1585214 | T/C | 0.08 | 0.3 | 0.5 | 0.3 | 2.29 × 10−6 | rs230499 | T/G | 0.06 | 0.3 | 0.5 | 0.4 | 5.60 × 10−9 |
| rs1585213 | C/T | 0.19 | 0.3 | 0.5 | 0.2 | 9.78 × 10−3 | rs230500 | A/G | 0.19 | 0.3 | 0.5 | 0.4 | 8.34 × 10−5 |
| rs1598856 | A/G | 0.28 | 0.4 | 0.5 | 0.3 | 4.82 × 10−2 | rs230502 | A/G | 0.25 | 0.1 | 0.5 | 0.2 | 9.77 × 10−4 |
| rs230535 | A/C | 0.25 | 0.4 | 0.5 | 0.4 | 6.11 × 10−3 | rs230503 | G/A | 0.25 | 0.3 | 0.5 | 0.4 | 1.96 × 10−3 |
| rs170731 | T/A | 0.11 | 0.3 | 0.5 | 0.4 | 1.30 × 10−6 | rs230538 | C/T | 0.17 | 0.2 | 0.5 | 0.3 | 4.38 × 10−5 |
| rs230534 | T/C | 0.08 | 0.3 | 0.5 | 0.4 | 3.36 × 10−8 | rs230539 | G/A | 0.06 | 0.3 | 0.5 | 0.3 | 3.24 × 10−8 |
| rs230533 | A/G | 0.17 | 0.4 | 0.5 | 0.4 | 8.10 × 10−5 | rs1598858 | A/G | 0.14 | 0.1 | 0.5 | 0.1 | 1.82 × 10−4 |
| rs230532 | T/A | 0.31 | 0.4 | 0.5 | 0.4 | 3.83 × 10−2 | rs1598859 | T/C | 0.28 | 0.1 | 0.5 | 0.1 | 3.44 × 10−2 |
| rs230531 | G/A | 0.03 | 0.3 | 0.5 | 0.4 | 2.20 × 10−9 | rs3774956 | C/T | 0.22 | 0.2 | 0.5 | 0.2 | 3.77 × 10−3 |
| rs230530 | A/G | 0.31 | 0 | 0.5 | 0.2 | 2.41 × 10−3 | rs4648045 | T/C | 0.19 | 0.2 | 0.5 | 0.1 | 3.80 × 10−3 |
| rs230529 | T/C | 0.11 | 0.1 | 0.5 | 0.3 | 2.09 × 10−7 | rs3774957 | A/T | 0.19 | 0.2 | 0.5 | 0.1 | 2.87 × 10−3 |
| rs230528 | G/T | 0.06 | 0.2 | 0.5 | 0.3 | 1.60 × 10−8 | rs1287 | G/A | 0.03 | 0.2 | 0.5 | 0.2 | 5.88 × 10−8 |
| rs230527 | A/G | 0.22 | 0.2 | 0.5 | 0.4 | 6.11 × 10−6 | rs7692606 | T/C | 0.19 | 0.2 | 0.5 | 0.1 | 2.81 × 10−3 |
| rs230526 | A/G | 0.22 | 0.1 | 0.5 | 0.3 | 7.88 × 10−5 | rs230546 | G/C | 0 | 0.8 | 0 | 1 | 1.00 × 10−3 |
| rs230525 | G/A | 0.19 | 0.3 | 0.5 | 0.4 | 3.08 × 10−5 | rs647417 | A/T | 0 | 0.8 | 0 | 1 | 8.16 × 10−5 |
| rs79651301 | G/A | 0.17 | 0 | 0.3 | 0.1 | 3.11 × 10−2 | rs647424 | G/T | 0 | 0.6 | 0 | 1 | 3.50 × 10−8 |
| rs4648004 | A/G | 0.22 | 0 | 0.4 | 0.1 | 6.08 × 10−3 | rs1020759 | C/T | 0 | 0 | 0.5 | 0.2 | 0.00 × 100 |
| rs230523 | C/T | 0 | 0.1 | 0.5 | 0.4 | 0.00 × 100 | rs3774959 | G/A | 0.11 | 0.1 | 0.5 | 0.1 | 4.48 × 10−5 |
| rs230522 | A/G | 0.11 | 0.3 | 0.5 | 0.4 | 5.24 × 10−7 | rs3774960 | C/T | 0.25 | 0.3 | 0.5 | 0.2 | 2.31 × 10−2 |
| rs118882 | T/C | 0.11 | 0.3 | 0.5 | 0.4 | 1.23 × 10−6 | rs4698858 | C/G | 0.28 | 0.2 | 0.5 | 0.2 | 3.62 × 10−2 |
