Capture and Ex-Situ Analysis of Environmental Biofilms in Livestock Buildings
Abstract
1. Introduction
2. Materials and Methods
2.1. Livestock Building, Coupons Disposition, and Sampling
2.2. Confocal Laser Scanning Microscopy
2.3. Enumeration of Bacteria Detached from Coupons
2.4. High-Throughput Sequencing of the 16S rRNA and Diversity Analysis
2.4.1. DNA Extraction, PCR, and Sequencing
2.4.2. Diversity Analysis Using Bioinformatics
2.5. Statistical Analysis
3. Results
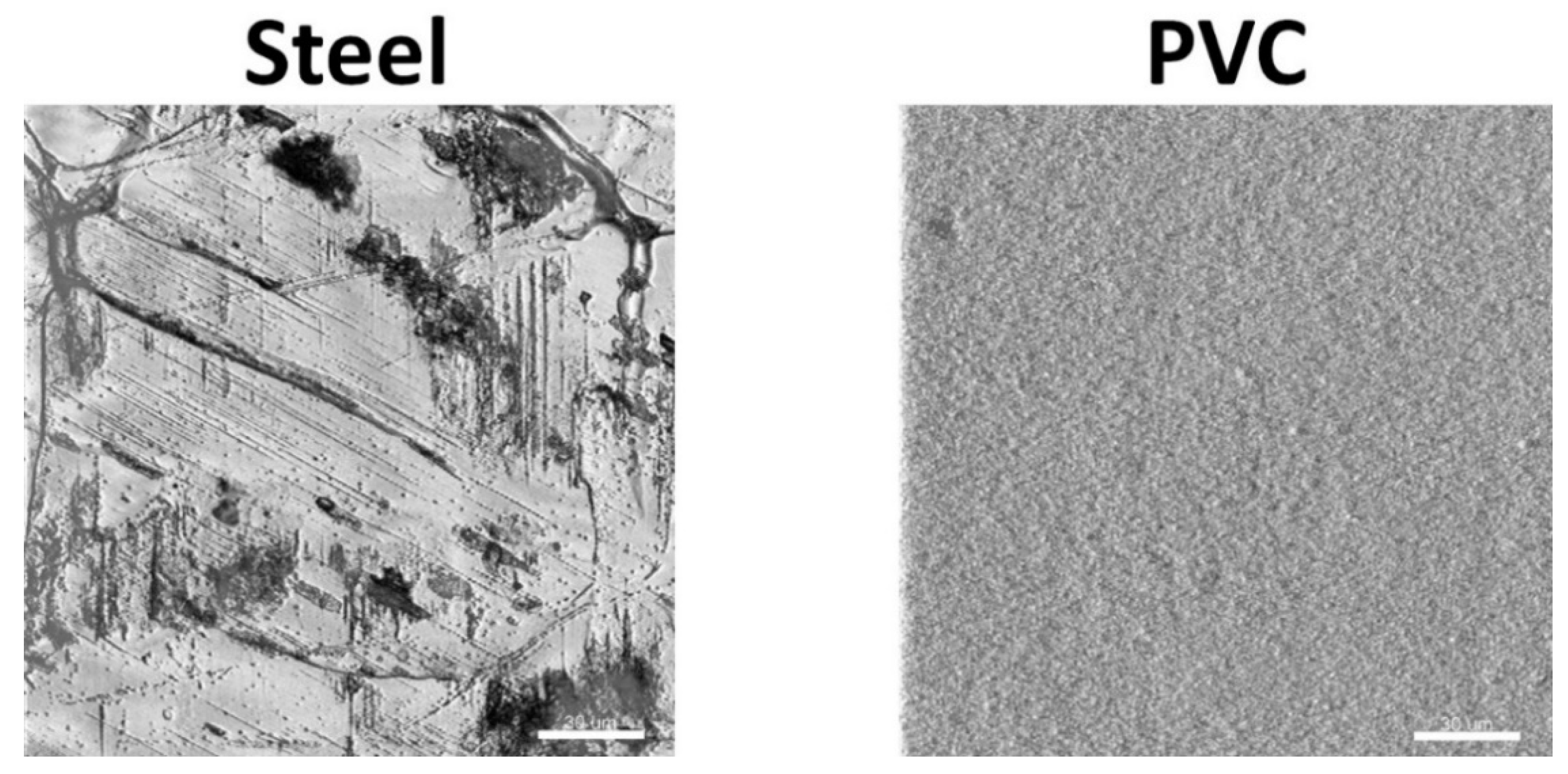
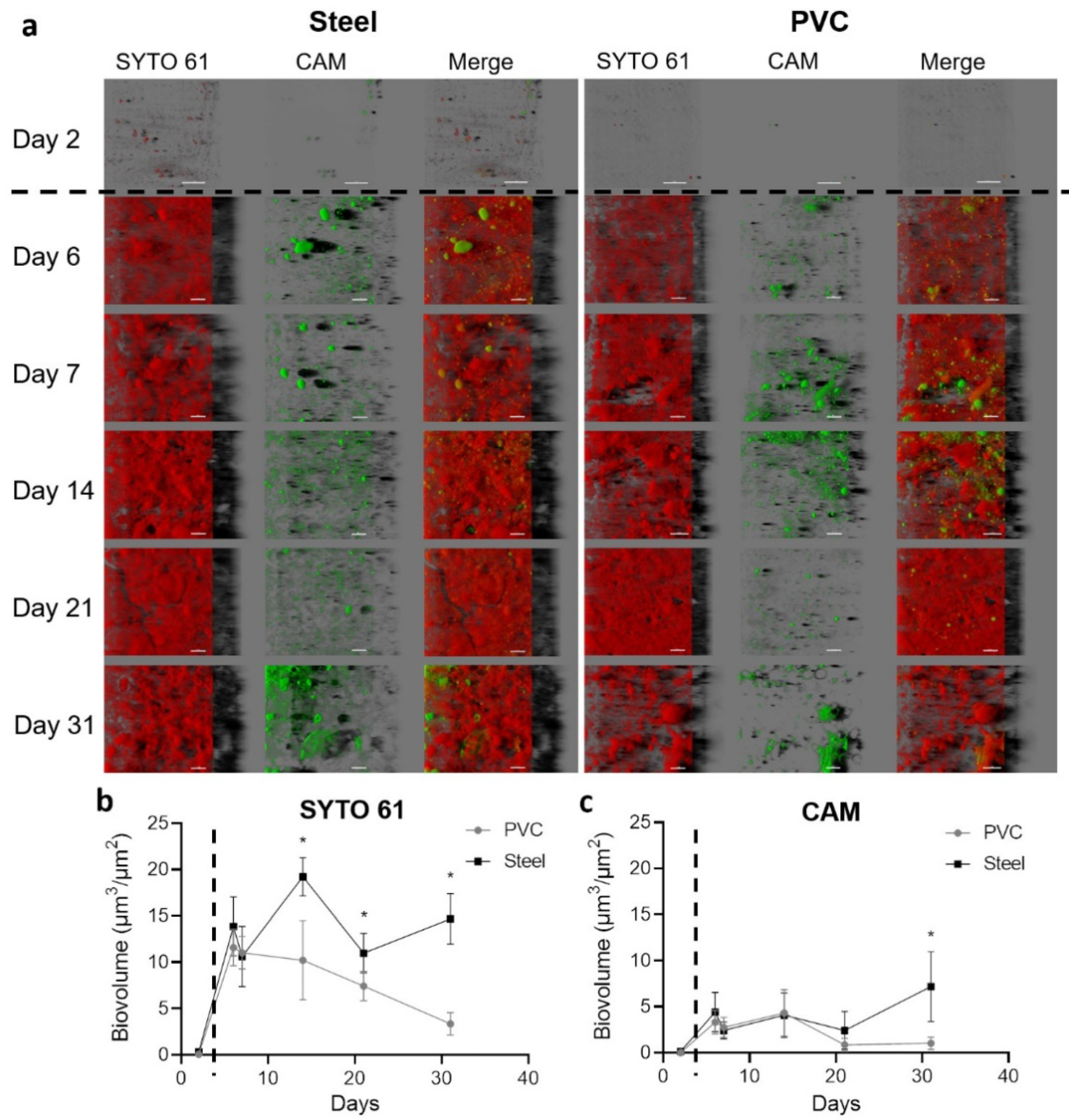
3.1. Coupons Are Colonized by a Densely Clustered Biofilm with Only a Minor Fraction of Cells Metabolically Active
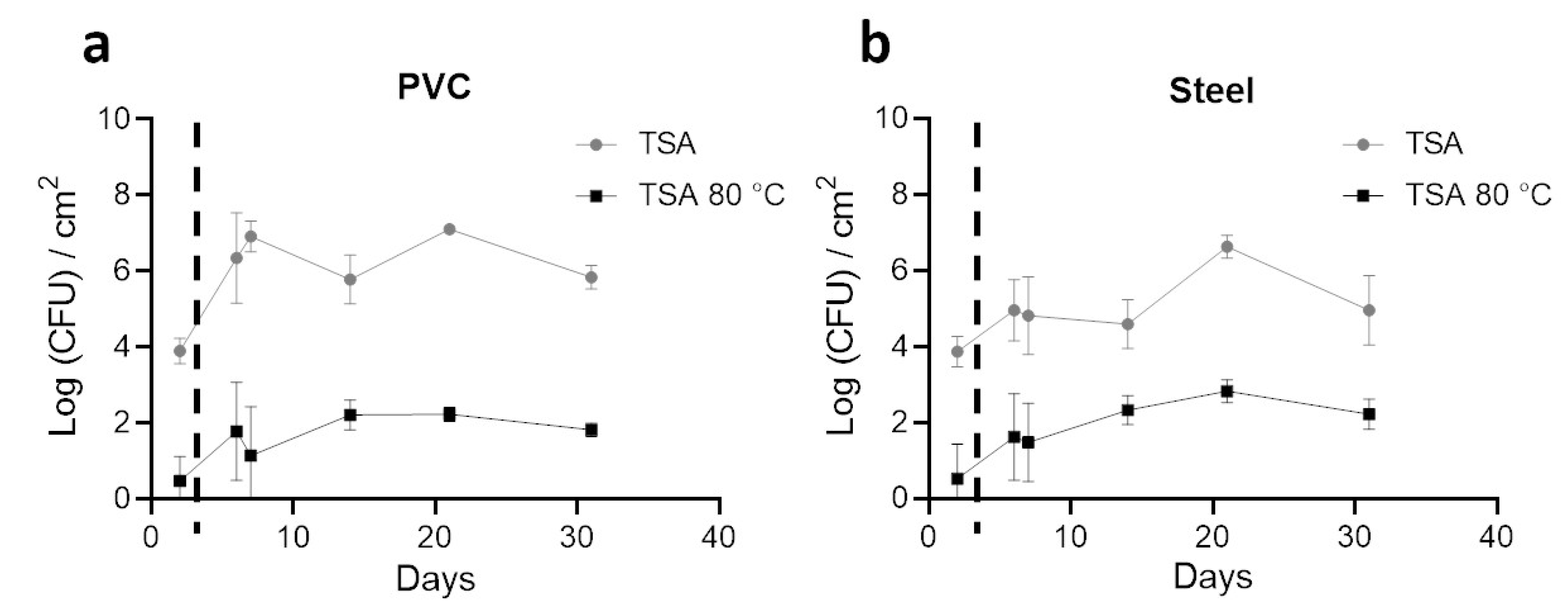
3.2. Enumeration of Aerobic Cultivable Bacteria from Coupons
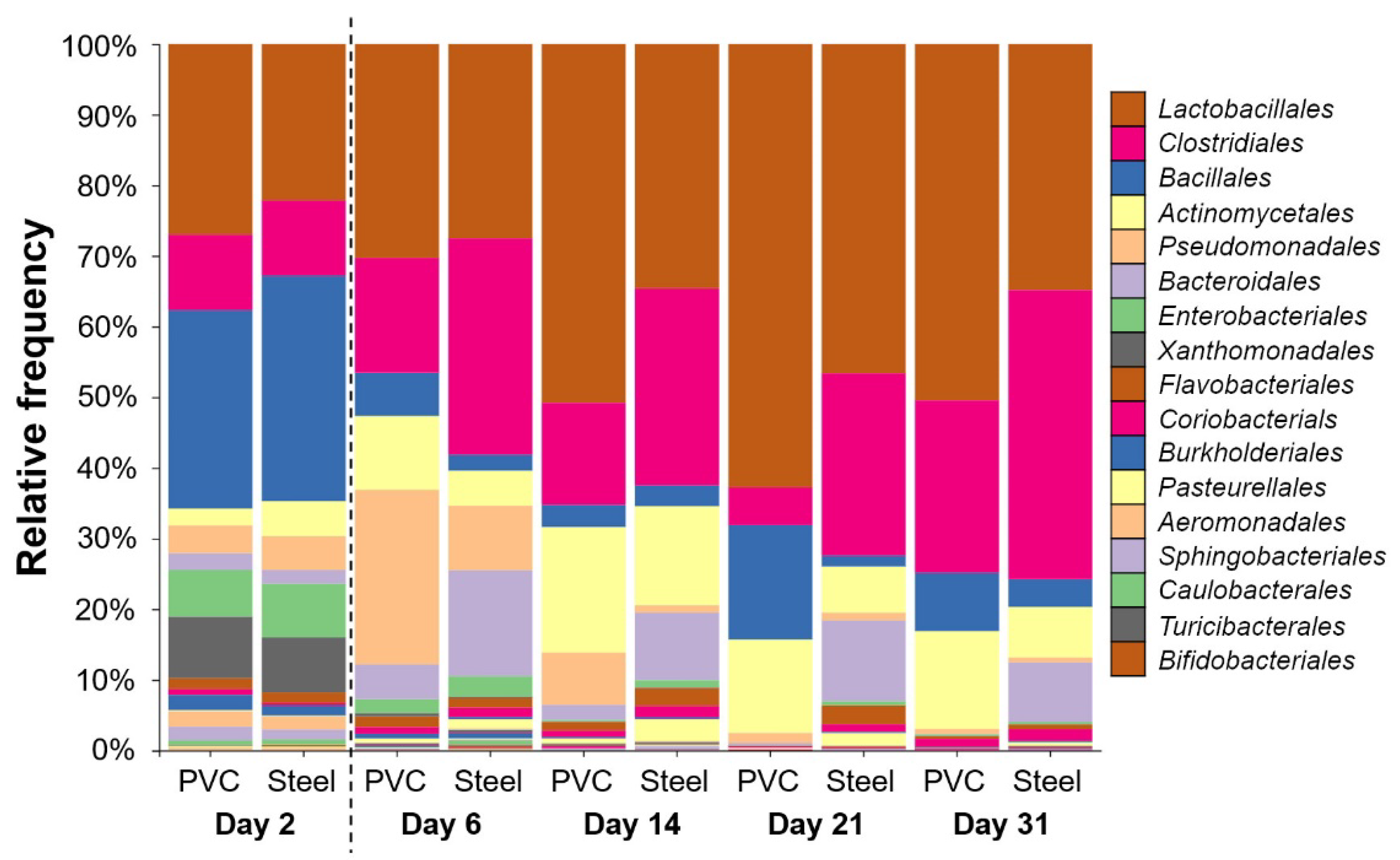
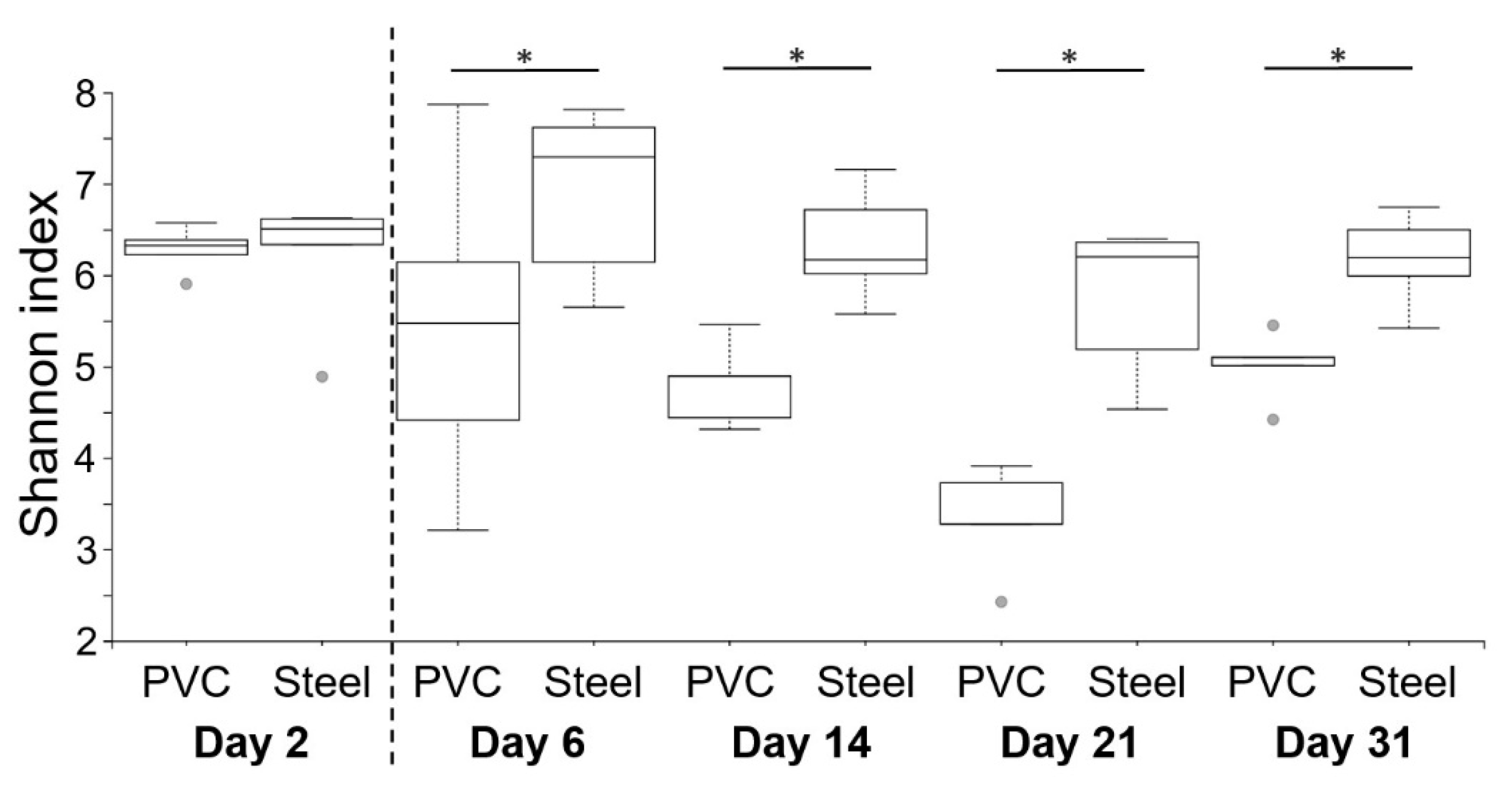
3.3. 16S rRNA High-Throughput Sequencing Analysis to Decipher the Dynamic of Biofilm Bacterial Diversity
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Godyń, D.; Nowicki, J.; Herbut, P. Effects of environmental enrichment on pig welfare—A review. Animals 2019, 9, 383. [Google Scholar] [CrossRef]
- Luyckx, K.Y.; Van Weyenberg, S.; Dewulf, J.; Herman, L.; Zoons, J.; Vervaet, E.; Heyndrickx, M.; De Reu, K. On-farm comparisons of different cleaning protocols in broiler houses. Poult. Sci. 2015, 94, 1986–1993. [Google Scholar] [CrossRef]
- Mannion, C.; Leonard, F.C.; Lynch, P.B.; Egan, J. Efficacy of cleaning and disinfection on pig farms in Ireland. Vet. Rec. 2007, 161, 371–375. [Google Scholar] [CrossRef]
- Misra, S.; van Middelaar, C.E.; Jordan, K.; Upton, J.; Quinn, A.J.; de Boer, I.J.M.; O’Driscoll, K. Effect of different cleaning procedures on water use and bacterial levels in weaner pig pens. PLoS ONE 2020, 15, e0242495. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wuertz, S. Bacteria and archaea on earth and their abundance in biofilms. Nat. Rev. Microbiol. 2019, 17, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C. The Perfect Slime. Colloids Surf. B Biointerface 2011, 86, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef] [PubMed]
- Bridier, A.; Piard, J.-C.; Pandin, C.; Labarthe, S.; Dubois-Brissonnet, F.; Briandet, R. Spatial organization plasticity as an adaptive driver of surface microbial communities. Front. Microbiol. 2017, 8, 1364. [Google Scholar] [CrossRef] [PubMed]
- Bridier, A.; Briandet, R.; Thomas, V.; Dubois-Brissonnet, F. Resistance of bacterial biofilms to disinfectants: A review. Biofouling 2011, 27, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Chamignon, C.; Guéneau, V.; Medina, S.; Deschamps, J.; Gil-Izquierdo, A.; Briandet, R.; Mousset, P.-Y.; Langella, P.; Lafay, S.; Bermúdez-Humarán, L.G. Evaluation of the probiotic properties and the capacity to form biofilms of various lactobacillus strains. Microorganisms 2020, 8, 1053. [Google Scholar] [CrossRef]
- Høiby, N.; Bjarnsholt, T.; Givskov, M.; Molin, S.; Ciofu, O. Antibiotic resistance of bacterial biofilms. Int. J. Antimicrob. Agents 2010, 35, 322–332. [Google Scholar] [CrossRef] [PubMed]