| rs230521 | C/G | 0.19 | 0.2 | 0.5 | 0.3 | 6.59 × 10−5 | rs7684888 | G/A | 0.03 | 0.1 | 0.4 | 0.1 | 2.34 × 10−5 |
| rs230520 | G/A | 0.22 | 0.3 | 0.5 | 0.4 | 6.27 × 10−4 | rs11722146 | G/A | 0.11 | 0.1 | 0.4 | 0.1 | 3.24 × 10−3 |
| rs230519 | T/C | 0.22 | 0.3 | 0.5 | 0.4 | 6.10 × 10−4 | rs12509403 | C/T | 0.08 | 0.1 | 0.4 | 0.1 | 7.08 × 10−4 |
| rs93059 | G/A | 0.17 | 0 | 0.5 | 0.2 | 1.51 × 10−5 | rs12509517 | G/C | 0.11 | 0.1 | 0.4 | 0.1 | 3.18 × 10−3 |
| rs230492 | A/G | 0.22 | 0.3 | 0.5 | 0.4 | 6.75 × 10−4 | rs3774967 | T/C | 0.14 | 0.1 | 0.4 | 0.1 | 1.04 × 10−2 |
| rs230493 | A/T | 0.22 | 0.3 | 0.5 | 0.4 | 2.89 × 10−4 | rs4699031 | A/G | 0.14 | 0.1 | 0.4 | 0.1 | 1.03 × 10−2 |
| rs35680095 | G/A | 0.06 | 0 | 0.2 | 0 | 3.66 × 10−2 | rs4235406 | T/C | 0.11 | 0.2 | 0.4 | 0.1 | 3.92 × 10−3 |
| rs230494 | G/A | 0.25 | 0.2 | 0.5 | 0.3 | 7.03 × 10−4 | rs9790601 | A/G | 0.06 | 0.1 | 0.4 | 0.1 | 9.49 × 10−5 |
| rs230495 | A/G | 0.31 | 0.2 | 0.5 | 0.3 | 5.60 × 10−3 | rs3774968 | A/G | 0.19 | 0.4 | 0.5 | 0.4 | 2.84 × 10−3 |
| rs230496 | G/A | 0.39 | 0.1 | 0.5 | 0.3 | 3.16 × 10−3 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirzo, S.M.; Kumar, A.; Sharma, N.K.; Li, L.; Balshaw, R.; Plummer, F.A.; Luo, M.; Liang, B. NFκB1 Polymorphisms Are Associated with Severe Influenza A (H1N1) Virus Infection in a Canadian Population. Microorganisms 2022, 10, 1886. https://doi.org/10.3390/microorganisms10101886
Mirzo SM, Kumar A, Sharma NK, Li L, Balshaw R, Plummer FA, Luo M, Liang B. NFκB1 Polymorphisms Are Associated with Severe Influenza A (H1N1) Virus Infection in a Canadian Population. Microorganisms. 2022; 10(10):1886. https://doi.org/10.3390/microorganisms10101886
Chicago/Turabian StyleMirzo, Suhrobjon Mullo, Anand Kumar, Naresh Kumar Sharma, Lin Li, Robert Balshaw, Francis A. Plummer, Ma Luo, and Binhua Liang. 2022. "NFκB1 Polymorphisms Are Associated with Severe Influenza A (H1N1) Virus Infection in a Canadian Population" Microorganisms 10, no. 10: 1886. https://doi.org/10.3390/microorganisms10101886
APA StyleMirzo, S. M., Kumar, A., Sharma, N. K., Li, L., Balshaw, R., Plummer, F. A., Luo, M., & Liang, B. (2022). NFκB1 Polymorphisms Are Associated with Severe Influenza A (H1N1) Virus Infection in a Canadian Population. Microorganisms, 10(10), 1886. https://doi.org/10.3390/microorganisms10101886