- Coenye, T. Response of sessile cells to stress: From changes in gene expression to phenotypic adaptation. FEMS Immunol. Med. Microbiol. 2010, 59, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Lerminiaux, N.A.; Cameron, A.D.S. Horizontal transfer of antibiotic resistance genes in clinical environments. Can. J. Microbiol. 2019, 65, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal gene transfer: Building the web of life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Taylor, K. Ten years of REACH—An animal protection perspective. Altern. Lab. Anim. 2018, 46, 347–373. [Google Scholar] [CrossRef]
- McEwen, S.A.; Collignon, P.J. Antimicrobial resistance: A one health perspective. Microbiol. Spectr. 2018, 6, ARBA-0009-2017. [Google Scholar] [CrossRef]
- Cross, A.R.; Baldwin, V.M.; Roy, S.; Essex-Lopresti, A.E.; Prior, J.L.; Harmer, N.J. Zoonoses under our noses. Microbes Infect. 2019, 21, 10–19. [Google Scholar] [CrossRef]
- Chlebicz, A.; Śliżewska, K. Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: A review. Int. J. Environ. Res. Public Health 2018, 15, 863. [Google Scholar] [CrossRef]
- Valentine, N.B.; Butcher, M.G.; Su, Y.-F.; Jarman, K.H.; Matzke, M.; Webb-Robertson, B.-J.; Panisko, E.A.; Seiders, B.A.B.; Wahl, K.L. Evaluation of sampling tools for environmental sampling of bacterial endospores from porous and nonporous surfaces. J. Appl. Microbiol. 2008, 105, 1107–1113. [Google Scholar] [CrossRef]
- Ismaïl, R.; Aviat, F.; Michel, V.; Le Bayon, I.; Gay-Perret, P.; Kutnik, M.; Fédérighi, M. Methods for recovering microorganisms from solid surfaces used in the food industry: A review of the literature. Int. J. Environ. Res. Public Health 2013, 10, 6169–6183. [Google Scholar] [CrossRef] [PubMed]
- Gomes, I.B.; Lemos, M.; Mathieu, L.; Simões, M.; Simões, L.C. The action of chemical and mechanical stresses on single and dual species biofilm removal of drinking water bacteria. Sci. Total Environ. 2018, 631–632, 987–993. [Google Scholar] [CrossRef] [PubMed]
- Stiefel, P.; Mauerhofer, S.; Schneider, J.; Maniura-Weber, K.; Rosenberg, U.; Ren, Q. Enzymes enhance biofilm removal efficiency of cleaners. Antimicrob. Agents Chemother. 2016, 60, 3647–3652. [Google Scholar] [CrossRef] [PubMed]
- Grand, I.; Bellon-Fontaine, M.-N.; Herry, J.-M.; Hilaire, D.; Moriconi, F.-X.; Naïtali, M. Possible Overestimation of Surface Disinfection Efficiency by Assessment Methods Based on Liquid Sampling Procedures as Demonstrated by in Situ Quantification of Spore Viability. Appl. Environ. Microbiol. 2011, 77, 6208–6214. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Goeres, D.M.; Loetterle, L.R.; Hamilton, M.A.; Murga, R.; Kirby, D.W.; Donlan, R.M. Statistical assessment of a laboratory method for growing biofilms. Microbiology 2005, 151, 757–762. [Google Scholar] [CrossRef] [PubMed]
- Douterelo, I.; Boxall, J.B.; Deines, P.; Sekar, R.; Fish, K.E.; Biggs, C.A. Methodological approaches for studying the microbial ecology of drinking water distribution systems. Water Res. 2014, 65, 134–156. [Google Scholar] [CrossRef]
- Deines, P.; Sekar, R.; Husband, P.S.; Boxall, J.B.; Osborn, A.M.; Biggs, C.A. A new coupon design for simultaneous analysis of in situ microbial biofilm formation and community structure in drinking water distribution systems. Appl. Microbiol. Biotechnol. 2010, 87, 749–756. [Google Scholar] [CrossRef]
- Douterelo, I.; Jackson, M.; Solomon, C.; Boxall, J. Microbial analysis of in situ biofilm formation in drinking water distribution systems: Implications for monitoring and control of drinking water quality. Appl. Microbiol. Biotechnol. 2016, 100, 3301–3311. [Google Scholar] [CrossRef]
- Krishnan, M.; Dahms, H.-U.; Seeni, P.; Gopalan, S.; Sivanandham, V.; Jin-Hyoung, K.; James, R.A. Multi metal assessment on biofilm formation in offshore environment. Mater. Sci. Eng. C Mater. Biol. Appl. 2017, 73, 743–755. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Rodríguez-Herrera, J.J.; Cabo, M.L. Tracking bacteriome variation over time in listeria monocytogenes-positive foci in food industry. Int. J. Food Microbiol. 2020, 315, 108439. [Google Scholar] [CrossRef]
- Moen, B.; Røssvoll, E.; Måge, I.; Møretrø, T.; Langsrud, S. Microbiota formed on attached stainless steel coupons correlates with the natural biofilm of the sink surface in domestic kitchens. Can. J. Microbiol. 2016, 62, 148–160. [Google Scholar] [CrossRef]
- Verschuren, L.M.G.; Calus, M.P.L.; Jansman, A.J.M.; Bergsma, R.; Knol, E.F.; Gilbert, H.; Zemb, O. Fecal microbial composition associated with variation in feed efficiency in pigs depends on diet and sex. J. Anim. Sci. 2018, 96, 1405–1418. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Lane, D.J. 16s/23s RRNA sequencing. In Nucleic Acid Techniques in Bacterial Systematics; John Wiley and Sons: New York, NY, USA, 1991; pp. 115–175. [Google Scholar]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed]
- Menees, T.S.; Radhakrishnan, R.; Ramp, L.C.; Burgess, J.O.; Lawson, N.C. Contact angle of unset elastomeric impression materials. J. Prosthet. Dent. 2015, 114, 536–542. [Google Scholar] [CrossRef] [PubMed]
- Joux, F.; Lebaron, P. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbes Infect. 2000, 2, 1523–1535. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Albenberg, L.; Esipova, T.V.; Judge, C.P.; Bittinger, K.; Chen, J.; Laughlin, A.; Grunberg, S.; Baldassano, R.N.; Lewis, J.D.; Li, H.; et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014, 147, 1055–1063.e8. [Google Scholar] [CrossRef]
- Das, T.; Sehar, S.; Manefield, M. The roles of extracellular DNA in the structural integrity of extracellular polymeric substance and bacterial biofilm development. Environ. Microbiol. Rep. 2013, 5, 778–786. [Google Scholar] [CrossRef] [PubMed]
- Golub, S.R.; Overton, T.W. Pellicle formation by escherichia coli K-12: Role of adhesins and motility. J. Biosci. Bioeng. 2021, 131, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.-C.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Wang, X.; Tsai, T.; Deng, F.; Wei, X.; Chai, J.; Knapp, J.; Apple, J.; Maxwell, C.V.; Lee, J.A.; Li, Y.; et al. Longitudinal investigation of the swine gut microbiome from birth to market reveals stage and growth performance associated bacteria. Microbiome 2019, 7, 109. [Google Scholar] [CrossRef]
- Shin, D.; Chang, S.Y.; Bogere, P.; Won, K.; Choi, J.-Y.; Choi, Y.-J.; Lee, H.K.; Hur, J.; Park, B.-Y.; Kim, Y.; et al. Beneficial roles of probiotics on the modulation of gut microbiota and immune response in pigs. PLoS ONE 2019, 14, e0220843. [Google Scholar] [CrossRef] [PubMed]
- Pollock, J.; Glendinning, L.; Smith, L.A.; Mohsin, H.; Gally, D.L.; Hutchings, M.R.; Houdijk, J.G.M. Temporal and nutritional effects on the weaner pig ileal microbiota. Anim. Microbiome 2021, 3, 58. [Google Scholar] [CrossRef]
- Fadeev, E.; Cardozo-Mino, M.G.; Rapp, J.Z.; Bienhold, C.; Salter, I.; Salman-Carvalho, V.; Molari, M.; Tegetmeyer, H.E.; Buttigieg, P.L.; Boetius, A. Comparison of two 16S RRNA primers (V3-V4 and V4-V5) for studies of arctic microbial communities. Front. Microbiol. 2021, 12, 637526. [Google Scholar] [CrossRef]
- Iwen, P.C.; Hinrichs, S.H.; Rupp, M.E. Utilization of the internal transcribed spacer regions as molecular targets to detect and identify human fungal pathogens. Med. Mycol. 2002, 40, 87–109. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.-C.; Million, M.; Hugon, P.; Armougom, F.; Raoult, D. Human gut microbiota: Repertoire and variations. Front. Cell. Infect. Microbiol. 2012, 2, 136. [Google Scholar] [CrossRef] [PubMed]
- Vartoukian, S.R.; Palmer, R.M.; Wade, W.G. Strategies for culture of “unculturable” bacteria. FEMS Microbiol. Lett. 2010, 309, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.-C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.-M.; Fournier, P.-E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guéneau, V.; Rodiles, A.; Piard, J.-C.; Frayssinet, B.; Castex, M.; Plateau-Gonthier, J.; Briandet, R. Capture and Ex-Situ Analysis of Environmental Biofilms in Livestock Buildings. Microorganisms 2022, 10, 2. https://doi.org/10.3390/microorganisms10010002
Guéneau V, Rodiles A, Piard J-C, Frayssinet B, Castex M, Plateau-Gonthier J, Briandet R. Capture and Ex-Situ Analysis of Environmental Biofilms in Livestock Buildings. Microorganisms. 2022; 10(1):2. https://doi.org/10.3390/microorganisms10010002
Chicago/Turabian StyleGuéneau, Virgile, Ana Rodiles, Jean-Christophe Piard, Bastien Frayssinet, Mathieu Castex, Julia Plateau-Gonthier, and Romain Briandet. 2022. "Capture and Ex-Situ Analysis of Environmental Biofilms in Livestock Buildings" Microorganisms 10, no. 1: 2. https://doi.org/10.3390/microorganisms10010002
APA StyleGuéneau, V., Rodiles, A., Piard, J.-C., Frayssinet, B., Castex, M., Plateau-Gonthier, J., & Briandet, R. (2022). Capture and Ex-Situ Analysis of Environmental Biofilms in Livestock Buildings. Microorganisms, 10(1), 2. https://doi.org/10.3390/microorganisms10010002






